Introduction
As a squamous cell carcinoma, nasopharyngeal
carcinoma (NPC) is derived from the epithelium of the nasopharynx.
Compared with the rest of the world, the incidence of NPC in China
is high (1), and therefore is a
serious public health problem in China. In regions covered by the
cancer registries in 2009, the crude incidence of NPC was
3.61/100,000 (5.08/100,000 in males and 2.10/100,000 in females;
4.19/100,000 in urban areas and 2.42/100,000 in rural areas)
(1). Due to the high incidence of
early metastasis, the rate of mortality is high in patients with
NPC. At present, radiotherapy is the first choice of therapy for
patients with NPC. However, despite improvements in the standards
of radiotherapy, the five-year survival rate remains to be ~50–60%
(2). Therefore, it is important
for clinicians and researchers to develop novel effective treatment
strategies.
The transcription factor nuclear factor-κB (NF-κB)
consists of 5 subunits; Rel (cRel), p65 (RelA, NF-κB3), RelB, p50
(NF-κB1) and p52 (NF-κB2) (3), and
the two most common dimers of NF-κB are p65 and p50 (3). In unstimulated cells, inhibitory κB
(IκB) is bound to NF-κB, which remains in an inactive form in the
cytoplasm (3). When cells are
stimulated by extracellular signals, IκB kinase complex (IKK)
phosphorylates IκB, exposing the nuclear localization sites of
NF-κB (3). Subsequently the free
NF-κB translocates into the nucleus where it binds with specific κB
sequences, inducing gene transcription (3). Previous histological studies have
indicated the importance of local inflammation in NPC tumorigenesis
(4). As a key inflammatory
signaling pathway, NF-κB has been demonstrated to be constitutively
active in NPC tissue by immunohistochemical staining (4). The constitutive activation of NF-κB
commonly results in malignant carcinoma cell proliferation in
various types of cancer cells and tissues, as the NF-κB signaling
pathway regulates a series of target genes involved in cell
proliferation, apoptosis, immune responses and transcription
(5).
MicroRNAs (miRNAs) are small non-coding RNAs of
20–25 nucleotides. miRNAs negatively regulate gene expression by
recognizing complementary sequences in the 3′-untranslated regions
(UTR) of target mRNAs, resulting in their degradation (6). In previous studies, the aberrant
expression of miRNAs has been associated with various types of
human cancer (7,8). In addition, as an important mediator,
miRNAs are accepted as key modulators and effectors of the NF-κB
signaling pathway. For example, miR-146a and miR-146b negatively
interact with interleukin-1 receptor-associated kinase 1 and tissue
necrosis factor receptor-associated factor 6 protein levels,
resulting in the activation of NF-κB (9). Furthermore, miR-199a has been
demonstrated to suppress IKKβ, which reduces the activity of NF-κB
signaling (10), whilst miR-200a
has been reported to regulate various signaling pathways through
targeting different genes, including epidermal growth factor
receptor, c-Met and ephrin type-A receptor 1 (11,12).
In the current study, the relative expression levels
of miR-200a were investigated in human NPC. In addition, the impact
of increased miR-200a levels on NPC cell proliferation and
migration was explored. The present study aimed to elucidate the
effect of miR-200a on NPC cell proliferation and the role of the
NF-κB signaling pathway in this process.
Materials and methods
Human samples and cell lines
A total of 40 samples of primary human NPC and 30
normal samples were collected at the People's Liberation Army 113th
hospital (Ningbo, China) and written informed consent was obtained
from all patients. Clinical information was obtained by reviewing
the medical records on radiographic images, by telephone or written
correspondence, and by reviewing death certificates. All specimens
had confirmed pathological diagnosis and were staged according to
the 1992 NPC staging system of China (13). The NPC cell lines (HNE1, CNE1 and
CNE2) and an immortalized nasopharyngeal epithelial cell line
(NP69) were purchased from the American Type Culture Collection
(Manassas, VA, USA) and cultured in Eagle's minimum essential
medium (MEM; GE Healthcare Life Sciences, Logan, UT, USA) with 10%
fetal bovine serum(FBS; GE Healthcare Life Sciences). The HEK293T
cells were cultured in 2 ml Dulbecco's modified Eagle's medium (GE
Healthcare Life Sciences) supplemented with 100 U/ml penicillin
(Beijing SolarBio Science & Technology Co., Ltd., Beijing,
China, Beijing, China), 100 U/ml streptomycin and 10% FBS.
Tumor necrosis factor-α (TNF-α)
treatment
CNE2 cells were seeded in the six-well plate at the
concentration of 1×106 cells/well. After 24 h, the CNE2
cells were treated with 10 ng/ml TNFα for 48 h. The relative levels
of miR-200a were then determined.
Transient transfection procedures
Shortly prior to transfection, 1.5×105
cells were seeded per well in a 6-well plate in 2 ml Dulbecco's
modified Eagle's medium containing serum and supplemented with 100
U/ml penicillin and 100 U/ml streptomycin. Prior to transfection,
the cells were incubated under normal growth conditions (typically
37°C and 5% CO2). Subsequently, miR-200a mimics,
miR-200a inhibitor or the miR negative control (Shanghai Genepharma
Co., Ltd., Shanghai, China) were pre-incubated with HiPerFect
transfection reagent (Qiagen China Co., Ltd., Shanghai, China) with
the final concentration of microRNA analogues at 100 nmol/l. The
sequence of miR-200a was as follows:
5′-CAGUGCAAUAGUAUUGUCAAAGC-3′.
siRNA transfection
Specific siRNA targeting p65 was purchased from
Shanghai Jima Co. (Shanghai, China). The siRNAs were transfected
into the cells using Vigofect transfection reagent (Vigorous
Biotechnology Beijing Co., Ltd., Beijing, China).
