Introduction
Oral cancer, particularly oral squamous cell
carcinoma (OSCC), is the most common type of head and neck cancer
worldwide, with ~540,000 new cases annually worldwide (1,2).
Despite surgery and chemotherapy being increasingly used to treat
OSCC, the 5-year survival rate of OSCC has not improved markedly
over previous years, due to late diagnosis, frequent loco-regional
recurrences at the primary site and metastasis to neck lymph nodes
following treatment (3,4). Therefore, it is imperative to
identify novel therapeutics to improve treatment of this
disease.
Cisplatin or cis-diammine-dichloroplatinum (II)
(CDDP), is a DNA damage-inducing chemotherapeutic drug, and is a
chemotherapeutic agent that is widely used in treatment of several
types of cancer, including testicular, ovarian, cervical, head and
neck, lung, oral and esophageal malignancies (5–7).
Despite CDDP being a potent anticancer agent for OSCC, its
application is limited due to its side effects, in particular
neurotoxicity, which is dose-limiting, and inherent and acquired
resistance (8–10). Therefore, novel and customized
treatment strategies are required to overcome chemotherapeutic drug
resistance and enhance its antitumor activity (11). Targeted therapies, which cause no
or minor side effects, may compensate for the limitations of
conventional chemotherapies.
It is well known that tumor suppressor genes (TSGs)
are important in the pathogenesis of human OSCC and other types of
cancer (12–14). The gene associated with
retinoid-interferon-induced mortality 19 (GRIM-19) is one of the
novel candidate TSGs located on human chromosome 19p13.1, and its
overexpression significantly increases cell death (15). Our previous study demonstrated that
the forced expression of GRIM-19 in OSCC cells significantly
inhibited cell proliferation and colony formation, and induced cell
apoptosis in vitro, which effectively suppressed tumor
growth in mouse models of human OSCC (16). In addition, it has been
demonstrated that the combination of conventional CDDP-based
chemotherapy and tumor suppressor gene therapy, including LKB1,
p53, NPRL2 and FUS1, may overcome cancer cell resistance to
chemotherapeutic drugs and enhance therapeutic efficacy in cancer
(17–20). However, to the best of our
knowledge, no reports are available regarding the combined effects
of exogenous expression of GRIM-19 tumor suppressor with any
chemotherapeutic drugs, including CDDP, on tumor suppression
activity. Therefore, the present study analyzed the potential
correlation between the expression of GRIM-19 by cationic liposome
(LP)-mediated gene transfer and the enhancement of CDDP sensitivity
in human OSCC cell lines. The effects of GRIM-19 on the enhancement
of low-dose CDDP-mediated antitumor activity in OSCC cell lines
were examined in vitro and in vivo, and the potential
molecular mechanisms of the combined treatment of cationic
LP-mediated GRIM-19 gene therapy and low-dose CDDP-based
chemotherapy were investigated.
Materials and methods
Cell line, plasmid and chemotherapeutic
drug
The HSC3 human oral squamous cell carcinoma cell
lines were purchased from the Cell Bank of Type Culture Collection
of Chinese Academy of Sciences, Shanghai Institute of Cell Biology,
Chinese Academy of Sciences (Shanghai China). The HSC3 cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) F-12 medium
(Invitrogen Life Technologies, Carlsbad, CA, USA) containing 10%
fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 1%
fungicide and penicillin/streptomycin (Biochrom, Ltd., Cambridge,
UK) at 37°C in a humidified atmosphere containing 5%
CO2. CDDP was obtained from Sigma-Aldrich. The
pVAX1-GRIM-19 (pGRIM-19) plasmid was constructed, as described in
our previous study (16). The
pVAX1 and pGRIM-19 plasmids were extracted and purified using an
Endofree Plasmid Giga kit (Qiagen, Chatsworth, CA, USA) and were
solubilized in Endo-free sterilized water for the subsequent
experiments. The present study was approved (no. JL2013568B) by the
ethics committee of Jilin University (Changchun, China).
Preparation of cationic LPs and plasmid
DNA complexes
The cationic LPs, composed of DOTAP/cholesterol
(Avanti Polar Lipids, Birmingham, AL, USA) used in the present
study were synthesized, as previously described (21). For cell transfection, the
pVAX/pVAX-LKB1 plasmid and LPs, diluted in equal volumes of DMEM,
were mixed to form DNA:LP complexes (LP-pVAX1 or LP-pGRIM-19),
according to their molecular weight ratio (1:6). For the animal
experiments, the LPs and plasmid DNA were diluted and mixed in 5%
dextrose, as previous described (17). The average particle size (150–300
nm) of the complexes was selected, according to previous study
(17).
Cell viability
To determine CDDP sensitivities, cell viability was
performed using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphe-nyltetrazolium bromide (MTT)
assay. Briefly, the HSC3 cells were seeded (5×103
cells/well) onto 96-well plates and transfected with the different
plasmid DNA complexes for 48 h at 37°C. Following transfection,
DMEM with serial concentrations (1.0–20.0 μM) of CDDP were
added and incubated for another 48 h at 37°C. The cell medium was
replaced with 20 μl MTT (Sigma-Aldrich) and incubated for
another 4 h, followed by the addition of 200 μl
solubilization solution containing dimethyl sulfoxide
(Sigma-Aldrich) into each well. The plates were maintained in a
dark room overnight, and the optical density of each sample was
measured at a 570 nm test wavelength using an ELISA multi-well
spectrophotometer (SpectraMax® M2/M2e; Molecular Devices
Corporation, Sunnyvale, CA, USA). The percentages of viable cells
were calculated based on the absorbency of the treated cells
relative to that of the untreated cells. The half maximal
inhibitory concentration (IC50) values of CDDP were also
calculated. To evaluate the inhibition on OSCC cell growth by the
combined treatment of LP-pGRIM-19 and low-dose CDDP, the half
maximal inhibitory concentration (IC50) of CDDP (10
μM) was used in all the following experiments in
vitro. The HSC3 cells were seeded (5×103 cells/well)
onto 96-well plates and transfected with either LP-pVAX1 or
LP-pGRIM-19, followed by treatment with the IC50 CDDP.
The cell viabilities following 48 h treatment were quantified using
an MTT assay, as above described. The visible colonies were then
counted under an IX51 inverted microscope (Olympus Corporation,
Tokyo, Japan).
Cell colony formation
The HSC3 cells were seeded into six-well culture
plates at 1×104 cells⁄well, and were transfected with
either LP-pVAX1 or LP-pGRIM-19, prior to treatment with
IC50 CDDP. The cells were incubated at 37°C for 10 days,
and the medium was replaced every 3 days. The colonies were fixed
with ice methanol for 30 min at room temperature and stained with
2% Giemsa (Sigma-Aldrich) for 10 min at room temperature. The
visible colonies were then counted.
