Introduction
As the most common type of primary malignant bone
tumor, osteosarcoma is typified by the formation of immature bone
or bone-like tissues from proliferating tumor cells. Approximately
80% of osteosarcoma cases are observed in the long bones of limbs,
with the majority at the metaphysis of knee joints, and the further
20% involving the axial skeleton and pelvis (1). The onset, progression, metastasis and
prognosis of malignant tumors are closely associated with the
expression of anti-apoptotic genes in tumor tissues (2). Under the protection of these genes,
tumor cells survive, continuously proliferate and metastasize,
leading to a poor prognosis. For example, the anti-apoptotic genes
livin and survivin are highly expressed in human osteosarcoma
tissues and serve important roles in the onset and progression of
disease (3,4).
RNA interference is a valuable tool in the study of
gene function, due to its high specificity and efficiency (5). Additionally, nanoparticles (NPs) that
carry genes are able to effectively deliver exogenous DNA into
cells and maintain high-level expression, thereby facilitating
research into gene function and gene therapy. Thus, attention has
been focused on developing gene-carrying NPs for cancer
therapy.
Therefore, in the current study MG-63 osteosarcoma
cells were transfected with monomethoxypolyethylene glycol-chitosan
(mPEG-CS) NPs carrying livin and survivin short hairpin RNAs
(shRNAs). The aim of the current study was to observe the effect of
knocking down livin and survivin on apoptosis, and to investigate
the association between the two genes and apoptosis.
Materials and methods
Reagents
The reagents used were as follows: Sodium
tripolyphosphate (TPP), Chengdu Kelong Chemical Co., Ltd. (Chengdu,
China); CS (degree of deacetylation, >90.0%), Shanghai Bo'ao
Biological Technology Co., Ltd. (Shanghai, China); mPEG (5000),
Sigma-Aldrich (St. Louis, MO, USA); Dulbecco's modified Eagle's
medium (DMEM) culture medium, Nanjing KeyGen Biotech Co., Ltd.
(Nanjing, China); fetal bovine serum (FBS), Hangzhou Sijiqing
Biological Engineering Materials Co., Ltd. (Hangzhou, China);
TRIzol reagent, Thermo Fisher Scientific, Inc. (Waltham, MA, USA);
reverse transcription-polymerase chain reaction kit, Takara Bio,
Inc. (Otsu, Japan); trypsin and
3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT), Roche Diagnostics (Basel, Switzerland); cell lysis buffer
and Hoechst staining kit, Beyotime Institute of Biotechnology
(Shanghai, China); Diaminobenzidine (DAB) Histochemistry kit
(Thermo Fisher Scientific, Inc.); rabbit anti-human livin (cat. no.
Ab5393) and survivin antibodies (cat. no. Ab76424) (1:1,000
dilution; Abcam, Cambridge, MA, USA), rabbit anti-human GAPDH
polyclonal antibody (1:900 dilution; cat. no. Ab9485; Abcam) and
horseradish peroxidase (HRP)-labeled goat anti-rabbit
immunoglobulin G antibody (1:900 dilution; cat. no. Ab97051;
Abcam).
Cell line
The MG-63 human osteosarcoma cell line was purchased
from the College of Life Sciences, Wuhan University (Wuhan,
China).
Synthesis of livin and survivin shRNA
plasmids
According to the gene sequences in GenBank
(http://www.ncbi.nlm.nih.gov/genbank/), the IDs for
livin and survivin were NM-022161 and NM-001168, respectively.
Livin and survivin shRNA plasmids were designed, synthesized and
sequenced by Obio Technology (Shanghai) Co., Ltd. (Shanghai,
China).
Preparation of mPEG-CS NPs
mPEG-CS NPs were prepared by ionic crosslinking as
described previously (6). In
brief, 50 mg CS was dissolved in 100 ml of 1% (V/V) acetic acid and
stirred for 30 min until completely dissolved, following which 1 ml
of 1% (V/V) mPEG solution was added and stirred for a further 2 h
to form the mPEG-CS solution. Subsequently, 1 mg/ml TPP solution
was slowly added whilst the solution was stirred until a visible
opalescence was observed, following which the solution was stirred
overnight at room temperature to produce an NP suspension. The
suspension was centrifuged at 17,860 × g for 10 min, washed twice
with sterile double-distilled water (ddH2O), dispersed
ultrasonically in fresh ddH2O with the same volume, and
stored at 4°C prior to use.
Preparation of gene-carrying NPs
Gene-carrying NPs were prepared by electrostatic
adsorption as described previously (7). Solutions (0.35 mg/ml, 2.5 µl
each) of recombinant livin and survivin shRNA plasmids were mixed
with 15 µl mPEG-CS NP solution, incubated in a 5°C water
bath for 20 min and vortexed for 30 min, forming an mPEG-CS-(livin
shRNA + survivin shRNA) NP suspension. Subsequently, mPEG-CS-livin
shRNA and mPEG-CS-survivin shRNA alone NP suspensions were obtained
via the same method.
Cell culture
MG-63 cells were cultured in DMEM containing 10%
FBS, into which streptomycin-penicillin double antibiotic solutions
(Sigma-Aldrich) were added to a final concentration of 100 U/ml.
The cells were then incubated in a 5% CO2 atmosphere at
37°C, digested with 0.25% trypsin and passaged, and cells in the
logarithmic growth phase were selected for investigation.