RNA extraction
Total RNA (2 µg) was extracted from cell
lines and human samples (5 mg) with TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
In order to detect and quantify mature miRNA-200a, a
TaqMan MicroRNA Reverse Transcription kit and a TaqMan MicroRNA
Assay were used according to the manufacturer's instructions
(Applied Biosystems Life Technologies, Foster City, CA, USA). U6
RNA was used for normalization. To quantify miRNA levels, 10 ng
total RNA was reverse transcribed using TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems Life Technologies) with
specific primers for miR-200a and U6. Subsequently, the PCR
amplifications were performed in 20 µl reaction volume
containing 10 µg TaqMan 2X Universal PCR Master Mix, 1
µl 20X TaqMan MicroRNA Assay mix (Applied Biosystems Life
Technologies) and 1.33 µl template cDNA in the same system
used for mRNA quantification. The thermal cycling conditions were
as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15
sec and 60°C for 1 min. Relative miRNA expression of miR-200a was
normalized against the endogenous control, U6 RNA, using the
comparative 2−ΔΔCt method (14). CFX Manager™ software (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used for quantification
analysis for mRNA and miRNA. For reverse transcription, the
specific primers are listed, as follows: (5′-3′): miR-200a,
CUGGAUUUCCCAGCUUGACUCUAACACUGUCUGGUAACGAUGUUCAAAGGUGACCCGC; U6,
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATATG. The primers
used for qPCR were as follows (5′-3′): miR-200a forward,
GCAAAGTGCATCCATTTTGTTTGT; U6 forward, GCGCGTCGTGAAGCGTTC; universal
reverse primer, GTGCAGGGTCCGAGGT.
Protein extraction, western blotting and
antibodies
Cellular proteins were extracted using RIPA buffer
[50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1% (v/v) NP-40, 0.1% (w/v)
SDS; Beijing SolarBio Science & Technology Co., Ltd.)
containing 1% (v/v) phenylmethanesulfonylfluoride (Beijing SolarBio
Science & Technology Co., Ltd.), 0.3% (v/v) protease inhibitor
(Sigma-Aldrich, St. Louis, MO, USA) and 0.1% (v/v) phosphorylated
proteinase inhibitor (Sigma-Aldrich). Lysates were centrifuged at
13,000 g at 4°C for 15 min and the supernatant was collected for
total protein analysis. A bicinchoninic acid protein assay kit
(Pierce Biotechnology, Inc., Rockford, IL, USA) was used to
determine the protein concentration. Equal amounts of protein (15
µg) were separated on an SDS-PAGE gel [10% (v/v)
polyacrylamide; Beijing SolarBio Science & Technology Co.,
Ltd.] and transferred onto a polyvinylidene fluoride membrane
(Merck Millipore, Darmstadt, Germany). Nonspecific binding was
blocked using 8% (w/v) milk in Tris-buffered saline-Tween 20 (TBST)
for 2 h at room temperature. The membranes were incubated with
primary antibodies against β-actin (8H10D10; mouse monoclonal; cat.
no. 3700; 1:3,000, Cell Signaling Technology, Inc., Danvers, MA,
USA), NF-κB p65 (D14E12) XP® Rabbit mAb (cat. no. 8242;
1:1,000; Cell Signaling Technology, Inc.), NF-κB1 p105/p50 antibody
(cat. no. 3035; 1:1,000; Cell Signaling Technology, Inc.), Cox2
(D5H5) XP® rabbit mAb (cat. no. 12282; 1:1,000; Cell
Signaling Technology, Inc.), MMP-2 (D8N9Y) rabbit mAb (cat. no.
13132; 1:1,000; Cell Signaling Technology, Inc.), human vascular
endothelial growth factor-165 (cat. no. 8065; 1:1,000; Cell
Signaling Technology, Inc.) Phosphorylated (p)-NF-κB p65 (Ser536;
93H1) rabbit mAb (cat. no. 3033; 1:1,000; Cell Signaling
Technology, Inc.), ICAM-2 (D7P2Q) rabbit mAb (cat. no. 13355;
1:1,000, Cell Signaling Technology, Inc.), MCP-1 antibody (cat. no.
2027; 1:1,000; Cell Signaling Technology, Inc.) and IκBα (44D4)
rabbit mAb (cat. no. 4812; 1:1,000; Cell Signaling Technology,
Inc.) overnight at 4°C. Following three washes with TBST, the
membranes were incubated in horseradish peroxidase (HRP)-conjugated
goat anti-rabbit (cat. no. ZB-2307; 1:5,000; Zhongshan Gold Bridge
Biotechnology Co., Ltd., Beijing, China) and anti-mouse (cat. no.
ZB-2305 1:5,000; Zhongshan Gold Bridge Biotechnology, Co., Ltd.) or
HRP-conjugated mouse anti-goat antibodies (cat. no. ZB-5305;
Beijing Zhongshan Jinqiao Biotechnology Co., Ltd., Beijing, China;
1:5,000) for 2 h at room temperature and then washed with TBST
three times (5 min each). The target proteins were subsequently
visualized using enhanced chemiluminescence (EMD Millipore,
Billerica, MA, USA) according to the manufacturer's instructions
and quantified using density analysis with ImageJ 4.0 software
(National Institutes of Health, Bethesda, MA, USA) normalized
against β-actin, and expressed as the fold-change, compared with
the control.