Cell apoptosis
Flow cytometry was used to detect cell apoptosis. In
brief, the HSC3 cells were transfected with either LP-pVAX1 or
LP-pGRIM-19, and were then treated with IC50 CDDP for 24
h. Following treatment, the cells were collected and fixed in 4%
paraformaldehyde (Sigma-Aldrich), permeabilized with 70% ethanol,
washed with phosphate-buffered saline (PBS) and stained with
propidium iodide (PI; Sigma-Aldrich) solution containing 40
μg/ml PI and 10 μg/ml DNase-free RNase A
(Sigma-Aldrich). Apoptosis assays were performed, according to the
manufacturer's instructions of the Annexin V-FITC Detection kit
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). The rates of
apoptosis were determined using CellQuest 2.7 software (BD
Biosciences, San Jose, CA, USA). In addition, the induction of
caspase-3, -8 and -9 activity in the HSC3 cells treated with
LP-pGRIM-19 and CDDP were measured using caspase-3, -8 and -9
colorimetric protease assay kits (EMD Millipore, Billerica, MA,
USA), according to the manufacturer's instructions. Briefly, the
HSC3 cells were seeded into 6-well plates at a density of
4×105 cells/well and cultured for 24 h, and collected by
centrifugation at 1500 rpm for 5 min at 4°C. Caspase-3, -8, and -9
expression levels were measured in the cell lysates using
caspase-3, -8, and -9 colorimetric protease assay kits according to
the manufacturer's instructions. The optical density was read using
a microplate reader (UM313565; Thermo Fisher Scientific Inc.,
Waltham, MA, USA) at 405 nm.
Invasion and migration assays
Cell invasion assays were performed using a QCM
ECMatrix Cell Invasion Assay kit (24-well; 8 μm; EMD
Millipore), according to the manufacturer's instructions. Briefly,
the HSC3 cells were treated with LP-pGRIM-19 or CDDP for 24 h, and
1×104 cells were seeded in the Transwell chamber (BD
Biosciences). DMEM supplemented with 20% FBS was added to the lower
chamber as the chemoattractant. Following incubation for 24 h, the
non-invading cells in the upper chamber were gently removed using a
cotton-tipped swab, and the invading cells in the lower chamber
were fixed with methanol and stained with 2% Giemsa solution. The
invasive ability was determined by the number of penetrating cells
under a Nikon phase-contrast microscope L150 (Nikon Corporation,
Tokyo, Japan) and counted in >10 fields of view at ×200
magnification in each well.
The in vitro migration assay was similar to
the invasion assay described above, however a non-Matrigel-coated
24-well Boyden Chamber (8 μm, Millipore) was used. A total
of 2×103 cells were added to the Transwell chamber, the
incubation duration was 24 h, and the subsequent steps were
consistent with the invasion assay. The number of cells migrated
were counted by counting the cells in 10 randomly-selected fields
per filter, under a Nikon phase-contrast microscope (Nikon
Corporation, Tokyo, Japan). Triplicate assays were performed for
each group of cells in the invasion and migration assays.
Western blot analysis
The cells were collected by trypsinization (5%
trypsin; Sigma-Aldrich) and lysed in radioimmunoprecipitation assay
lysis buffer (Sigma-Aldrich) with the addition of protease
inhibitors (Roche Diagnostics GmbH, Mannheim, Germany) and
phosphatase inhibitors (Sigma-Aldrich) for 30 min on ice. The
protein quantity was analyzed using Bradford reagent (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The cell extracts (20
μg protein) were separated by 10% SDS-PAGE (Sigma-Aldrich)
and transferred onto nitrocellulose membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked with 5% dry milk in
PBS, and incubated overnight at 4°C with the following antibodies:
Mouse monoclonal anti-human GRIM-19 (1:2,000; cat. no. sc-365045;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA), mouse monoclonal
anti-human signal transducer and activator of transcription 3
(Stat3; 1:1,000; cat. no. sc-8019; Bedford, MA, USA), rabbit
monoclonal anti-human matrix metalloproteinase (MMP-2; 1:1,000;
cat. no. sc-13132; Cell Signaling Technology, Inc., Danvers, MA,
USA), mouse monoclonal anti-human MMP-9 (1:2,000; cat. no.
sc-21773; Santa Cruz Biotechnology, Inc.), mouse monoclonal
anti-human vascular endothelial growth factor (VEGF; 1:2,000; cat.
no. sc-53462; Santa Cruz Biotechnology), a mouse monoclonal
anti-human phosphorylated (p-)p53 (1:3,000; cat. no. sc-126; Santa
Cruz Biotechnology, Inc.), mouse monoclonal anti-human Bcl-2
(1:1,000; cat. no. sc-7382; Santa Cruz Biotechnology, Inc.) and
mouse monoclonal anti-human cyclin D1 (1:1,000; cat. no. sc-450;
Santa Cruz Biotechnology, Inc.). Mouse monoclonal anti-human
β-actin (1:10,000; cat. no. sc-8432; Santa Cruz Biotechnology,
Inc.) was used as a loading control. Following incubation with the
primary antibodies, the membranes were washed twice in PBS and
incubated with horseradish peroxidase-conjugated goat anti-mouse
antibodies (1:5,000; cat. no. sc-2005; Santa Cruz Biotechnology,
Inc.) or goat anti-rabbit antibodies (1:5,000; cat. no. sc-2004;
Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. The
proteins were detected by the enhanced protein bands, which were
visualized using enhanced chemiluminescence reagent
(Sigma-Aldrich). The integrated density value (IDV) was analyzed
using a computerized image analysis system (Fluor Chen 2.0; Bio-Rad
Laboratories, Inc.) and normalized with that of β-actin.
Tumor growth in vivo
A total of 60 female BALB nude mice aged 4–6 weeks
(18–20 g) were purchased from the Institute of Laboratory Animal
Science, Jilin University (Changchun, China). A total of
2×106 (100 μl) HSC3 cells suspended in 100
μl PBS were subcutaneously injected into the left abdominal
wall, and the size of the resulting tumor was measured daily for 7
days following injection. The tumor volume was calculated as
follows: 0.5236 × width2 × length. At ~20 days following
inoculation of the HSC3 cells, the average tumor volume measured up
to 100 mm3. These tumor-bearing nude mice were randomly
divided into the following six groups (n=10group): (i) control
group; (ii) LP-pVAX1 group; (iii) CDDP group; (iv) LP-pGRIM-19
group; (v) CDDP+LP-pGRIM-19 group; (vi) CDDP+LP-pGRIM-19 group. In
the control group, nude mice were injected with 100 μl PBS.
Other mice were administered with the LP-pVAX1 or LP-pGRIM-19
complexes by intravenous injection at a dose of 20 μg/mouse,
and/or low-dose CDDP (2 mg/kg−1/mouse) by
intraperitoneal injection once a week for 21 days, respectively.
The mice were sacrificed 7 days following the final plasmid
injection. Tumor tissue was excised, the volume measured volume and
weighed. In addition, splenic tissues were collected and cultured
for a splenocyte surveillance investigation using an MTT assay, as
described previously (22).
Briefly, the splenic tissue samples were collected from the mice,
and single-cell spleen suspensions were added to serum-free DMEM by
filtering the suspension through a sieve mesh with the aid of a
glass homogenizer, in order to exert gentle pressure on the spleen
fragments.
Statistical analysis
Data from at least three independent experiments are
expressed as the mean ± standard deviation. Statistical comparison
of more than two groups was performed using one-way analysis of
variance followed by Tukey's post-hoc test. GraphPad Prism 5.01
software (GraphPad Software, Inc., San Diego, CA, USA) and
SPSS® 16.0 (SPSS, Inc., Chicago, IL, USA) for
Windows® were used for statistical analyses. P<0.05
was considered to indicate a statistically significant
difference.
Results
Treatment with a combination of
LP-pGRIM-19 and low-dose CDDP enhances inhibition of cell
proliferation and cell colony formation
To further determine whether the exogenous
expression of GRIM-19 sensitizes the response of OSCC cells to
CDDP, the present study analyzed the sensitivities of the HSC3
cells to CDDP following treatment with different concentrations of
CDDP alone, or combined with LP-pGRIM-19 or LP-pVAX1 transfection,
respectively. Compared with CDDP treatment, the IC50
concentrations of CDDP in the HSC3 cells decreased between
7.02±0.46 and 3.85±0.41 μM when the cells were treated with
LP-pGRIM-19+CDDP (Fig. 1A), which
suggested that the exogenous expression of GRIM-19 enhanced the
sensitivity of the HSC3 cell lines to CDDP. Based on these results
the respective IC50 concentrations of CDDP were selected
for further treatments in the present study.
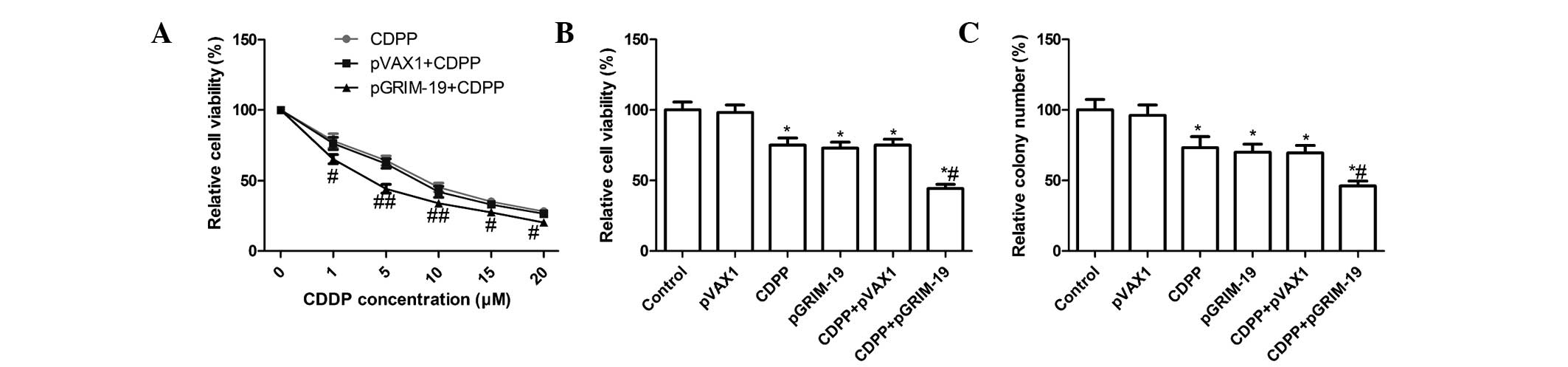 | Figure 1Combined effect of the exogenous
expression of GRIM-19 and treatment with CDDP on tumor cell
proliferation and clonogenicity in HSC3 cells. (A) HSC3 cells were
transfected with LP-pGRIM-19 and treated with various doses of
CDDP. Following 48 h treatment, cell viability was analyzed using
an MTT assay, and the IC50 values were calculated. (B)
HSC3 cells were transfected with LP-pGRIM-19 and then treated with
the IC50 concentration of CDDP for 48 h. Cell viability
was analyzed using an MTT assay. (C) Effects of of LP-pGRIM-19 and
low dose CDDP on colony formation of the HSC3 cells were determined
following treatment with low-dose CDDP and LP-pGRIM-19, alone and
in combination. Data are expressed as the mean ± standard
deviation. *P<0.05, vs. control;
#P<0.05, vs. CDDP. GRIM-19, gene associated with
retinoid-interferon-induced mortality 19; CDDP, cisplatin; LP,
liposome; IC50, half maximal inhibitory concentration;
p, plasmid; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. |
To determine whether the exogenous expression of
GRIM-19 enhances low-dose CDDP-mediated cell proliferation
inhibition, the HSC3 cells were transiently transfected with
LP-pGRIM-19 and, following transfection, the cells were treated
with low-dose CDDP at the IC50 concentration in The HSC3
cells. Significantly enhanced inhibition of cell proliferation
following treatment with LP-pGRIM-19+low-dose CDDP was observed in
the HSC3 cells, compared with treatment with either low-dose CDDP
or LP-pGRIM-19 alone (P<0.05; Fig.
1B).
Subsequently, the effects of the combination of
LP-pGRIM-19 and low-dose CDDP on colony formation of the HSC3 cells
were analyzed. As shown in Fig.
1C, the combination of LP-pGRIM-19 and low-dose CDDP
significantly inhibited the colony formation of the HSC3 cells,
compared with treatment with either low-dose CDDP or LP-pGRIM-19
alone (P<0.05; Fig. 1C).