Luciferase target assay
TargetScan (http://www.targetscan.org/) was used to determine the
potential binding sites of miR-200a on the 3′UTR of IκBα. For the
luciferase assay, the 3′UTR of IκBα, including the binding site for
miR-200a, was amplified from CNE2 cells using the following
primers: IκBα-F, 5′-AAGGAGGAGGGCAGAATCAT-3′; IκBα-R,
5′-ATCTGCATGGTGATGTTGGA-3′.
The PCR product was digested with XbaI (New
England Biolabs, Beverly, MA, USA) and cloned into the reporter
plasmid pGL3 (Promega Corporation, Madison, WI, USA) downstream of
the luciferase reporter gene. The modified firefly luciferase
vector (500 ng/µl; Promega Corporation) was transfected into
HEK293 cells (2×105 cells/ml; as previously described).
Firefly and Renilla luciferase activities were measured 48 h
following transfection with the Dual-Luciferase Reporter Assay
System (Promega Corporation). Firefly activity was normalized to
Renilla activity to control the transfection efficiency.
Dihydroethidium (DHE) staining
The cells were cultured in six-well chamber slides
at a density of 1×106 cells/well, washed with PBS three
times (5 min/wash) and incubated with ROS Fluorescent Probe-DHE (10
µM; Vigorous Biotechnology Beijing Co., Ltd.) in serum-free
DMEM F-12 medium for 30 min at 37°C in the dark. Following
incubation, the slides were fixed in 4% paraformaldehyde for 30 min
at room temperature, were washed again and mounted.
Immunofluorescence images were captured using fluorescence
microscopy (Leica CM3000; Leica Microsystems GmbH, Wetzlar,
Germany).
Dimethyl thiazolyl diphenyl tetrazolium
(MTT) assay
To evaluate the effect of miR-200a on cell
proliferation, cells were seeded 5,000 cells/well in 100 µl
MEM in 96-well plates and transfected with miR-200a mimics (50 nM)
and negative control-miRNA mimics (50 nM), as described above. At
24, 48 and 72 h post-transfection, 20 µl MTT reagent
(Beijing SolarBio Science & Technology Co., Ltd.) was added to
the wells, which were then incubated for 4 h at 37°C. Following
removal of the medium, 200 µl dimethyl sulfoxide was added
to dissolve the formazan and the absorbance was measured at 550 nm.
Wells containing only CNE2 cells, which served as blanks.
Statistical analysis
Data are presented as the mean ± standard error from
3 independent experiments. Statistical analysis was conducted with
Student's t-test using GraphPad Prism 5 software (National
Institutes of Health). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-200a is upregulated in human NPC
cells and tissue samples
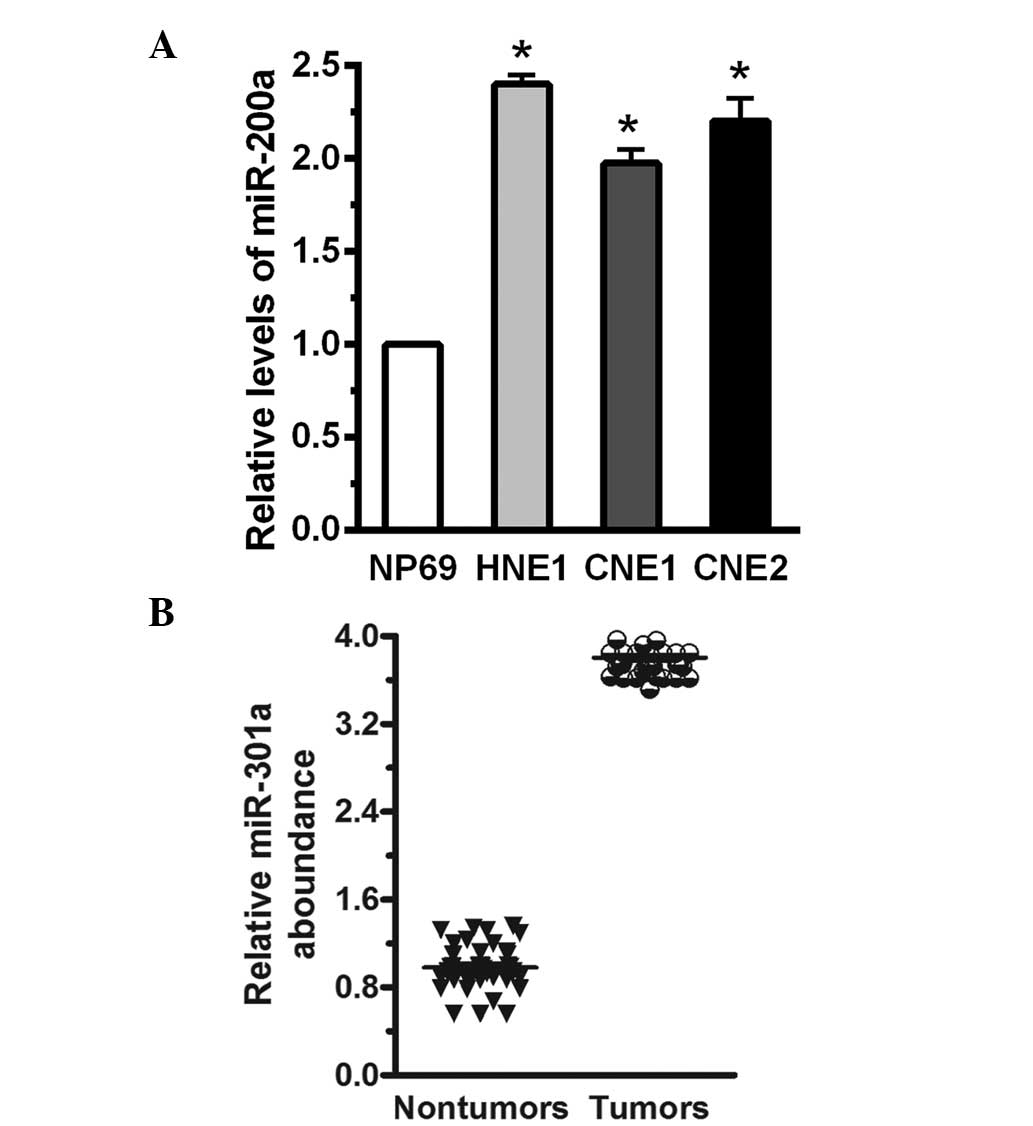
The relative levels of miR-200a in human NPC cells
and tissue samples were measured in human NPC cells and patient
tissue samples using RT-qPCR. Compared with NP69 immortalized
nasopharyngeal epithelial cells, miR-200a levels were increased by
greater than 2.38-fold in HNE1, CNE1 and CNE2 human NPC cells, and
miR-200a expression levels were normalized to U6 (P<0.05;
Fig. 1A). The expression levels of
miR-200a were measured in 40 human NPC cancer specimens compared
with 30 normal tissue samples. Compared with the normal tissue, the
mean expression level of miR-200a was increased by greater than
3-fold (P<0.05; Fig. 1B). This
indicated that miR-200a was significantly increased in the human
NPC cells and tumor tissue.