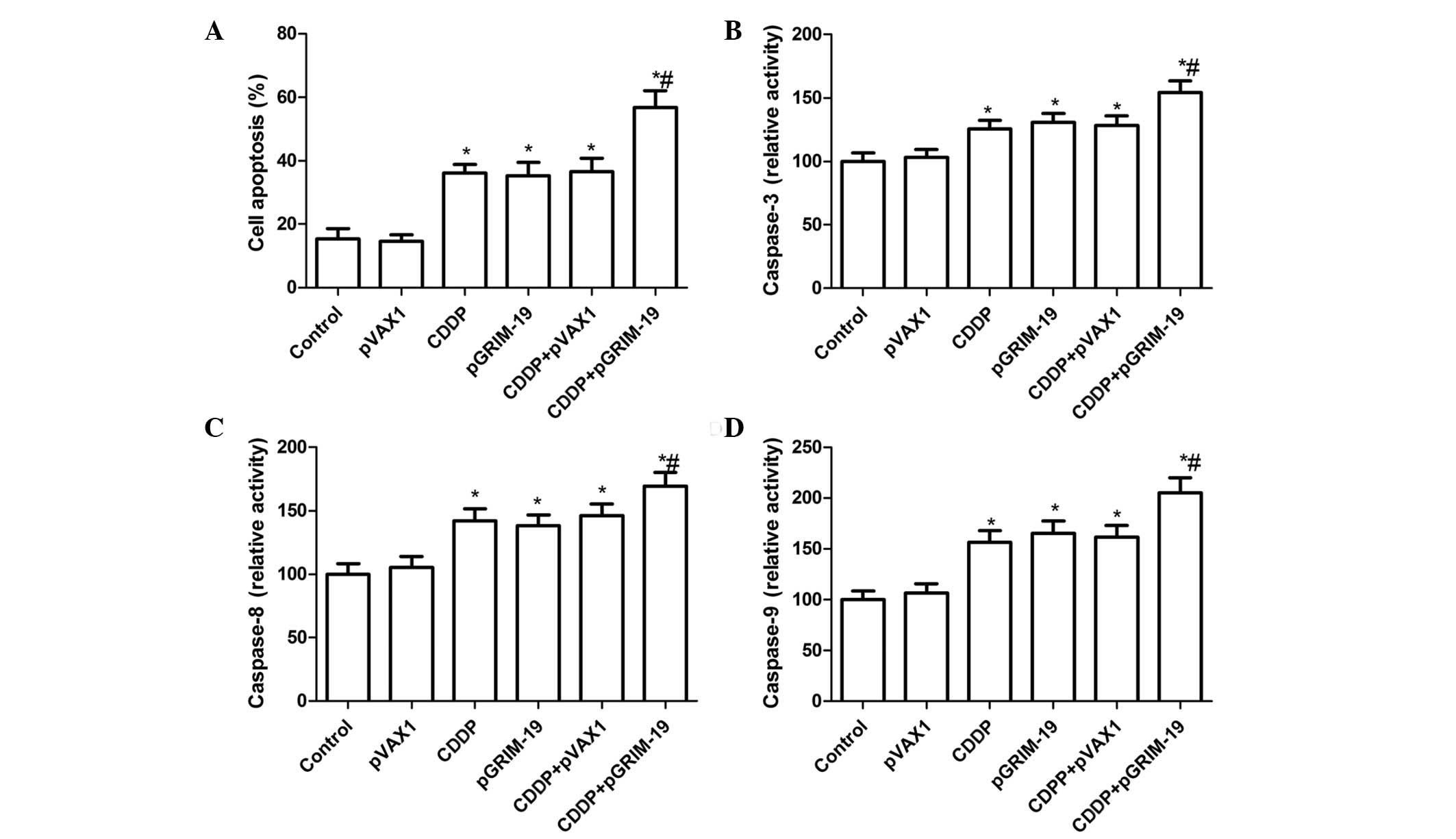
Treatment with a combination of
LP-pGRIM-19 and low-dose CDDP enhances the induction of HSC3 cell
apoptosis
To evaluate whether the exogenous expression of
GRIM-19 enhances CDDP-induced cell apoptosis, the HSC3 cells were
treated with low-dose CDDP and LP-pGRIM-19 either alone or in
combination, and cell apoptosis was detected using flow cytometry.
Treatment with LP-pGRIM-19+low-dose CDDP led to a significant
increase in apoptosis, compared with either low-dose CDDP or
LP-pGRIM-19 treatment alone (P<0.05), as shown Fig. 2A. In addition, no significance
different was observed between the low-dose CDDP- and
LP-pGRIM-19-only treatment groups in the induction of OSCC
apoptosis.
In order to examine the possible mechanism of the
pro-apoptotic effect of the combination of LP-pGRIM-19 and low-dose
CDDP, the activities of caspase-3, -8 and-9 were investigated. The
results (Fig. 2B–D) indicated that
the combination of LP-pGRIM-19 and low-dose CDDP significantly
increased the activities of caspase-3, -8 and -9 in the HSC3 cells,
relative to either CDDP or LP-pGRIM-19 treatment alone
(P<0.05).
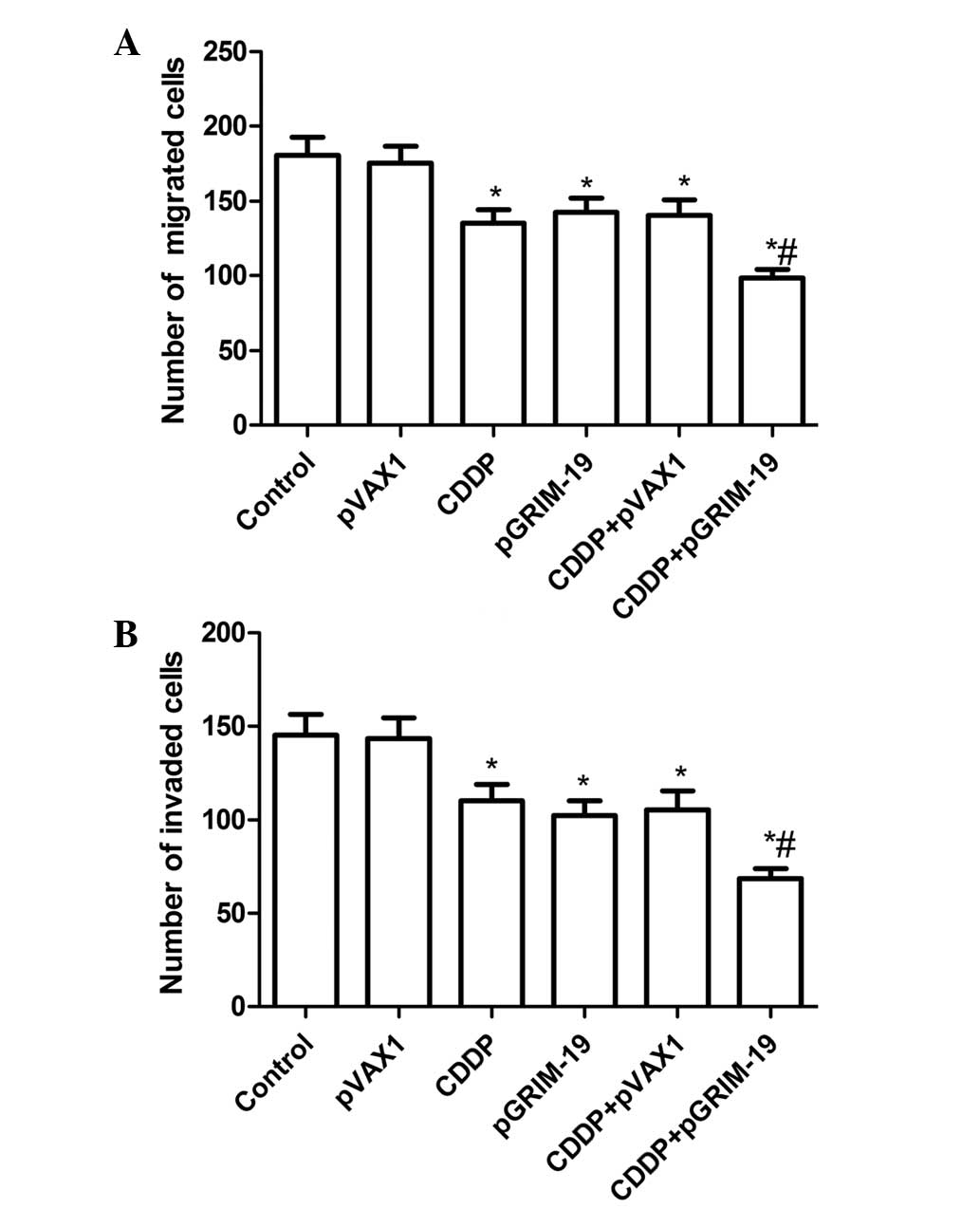
Treatment with a combination of
LP-pGRIM-19 and low-dose CDDP enhances the inhibition of cell
migration and invasion
The present study aimed to determine whether the
combination of LP-pGRIM-19 and low-dose CDDP affected cell
vitality, demonstrated by migration or invasion activity.