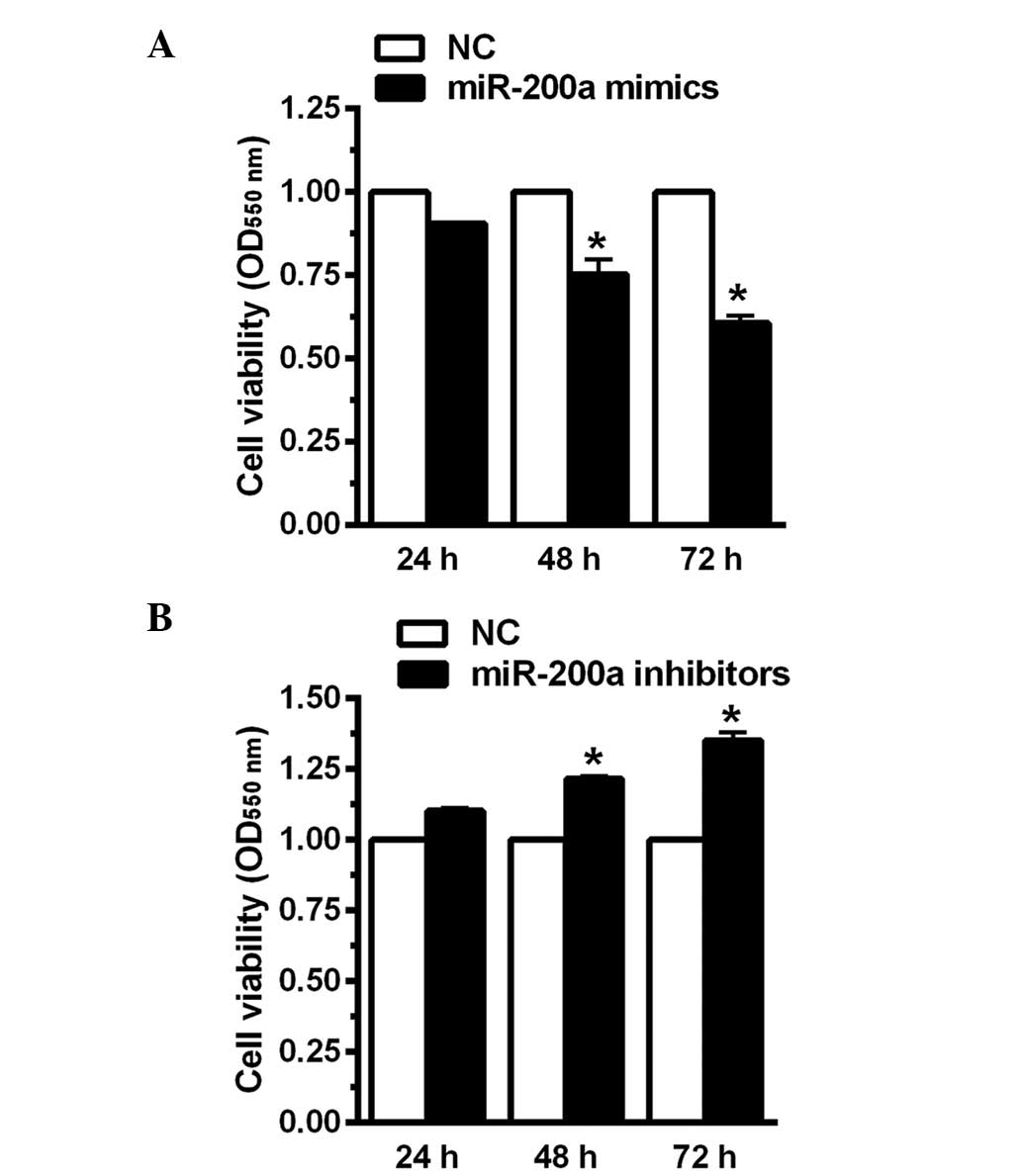
Downregulation of miR-200a increases CNE2
cell viability
In order to investigate the effect of miR-200a on
CNE2 cell viability, CNE2 cells were transfected with miR-200a
mimics, miR-200a inhibitors or the negative control for 24, 48 and
72 h. In the current study, the mimics were analogues that enhanced
miR-200a expression levels while inhibitors were analogues that
reduced the expression of miR-200a. The MTT assay indicated that
when miR-200a mimics were transfected into CNE2 human NPC cells,
cell viability was significantly decreased by 23 and 35% at 48 and
72 h, respectively (Fig. 2A).
However, when miR-200a expression was inhibited, cell viability was
enhanced by 17 and 36% at 48 and 72 h, respectively (Fig. 2B). These results indicated that
miR-200a may increase CNE2 cell viability.
miR-200a activates the NF-κB signaling
pathway
In NPC, the NF-κB signaling pathway is
constitutively activated (15). In
the current study, CNE2 cells were treated with 10 ng/µl
TNF-α for 48 h, following which the relative levels of miR-200a
were measured. As presented in Fig.
3A, the relative level of miR-200a was increased by greater
than 4.8-fold with TNF-α treatment. To address whether miR-200a
contributes to NF-κB activation, CNE2 cells were transfected with
miR-200a mimics. Western blot analysis indicated that when miR-200a
was overexpressed, NF-κB was significantly activated. As presented
in Fig. 3, compared with the
negative control, the p-NF-κB/NF-κB ratio was increased by greater
than 2.36-fold (Fig. 3B). Vascular
cell adhesion molecule (VCAM), intercel-lular adhesion molecule
(ICAM) and monocyte chemoattractant protein-1 (MCP-1) are important
adhesion molecules that are aberrantly increased in various types
of cancer (16). To further
investigate the alterations in NF-κB activation, the protein levels
of VCAM, ICAM and MCP-1 were measured. As presented in Fig. 3B, the relative levels of VCAM, ICAM
and MCP-1 were significantly increased. Together, these data
indicate that miR-200a contributed to TNF-α-stimulated NF-κB
activation. To investigate the effect of NF-κB on cell
proliferation, a small interfering RNA (siRNA) targeting NF-κB was
used. Following treatment with the siRNA against NF-κB, the protein
levels of NF-κB were reduced, as were the expression levels of
VCAM, ICAM and MCP-1. Additionally, this effect was observed in the
cells transfected with miR-200a mimics (Fig. 3C). VCAM was increased by 2.1-fold
following the addition of mimics, although the reduction in MCP-1
was less marked. Furthermore, as presented in Fig. 3D, with NF-κB knockdown, cell
viability was significantly reduced even in the cells transfected
with miR-200a mimics. These data suggest that miR-200a enhances
cell proliferation through the activation of the NF-κB signaling
pathway.
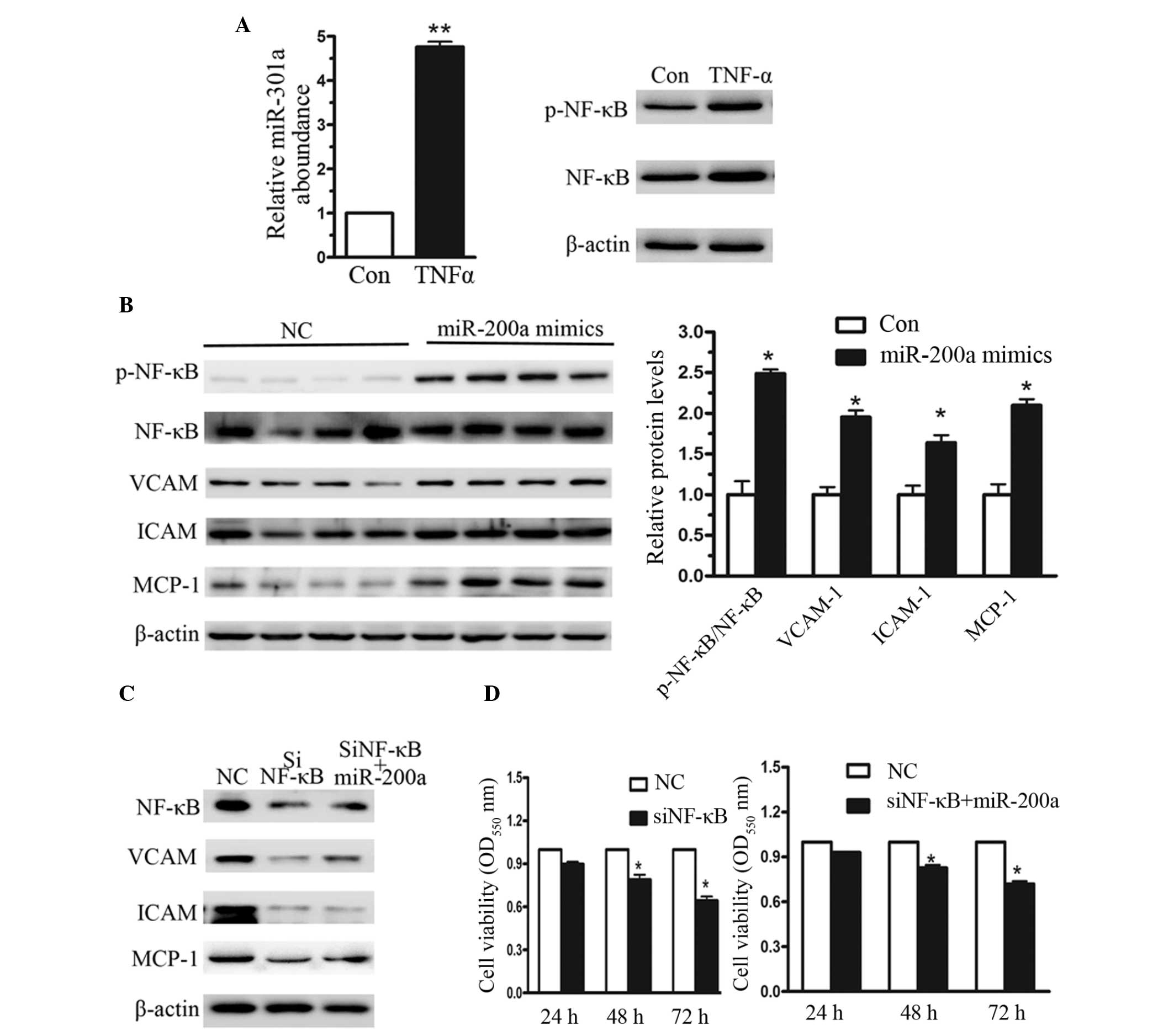 | Figure 3NF-κB signaling pathway activation
with overexpression of miR-200a in CNE2 cells. (A) Reverse
transcription-quantitative polymerase chain reaction was used to
measure the relative levels of miR-200a when CNE2 cells were
treated with 10 ng/µl TNFα for 48 h. (B) Western blot
analysis of NF-κB activation and its downstream regulators
following miR-200a overexpression. (C) Western blot analysis of an
short interfering RNA targeting NF-κB. (D) MTT assay indicated a
reduced rate of cell proliferation in cells cotransfected with
si-NF-κB and miR-200a mimics. Data represent the mean ± standard
error, n=3 independent experiments. *P<0.05 vs.