Therefore, cell migration and invasion assays were performed using
a Transwell assay. As shown in Fig.
3A, the combination of LP-pGRIM-19 and low-dose CDDP
significantly decreased the migration of HSC3 cells, compared with
the single LP-pGRIM-19 and CDDP treatment grou[s(P<0.05;
Fig. 3A). The ability of this
combination to reduce the invasiveness of HSC3 cells was
subsequently investigated. The Transwell matrix penetration assay,
which was coated with Matrigel revealed that the combination of
LP-pGRIM-19 and low-dose CDDP significantly reduced the
invasiveness of the HSC3 cells, compared with either CDDP or
LP-pGRIM-19 treatment alone (P<0.05; Fig. 3B).
Combination of LP-pGRIM-19 and low-dose
CDDP has a synergistic effect on relevant tumor effector molecules
in vitro
To elucidate the molecular mechanisms responsible
for the induction of synergistic growth inhibition and apoptosis of
OSCC cells by LP-pGRIM-19+low-dose CDDP, the expression levels of
Bcl-2, cyclinD1, Stat3 and p-p53 were analyzed in cells treated
with low-dose CDDP and LP-pGRIM-19 alone and in combination, using
western blot analysis. The combination of LP-pGRIM-19 and low-dose
CDDP resulted in further upregulation of the protein level of p-p53
and downregulation of the protein levels of Bcl-2, cyclinD1 and
Stat3 in the HSC3 cells, compared with the LP-pGRIM-19 or low-dose
CDDP alone treatment groups (Fig.
4).
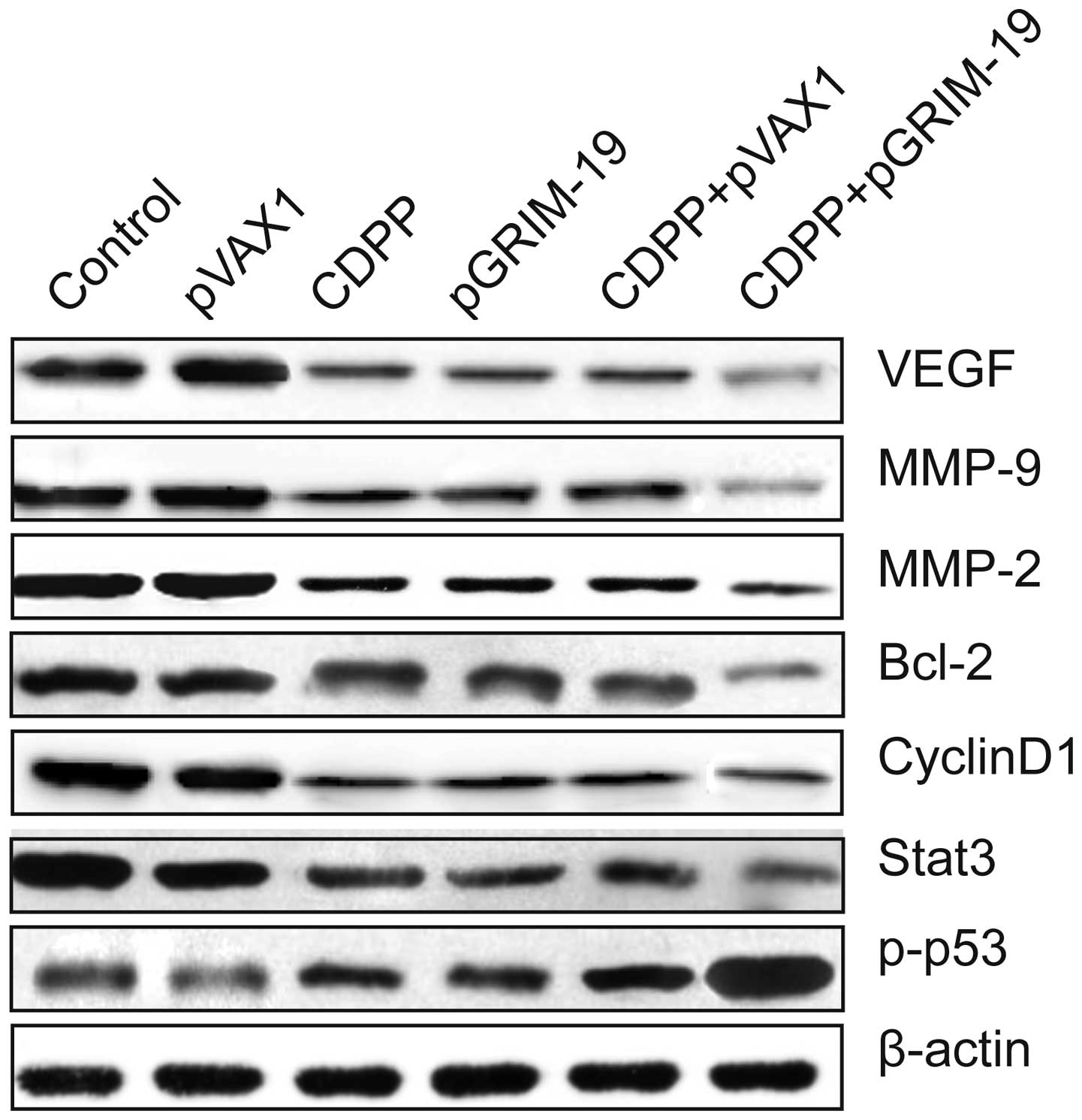 | Figure 4Combined effect of exogenous
expression of GRIM-19 and treatment with CDDP on proteins
associated with apoptosis and invasion. Protein expression levels
of p-p53, Stat3, Bcl-2, cyclin D1, MMP-2, MMP-9 and VEGF were
analyzed using western blot assay with specific antibodies
following treatment with low-dose CDDP and LP-pGRIM-19, alone and
in combination. β-actin was used as an internal control. GRIM-19,
gene associated with retinoid-interferon-induced mortality 19;
CDDP, cisplatin; LP, liposome; IC50, half maximal inhibitory