control. NF-κB, nuclear factor-κB; miR-200a, microRNA-200a; TNFα,
tissue necrosis factor α; si-NF-κB, short interfering NF-κB; Con,
control; p-NF-κB, phosphorylated NF-κB; VCAM, vascular cell
adhesion molecule; ICAM, intercellular adhesion molecule; MCP-1,
monocyte chemoattractant protein-1. |
IκBα is the host gene of miR-200a
To further investigate whether NF-κB was activated
by miR-200a overexpression, immunofluorescence was used. As
presented in Fig. 4A, increased
NF-κB (p65) protein levels were observed when CNE2 cells were
transfected with miR-200a mimics. In previous studies, numerous
miRNAs have been reported to activate NF-κB, for instance, miR-200a
targeted NF-κB-repressing factor and correlated with altered NF-κB
activation (17,18). In the current study, the target
gene of miR-200a was identified using a bio-informatics database.
TargetScan predicted miR-200a to target position 356–362 in the
3′UTR in IκBα (Fig. 4B). IκB is an
NF-κB inhibitory protein, which in unstimulated cells is bound to
p65 and P50, resulting in the inactivation of NF-κB in the
cytoplasm (19). Following the
activation of IKK, IκBα is degraded and the two subunits of NF-κB
translocate from the cytoplasm to the nucleus, thereby inducing the
downstream signaling pathway (20). Therefore, the effect of miR-200a on
IκBα expression was investigated. When CNE2 cells were transfected
with miR-200a mimics for 4 h, the expression levels of IκBα were
significantly reduced compared with the negative control (Fig. 4B). Furthermore, 48 h following
transfection with miR-200a in CNE2 cells, the expression level of
IκBα was reduced by 56%. However, when miR-200a was inhibited in
CNE2 cells, the expression level of IκBα was increased by almost
1-fold (Fig. 4C). A luciferase
reporter assay was used to investigate the effect of miR-200a on
the 3′-UTR of IκBα, and indicated that miR-200a significantly
reduced IκBα-3′-UTR-luciferase reporter activity (Fig. 4D). These data indicated that
miR-200a induced NF-κB activation, predominantly by targeting IκBα
in the human NPC cells.
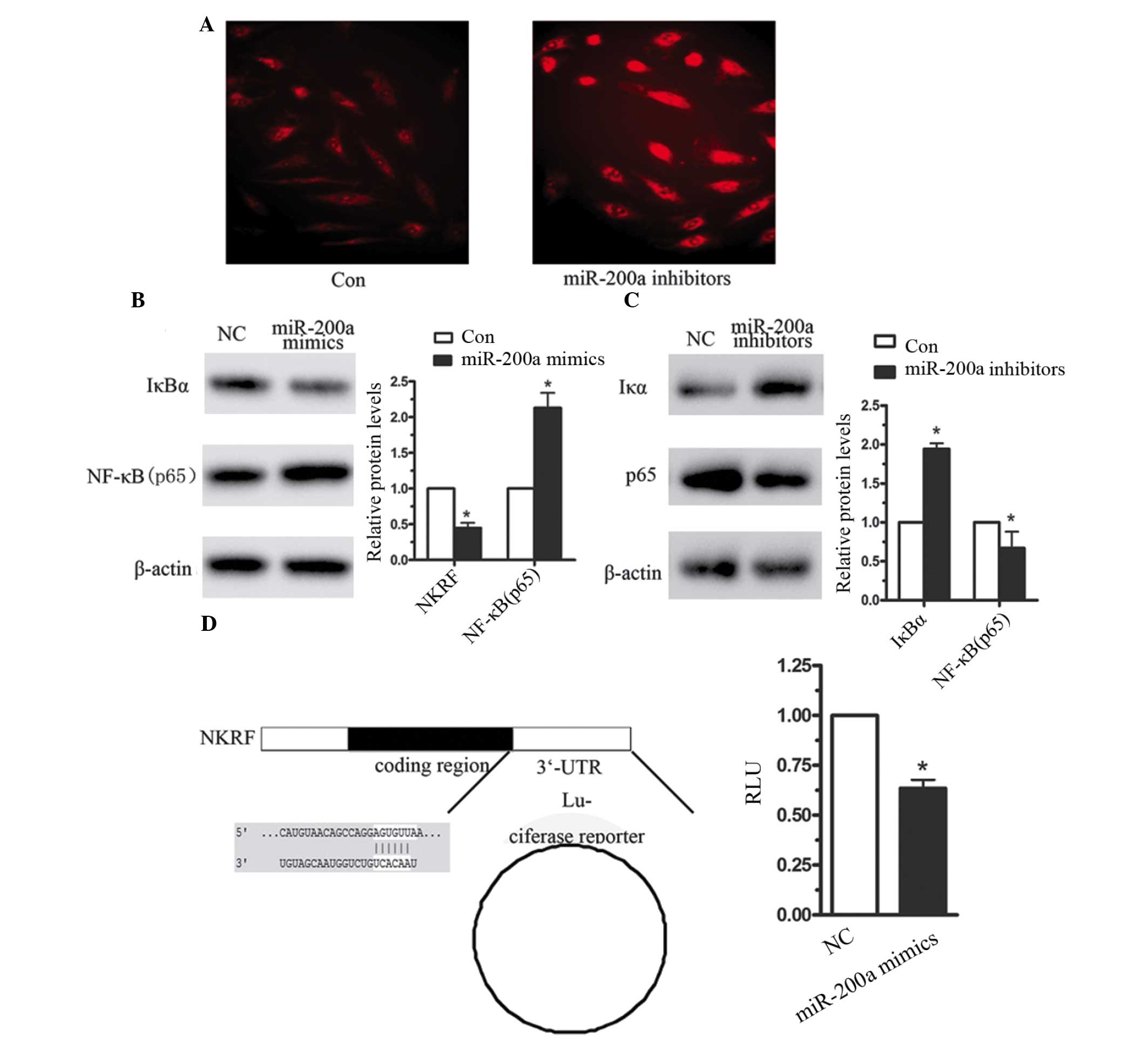 | Figure 4miR-200a targets IκBα in CNE2 cells.