concentration; p, plasmid; p-p53, phosphorylated p53; Stat3, signal
transducer and activator of transcription 3; Bcl-2, B cell lymphoma
2; MMP, matrix metalloproteinase; VEGF, vascular endothelial growth
factor. |
To further investigate the potential mechanisms
underlying the effects of the combination of LP-pGRIM-19 and
low-dose CDDP on cell invasion and migration, relevant effector
molecules, including MMP-2, MMP-9 and VEGF were examined using
western blot analysis. It was found that the protein expression
levels of MMP-2, MMP-9 and VEGF were downregulated in the HSC3
cells treated with of LP-pGRIM-19+low-dose CDDP, compared with
treatment with either CDDP or LP-pGRIM-19 alone (Fig. 4)
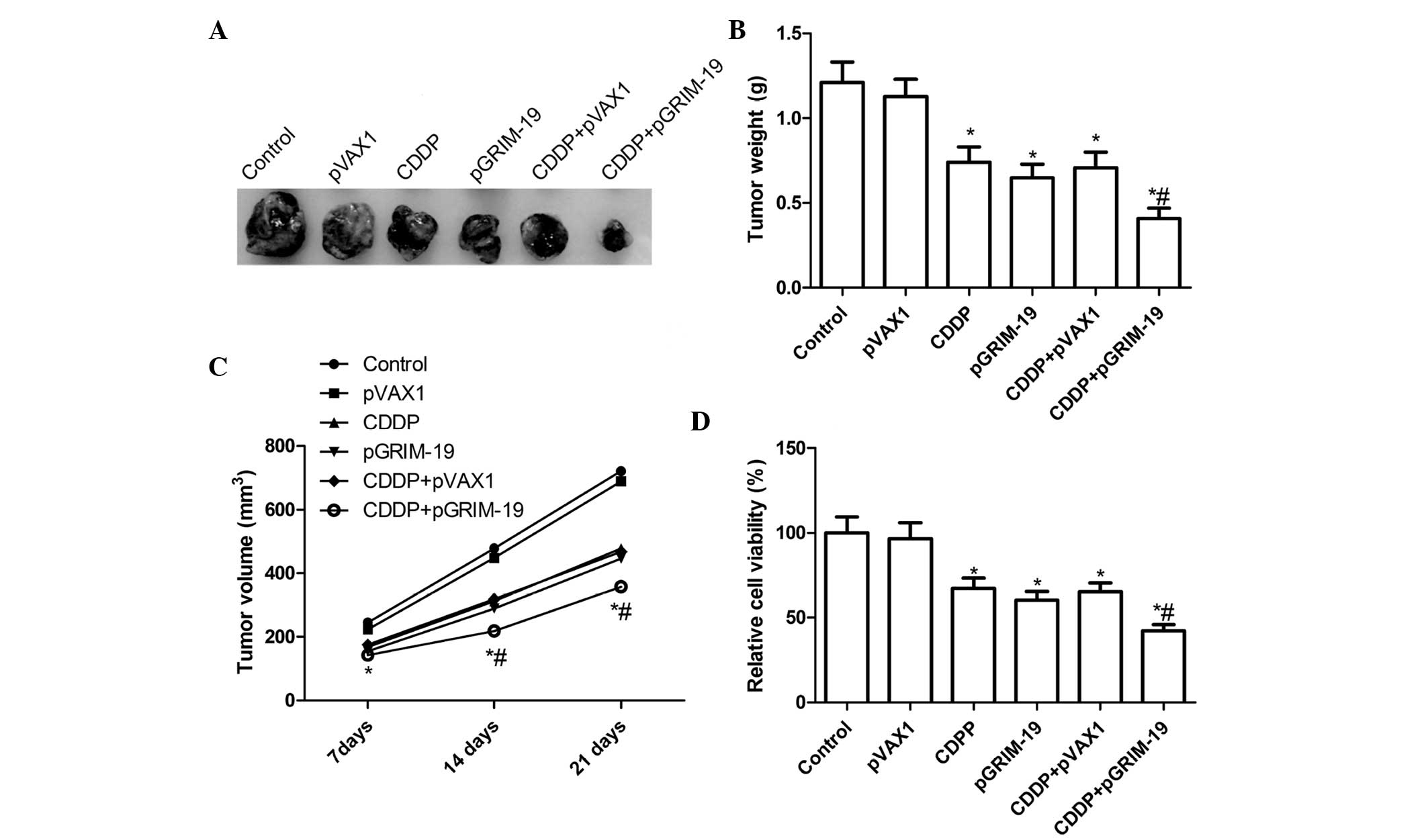
Treatment with a combination of
LP-pGRIM-19 and low-dose CDDP enhances tumor growth inhibition in
vivo
To determine whether GRIM-19 gene therapy enhances
CDDP-mediated antitumor activity in vivo, the present study
evaluated the effects of GRIM-19 combined with low-dose CDDP on
tumor regression in female BALB mice bearing HSC3 tumors. The mice
were sacrificed and tumor tissues were removed 21 days following
treatment, and the tumor weights were measured. The tumor weights
of the of LP-pGRIM-19+low-dose CDDP group were lower than those of
the control group and the CDDP-only and LP-pGRIM-19-only groups
(P<0.05; Fig. 5A and B). In
addition, the growth in tumor volume in the combination group was
significantly slower in the HSC3 tumor cells, compared with the
cispatin-only and LP-pGRIM-19-only groups (P<0.05; Fig. 5C). Subsequently, MTT assays were
used to examine the modulation of splenocyte proliferation and to
demonstrate the antitumor activities of this combination in
vivo. As shown in Fig. 5D,
splenocyte cell proliferation in the combination treatment group
decreased significantly, compared with the relative to
cispatin-only and LP-pGRIM-19-only groups (P<0.05). These
results suggested that GRIM-19 was responsible for the enhancement
of low-dose CDDP-induced antitumor activity in vivo.
Discussion
Drug resistance is a major cause of treatment
failure in patients with OSCC. The combination of conventional
CDDP-based chemotherapy and tumor suppressor gene therapy may
overcome cancer cell resistance to chemotherapeutic drugs and
enhance the therapeutic efficacy in cancer (17–20).
However, the role of GRIM-19, a tumor suppressor, in the response
to chemotherapy, has not been reported to date, to the best of our
knowledge. Therefore, the present study assessed the functional
effects of GRIM-19 on the effects of CDDP in OSCC. The results of
this investigation demonstrated that exogenously introduced
expression of GRIM-19 sensitized the response of OSCC cells to CDDP
treatment. Treatment with LP-GRIM-19 in combination with the
chemotherapeutic drug, CDDP, markedly inhibited tumor cell
proliferation, clonogenicity, migration and invasion, and induced
apoptosis in vitro. The present study also demonstrated that
co-treatment with LP-GRIM-19 and CDDP synergistically suppressed
tumor growth in a human OSCC tumor xenograft mouse model. To the
best of our knowledge, the present study is the first to report on
the combined introduction of the GRIM-19 tumor suppressor and the
conventional chemotherapeutic agent, CDDP, in human OSCC cells.
Apoptosis is the primary mechanism causing several
malignant cells to die when subjected to chemotherapy or
radiotherapy, and it has been demonstrated that mutations in
apoptotic pathways and decreased susceptibility to apoptosis are
associated with cellular resistance to chemotherapeutic drugs and
radiation treatment (23–25). The activation of p53 by
CDDP-induced DNA damage has been reported to have various effects
on cellular sensitivity towards CDDP (26), whereas downregulation in the
expression of Bcl-2 in cancer cells significantly increases the
sensitivity of cancer cells to CDDP (27). Our previous study demonstrated that
exogenously introduced expression of GRIM-19 in OSCC cells induced
cell apoptosis (16). In the
present study, the result revealed that the exogenously introduced
expression of GRIM-19 sensitized the response of the OSCC cells to
CDDP treatment, and that co-treatment with LP-GRIM-19 and CDDP
synergistically induced cell apoptosis, increased the activities of
caspase-3, -8 and-9, increased the expression of p-p53 and
decreased the expression of Bcl-2. These results suggested that the
overexpression of GRIM-19 in CDDP-treated OSCC cells may overcome
the cellular resistance and enhance the cellular response to the
chemotherapy, by facilitating drug-induced apoptosis, and by
upregulating and downregulating the expression levels of p53 and
Bcl-, respectively.
The altered expression of Stat3 has been reported to
be key in carcinogenesis by promoting cell proliferation,
differentiation and cell cycle progression, as well as inhibiting
apoptosis via the incessant induction of pro-growth genes,
including cyclin D1, Bcl-2, VEGF and MMP-2 (28–31).
The anticancer effects of CDDP can be enhanced by downregulating
the expression of Stat3 using Stat3 inhibitors or small interfering
RNA (32,33). GRIM-19, a Stat3 inhibitor, has been
found to upregulate the expression of GRIM-19 and inhibit Stat3
activation in different types of cancer (28,34–36),
due to the binding of GRIM-19 to the Stat3 gene and inhibiting its
transcription. In the present study, exogenously introduced
expression of GRIM-19 sensitized the response of OSCC cells to CDDP
treatment, and the combination of LP-pGRIM-19 and low-dose CDDP
resulted in further upregulation of p-p53, and downregulation of
Stat3, Bcl-2, cyclin D1, MMP-2, MMP-9 and VEGF at the ptotein
levels in HSC3 cells, compared with either LP-pGRIM-19 or low-dose
CDDP treatment alone. These results suggested that the enhanced
sensitivity of OSCC cells to CDDP by GRIM-19 may be through
inhibition of the Stat3 signaling pathway.
In conclusion, the present study demonstrated for
the first time, to the best of our knowledge, that treatment with
LP-pGRIM-19 in combination with a low dose of CDDP markedly
inhibited OSCC tumor cell proliferation, clonogenicity, migration
and invasion, and induced apoptosis in vitro, as well as
suppressing OSCC growth in vivo. The enhanced sensitivity of
OSCC cells to CDDP by GRIM-19 was associated with the upregulation
of p-p53 and downregulation of Bcl-2, VEGF, MMP-2 and MMP-9 at the
protein levels, which are involved in the activation of Stat3.
These findings provide novel insight into the molecular mechanisms
of GRIM-19-mediated tumor suppression and suggest that the
combination of GRIM-19 gene therapy with low-dose CDDP-based
chemotherapy may be a potent therapeutic strategy for the treatment
of human OSCC.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jerjes W, Upile T, Petrie A, Riskalla A,
Hamdoon Z, Vourvachis M, Karavidas K, Jay A, Sandison A, Thomas GJ,
et al: Clinicopathological parameters, recurrence, locoregional and
distant metastasis in 115 T1-T2 oral squamous cell carcinoma
patients. Head Neck Oncol. 2:92010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forastiere A, Koch W, Trotti A and
Sidransky D: Head and neck cancer. New Engl J Med. 345:1890–1900.
2001. View Article : Google Scholar
|
|
5
|
O'Dwyer PJ, Stevenson JP and Johnson SW:
Clinical pharmacokinetics and administration of established
platinum drugs. Drugs. 59(Suppl 4): 19–27. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baruah H, Barry CG and Bierbach U:
Platinum-intercalator conjugates: From DNA-targeted cisplatin
derivatives to adenine binding complexes as potential modulators of
gene regulation. Curr Top Med Chem. 4:1537–1549. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shimanishi M, Ogi K, Sogabe Y, Kaneko T,
Dehari H, Miyazaki A and Hiratsuka H: Silencing of GLUT-1 inhibits
sensitization of oral cancer cells to cisplatin during hypoxia. J
Oral Pathol Med. 42:382–388. 2013. View Article : Google Scholar
|
|
8
|
Perez RP: Cellular and molecular
determinants of cisplatin resistance. Eur J Cancer. 34:1535–1542.
1998. View Article : Google Scholar
|
|
9
|
Rabik CA and Dolan ME: Molecular
mechanisms of resistance and toxicity associated with platinating
agents. Cancer Treat Rev. 33:9–23. 2007. View Article : Google Scholar :
|
|
10
|
Cohen SM and Lippard SJ: Cisplatin: From
DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol.
67:93–130. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johnson DH: Evolution of cisplatin-based
chemotherapy in non-small cell lung cancer: A historical
perspective and the eastern cooperative oncology group experience.