(A) Immunofluorescence analysis of NF-κB expression in CNE2 cells
following transfection with miR-200a inhibitors or the negative
control. (B) Western blot analysis of IκBα, following transfection
of CNE2 cells with (B) miR-200a mimics, (C) miR-200a inhibitors and
the negative control. (D) A luciferase reporter assay was used to
investigate the effect of miR-200a on IκBα-3′-UTR. Data are
presented as the mean ± standard error, n=3 independent
experiments. *P<0.05 vs. control. miR-200a,
microRNA-200a; IκB, inhibitory κB; NF-κB, nuclear factor-κB; UTR,
untranslated region; NC, negative control, NKRF, NF-κB repressing
factor. |
Discussion
MicroRNAs have been widely demonstrated to regulate
various cellular processes, in particular cancer development and
progression (21). NPC is common
in men and women and several studies have indicated abnormal miRNA
levels in NPC (22,23). The current study reported that
miR-200a was upregulated in CNE2 human NPC cells and suggests an
oncogenic role for miR-200a in NPC progression.
In order to investigate the association between
miR-200a and human NPC, the relative levels of miR-200a were
measured in human NPC cells and tissue samples. This indicated that
miR-200a was upregulated in human NPC. Furthermore, the MTT assay
demonstrated that miR-200a is able to activate CNE2 cell
proliferation. TNF-α treatment was observed to induce increased
expression of miR-200a, with a 4-fold increase in CNE2 cells. In
addition, NF-κB was activated when CNE2 cells were transfected with
miR-200a mimics for 48 h, and upregulation of the downstream
regulators of NF-κB signaling was observed, including that of VCAM,
ICAM and MCP-1. The activation of the NF-κB signaling pathway was
further validated via the investigation of the relative levels of
IκBα using western blotting and a luciferase reporter assay.
Together, these data indicated that miR-200a induced NF-κB
activation through the targeting IκBα.
Dysregulation of the NF-κB signaling pathway is well
characterized in cancer cell proliferation, angiogenesis, migration
and invasion (24,25); NF-κB signaling has been observed to
be significantly activated in glioma, and the NF-κB pathway has
been reported to significantly induce cancer cell proliferation and
invasion in thyroid cancer. The current study provided further
evidence of NF-κB activation in human NPC and investigated the
association with miR-200a. These data indicated that NF-κB and its
downstream regulator proteins were positively regulated by
miR-200a. In addition, the current study demonstrated that IκBα is
a target gene for miR-200a. IκBα has been demonstrated to repress
NF-κB translation through binding with specific negative regulatory
elements (26).
In conclusion, increased levels of miR-200a were
observed in the current study in human NPC tissue samples and cell
lines. In addition, miR-200a was demonstrated to enhance the
proliferation of CNE2 cells. Furthermore, miR-200a activated the
NF-κB signaling pathway through targeting IκBα, a negative
regulator of NF-κB.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of Ningbo (grant nos. 2013A610215 and
2014A610230).
References
|
1
|
Xu ZJ, Zheng RS, Zhang SW, Zou XN and Chen
WQ: Nasopharyngeal carcinoma incidence and mortality in China in
2009. Chin J Cancer. 32:453–460. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou W, Feng X, Ren C, Jiang X, Liu W,
Huang W, Liu Z, Li Z, Zeng L, Wang L, et al: Overexpression of
BCAT1, a c-Myc target gene, induces cell proliferation, migration
and invasion in nasopharyngeal carcinoma. Mol Cancer. 12:532013.