Chest. 117(Suppl 1): S133–S137. 2000. View Article : Google Scholar
|
|
12
|
Maitra A, Wistuba II, Washington C,
Virmani AK, Ashfaq R, Milchgrub S, Gazdar AF and Minna JD:
High-resolution chromosome 3p allelotyping of breast carcinomas and
precursor lesions demonstrates frequent loss of heterozygosity and
a discontinuous pattern of allele loss. Am J Pathol. 159:119–130.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zabarovsky ER, Lerman MI and Minna JD:
Tumor suppressor genes on chromosome 3p involved in the
pathogenesis of lung and other cancers. Oncogene. 21:6915–6935.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lerman MI and Minna JD: The 630-kb lung
cancer homozygous deletion region on human chromosome 3p21.3:
Identification and evaluation of the resident candidate tumor
suppressor genes. The international lung cancer chromosome 3p213
tumor suppressor gene consortium. Cancer Res. 60:6116–6133.
2000.PubMed/NCBI
|
|
15
|
Angell JE, Lindner DJ, Shapiro PS, Hofmann
ER and Kalvakolanu DV: Identification of GRIM-19, a novel cell
death-regulatory gene induced by the interferon-beta and retinoic
acid combination, using a genetic approach. J Biol Chem.
275:33416–33426. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li M, Li Z, Liang C, Han C, Huang W and
Sun F: Upregulation of GRIM-19 suppresses the growth of oral
squamous cell carcinoma in vitro and in vivo. Oncolo Rep.
32:2183–2190. 2014.
|
|
17
|
Ou W, Ye S, Yang W, Wang Y, Ma Q, Yu C,
Shi H, Yuan Z, Zhong G, Ren J, et al: Enhanced antitumor effect of
cisplatin in human NSCLC cells by tumor suppressor LKB1. Cancer
Gene Ther. 19:489–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nemunaitis J, Swisher SG, Timmons T,
Connors D, Mack M, Doerksen L, Weill D, Wait J, Lawrence DD, Kemp
BL, et al: Adenovirus-mediated p53 gene transfer in sequence with
cisplatin to tumors of patients with non-small-cell lung cancer. J
Clin Oncol. 18:609–622. 2000.PubMed/NCBI
|
|
19
|
Ueda K, Kawashima H, Ohtani S, Deng WG,
Ravoori M, Bankson J, Gao B, Girard L, Minna JD, Roth JA, et al:
The 3p21.3 tumor suppressor NPRL2 plays an important role in
cisplatin-induced resistance in human non-small-cell lung cancer
cells. Cancer Res. 66:9682–9690. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng WG, Wu G, Ueda K, Xu K, Roth JA and
Ji L: Enhancement of antitumor activity of cisplatin in human lung
cancer cells by tumor suppressor FUS1. Cancer Gene Ther. 15:29–39.
2008. View Article : Google Scholar
|
|
21
|
Chen X, Wang X, Wang Y, Yang L, Hu J, Xiao
W, Fu A, Cai L, Li X, Ye X, et al: Improved tumor-targeting drug
delivery and therapeutic efficacy by cationic liposome modified
with truncated bFGF peptide. J Control Release. 145:17–25. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Li Z and Wang K: Combining
sorafenib with celecoxib synergistically inhibits tumor growth of
non-small cell lung cancer cells in vitro and in vivo. Oncol Rep.
31:1954–1960. 2014.PubMed/NCBI
|
|
23
|
Lowe SW, Ruley HE, Jacks T and Housman DE:
p53-dependent apoptosis modulates the cytotoxicity of anticancer
agents. Cell. 74:957–967. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu JR, Opipari AW, Tan L, Jiang Y, Zhang
Y, Tang H and Nuñez G: Dysfunctional apoptosome activation in
ovarian cancer: Implications for chemoresistance. Cancer Res.
62:924–931. 2002.PubMed/NCBI
|
|
25
|
Wurstle ML, Zink E, Prehn JH and Rehm M:
From computational modelling of the intrinsic apoptosis pathway to
a systems-based analysis of chemotherapy resistance: Achievements,
perspectives and challenges in systems medicine. Cell Death Dis.
5:e12582014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niedner H, Christen R, Lin X, Kondo A and
Howell SB: Identification of genes that mediate sensitivity to
cisplatin. Mol Pharmacol. 60:1153–1160. 2001.PubMed/NCBI
|
|
27
|
Park SA, Park HJ, Lee BI, Ahn YH, Kim SU
and Choi KS: Bcl-2 blocks cisplatin-induced apoptosis by
suppression of ERK-mediated p53 accumulation in B104 cells. Brain
Res Mol Brain Res. 93:18–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Gao L, Li Y, Lin G, Shao Y, Ji K,
Yu H, Hu J, Kalvakolanu DV, Kopecko DJ, et al: Effects of
plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PC-3
M tumor cell growth. Clin Cancer Res. 14:559–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Turkson J: STAT proteins as novel targets
for cancer drug discovery. Expert Opin Ther Targets. 8:409–422.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Niu G, Wright KL, Huang M, Song L, Haura
E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al:
Constitutive Stat3 activity up-regulates VEGF expression and tumor
angiogenesis. Oncogene. 21:2000–2008. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F,
Sawaya R and Huang S: Stat3 activation regulates the expression of
matrix metalloproteinase-2 and tumor invasion and metastasis.
Oncogene. 23:3550–3560. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kumar B, Yadav A, Hideg K, Kuppusamy P,
Teknos TN and Kumar P: A novel curcumin analog (H-4073) enhances
the therapeutic efficacy of cisplatin treatment in head and neck
cancer. PloS One. 9:e932082014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han Z, Feng J, Hong Z, Chen L, Li W, Liao
S, Wang X, Ji T, Wang S, Ma D, et al: Silencing of the STAT3
signaling pathway reverses the inherent and induced chemoresistance
of human ovarian cancer cells. Biochem Biophys Res Commun.
435:188–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Li M, Wei Y, Feng D, Peng C, Weng
H, Ma Y, Bao L, Nallar S, Kalakonda S, et al: Downregulation of
GRIM-19 expression is associated with hyperactivation of
STAT3-induced gene expression and tumor growth in human cervical
cancers. J Interferon Cytokine Res. 29:695–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hao H, Liu J, Liu G, Guan D, Yang Y, Zhang
X, Cao X and Liu Q: Depletion of GRIM-19 accelerates hepatocellular
carcinoma invasion via inducing EMT and loss of contact inhibition.
J Cell Physiol. 227:1212–1219. 2012. View Article : Google Scholar
|
|
36
|
Okamoto T, Inozume T, Mitsui H, Kanzaki M,
Harada K, Shibagaki N and Shimada S: Upregulation of GRIM-19 in
cancer cells suppresses STAT3-mediated signal transduction and
cancer growth. Mol Cancer Ther. 9:2333–2343. 2010. View Article : Google Scholar : PubMed/NCBI
|