View Article : Google Scholar
|
|
3
|
Burkitt MD, Williams JM, Duckworth CA,
O'Hara A, Hanedi A, Varro A, Caamaño JH and Pritchard DM: Signaling
mediated by the NF-κB sub-units NF-κB1, NF-κB2 and c-Rel
differentially regulate Helicobacter felis-induced gastric
carcinogenesis in C57BL/6 mice. Oncogene. 32:5563–5573. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chung GT, Lou WP, Chow C, To KF, Choy KW,
Leung AW, Tong CY, Yuen JW, Ko CW, Yip TT, et al: Constitutive
activation of distinct NF-κB signals in EBV-associated
nasopharyngeal carcinoma. J Pathol. 231:311–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hamoudi RA, Appert A, Ye H,
Ruskone-Fourmestraux A, Streubel B, Chott A, Raderer M, Gong L,
Wlodarska I, DeWolf-Peeters C, et al: Differential expression of
NF-kappaB target genes in MALT lymphoma with and without chromosome
translocation: Insights into molecular mechanism. Leukemia.
24:1487–1497. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, Forte N, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1086–1099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fitzgerald TL, Lertpiriyapong K, Cocco L,
Martelli AM, Libra M, Candido S, Montalto G, Cervello M, Steelman
L, Abrams SL and McCubrey JA: Roles of EGFR and KRAS and their
downstream signaling pathways in pancreatic cancer and pancreatic
cancer stem cells. Adv Biol Regul. 59:65–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dang K and Myers KA: The role of
hypoxia-induced miR-210 in cancer progression. Int J Mol Sci.
16:6353–6372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai L, Gu L and Di W: MiR-199a attenuates
endometrial stromal cell invasiveness through suppression of the
IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol Hum
Reprod. 18:136–145. 2012. View Article : Google Scholar :
|
|
11
|
Zhen Q, Liu J, Gao L, Liu J, Wang R, Chu
W, Zhang Y, Tan G, Zhao X and Lv B: MicroRNA-200a Targets EGFR and
c-Met to Inhibit Migration, Invasion, and Gefitinib Resistance in
Non-Small Cell Lung Cancer. Cytogenet Genome Res. 146:1–8. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsouko E, Wang J, Frigo DE, Aydodu E and
Williams C: miR-200a inhibits migration of triple-negative breast
cancer cells through direct repression of the EPHA2 oncogene.
Carcinogenesis. 36:1051–1060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hong MH, Mai HQ, Min HQ, Ma J, Zhang EP
and Cui NJ: A comparison of the Chinese 1992 and fifth-edition
International Union Against Cancer staging systems for staging
nasopharyngeal carcinoma. Cancer. 89:242–247. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo J, Li M, Meng X, Sui J, Dou L, Tang W,
Huang X, Man Y, Wang S and Li J: MiR-291b-3p induces apoptosis in
liver cell line NCTC1469 by reducing the level of RNA-binding
protein HuR. Cell Physiol Biochem. 33:810–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kan R, Shuen WH, Lung HL, Cheung AK, Dai
W, Kwong DL, Ng WT, Lee AW, Yau CC, Ngan RK, Tung SY and Lung ML:
NF-κB p65 Subunit Is Modulated by Latent Transforming Growth
Factor-β Binding Protein 2 (LTBP2) in Nasopharyngeal Carcinoma
HONE1 and HK1 Cells. PLoS One. 10:e01272392015. View Article : Google Scholar
|
|
16
|
Astarci E, Sade A, Cimen I, Savaş B and
Banerjee S: The NF-κB target genes ICAM-1 and VCAM-1 are
differentially regulated during spontaneous differentiation of
Caco-2 cells. FEBS J. 279:2966–2986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu Z and Li Y, Takwi A, Li B, Zhang J,
Conklin DJ, Young KH, Martin R and Li Y: miR-301a as an NF-κB
activator in pancreatic cancer cells. EMBO J. 30:57–67. 2011.
View Article : Google Scholar :
|
|
18
|
Bao C, Li Y, Huan L, Zhang Y, Zhao F, Wang
Q, Liang L, Ding J, Liu L, Chen T, Li J, Yao M, Huang S and He X:
NF-κB signaling relieves negative regulation by miR-194 in
hepatocellular carcinoma by suppressing the transcription factor
HNF–1α. Sci Signal. 8:ra752014. View Article : Google Scholar
|
|
19
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, Liang L, Li J and He XP:
Genome-wide screening reveals that miR-195 targets the TNF-α/NF-κB
pathway by down-regulating IκB kinase alpha and TAB3 in
hepatocellular carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Olarerin-George AO, Anton L, Hwang YC,
Elovitz MA and Hogenesch JB: A functional genomics screen for
microRNA regulators of NF-kappaB signaling. BMC Biol. 11:192013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu N, Cui RX, Sun Y, Guo R, Mao YP, Tang
LL, Jiang W, Liu X, Cheng YK, He QM, et al: A four-miRNA signature
identified from genome-wide serum miRNA profiling predicts survival
in patients with nasopharyngeal carcinoma. Int J Cancer.
134:1359–1368. 2014. View Article : Google Scholar
|
|
23
|
Zhao Y, Chen X, Jing M, Du H and Zeng Y:
Expression of miRNA-146a in nasopharyngeal carcinoma is upregulated
by Epstein-Barr virus latent membrane protein 1. Oncol Rep.
28:1237–1242. 2012.PubMed/NCBI
|
|
24
|
Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C,
Wu J, Hu B, Cheng SY, Li M and Li J: TGF-β induces miR-182 to
sustain NF-κB activation in glioma subsets. J Clin Invest.
122:3563–3578. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pozdeyev N, Berlinberg A, Zhou Q, Wuensch
K, Shibata H, Wood WM and Haugen BR: Targeting the NF-κB pathway as
a combination therapy for advanced thyroid cancer. PLoS One.
10:e01349012015. View Article : Google Scholar
|
|
26
|
Nourbakhsh M, Oumard A, Schwarzer M and
Hauser H: NRF, a nuclear inhibitor of NF-kappaB proteins silencing
interferon-beta promoter. Eur Cytokine Netw. 11:500–501. 2000.
|