Grouping and cell transfection
One day prior to transfection, MG-63 cells in the
logarithmic growth phase were seeded into 6-well plates at a
density of 2×105 cells/well, and cultured in DMEM
without double antibiotic solution or serum in a 5% CO2
atmosphere at 37°C. When 80% confluent, the cells were divided into
four groups and transfected as follows: Livin + survivin
interference group, transfected with mPEG-CS-(livin shRNA +
survivin shRNA) NPs; livin interference group, transfected with
mPEG-CS-livin shRNA NPs; survivin interference group, transfected
with mPEG-CS-survivin shRNA NPs; and negative control group,
transfected with mPEG-CS-blank plasmid NPs. Cells were transfected
according to the instructions of the Lipofectamine 2000 kit (Thermo
Fisher Scientific, Inc.) at 40–60% confluence. Following 4–6 h of
transfection, the cells were cultured in complete DMEM.
Measurement of livin and survivin mRNA
expression in MG-63 cells by reverse transcription-polymerase chain
reaction (RT-PCR)
Following 48 h of transfection, the culture medium
was discarded, and 1 ml TRIzol reagent was added to extract total
DNA according to the manufacturer's instructions. cDNA synthesis
from total RNA (1 µg) was performed in a reaction volume of
20 µl containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM
MgCl2, 5 mM dithiothreitol, 0.5 mM deoxynucleoside
triphosphate mix, 200 units SuperScript™III reverse transcriptase
(all from Thermo Fisher Scientific) and 1 µl random primers
(hexanucleotide mix, 10X; Roche Applied Science, Mannheim,
Germany). Initially, RNA was denatured at 65°C for 5 min, followed
by addition of the reaction mixture and reverse transcription at
50°C for 50 min. The reaction was stopped by denaturing the enzyme
at 70°C for 15 min. The primers used for PCR amplification and the
length of the amplified fragments were as follows: Livin forward,
5′-TAA AGA CAG TCC AAG TGC CT-3′ and reverse, 5′-TGA TGG CCT GTG
TGG AAG AA-3′, length of amplified fragment was 348 base pairs
(bp); survivin forward, 5′-ACC ACA GTC CAT GCC ATC AC-3′ and
reverse, 5′-GTT CTT GGC TTT CTC TGT CC-3′, length of amplified
fragment was 300 bp; and GAPDH forward, 5′-ACC ACA GTC CAT GCC ATC
AC-3′ and reverse, 5′-TGC TGT AGC CAA ATT CGT TG-3′, length of
amplified fragment was 450 bp. The primers were supplied by Thermo
Fisher Scientific. RT-PCR was conducted as follows: Reverse
transcription at 50°C for 30 min, cycling at 95°C for 15 min,
denaturation at 94°C for 30 sec, annealing at 57°C for 30 sec,
extension at 72°C for 30 sec (30 cycles in total), and extension at
72°C for 10 min. PCR products were subjected to agarose gel
electrophoresis.
Measurement of livin and survivin protein
expression in MG-63 cells by western blotting
Following 48 h of transfection, the culture medium
was discarded, and cells were washed twice with 4°C
phosphate-buffered saline (PBS), 150 µl pre-cooled cell
lysis buffer was added and incubated on ice for 30 min, prior to
centrifugation at 4°C and 13,980 × g for 15 min. Subsequently, the
supernatant was collected as the protein sample and quantified
using a BCA kit (Sigma-Aldrich). Proteins were denatured, subjected
to sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(Thermo Fisher Scientific), electronically transferred onto a
nitrocellulose membrane and then blocked for 3 h at 37°C using
blocking buffer (Sigma-Aldrich). Membranes were incubated with
primary antibodies overnight at 4°C, washed three times with PBS
and then incubated with secondary antibodies at room temperature
for 2 h. After several washes with PBS, the membranes were
incubated with Luminata Forte Western HRP Substrate (EMD-Millipore,
Billerica, MA, USA) for 3 min or Western Bright (Advansta, Inc.,
Menlo Park, CA, USA) diluted 1:1 with water for 30 sec. Images were
captured under red safe light on X-ray Film (Fujifilm, Tokyo,
Japan) using an X-ray developing unit (Agfa, Köln, Germany) for 10
min. The DAB kit was used for color development. Relative protein
expression levels were calculated from the ratios of livin/β-actin
and survivin/β-actin.
Measurement of the effect of RNA
interference on the inhibition of MG-63 cell proliferation using an
MTT assay
Following transfection, the cells were cultured for
24, 48 and 72 h. MTT solution (10 µl, 5 mg/ml) was added to
each well and cells were incubated at 37°C for 4 h. Subsequently,
the supernatant was removed, 10 µl dimethyl sulfoxide was
added to each well and the cells were agitated for 10 min at 37°C.
Absorbance (A) of each well was measured with a Standard
Filter-Microplate Photometer (wavelength at 490 nm; Applied
Biosciences, Thermo Fisher Scientific). The inhibitory rate was
calculated as follows: (IR%) = (1 − Aexperiment
group/Ablank control group) × 100. The values of
each group are presented as the mean ± standard deviation (SD).
Effect of RNA interference on MG-63 cell
apoptosis using Hoechst staining
Following 48 h of transfection, the cells were fixed
according to the instructions of the Hoechst staining kit.
Following the removal of the fixing solution, cells were stained
whilst agitated at room temperature for 5 min. Subsequently, the
staining solution was removed and the microscope slide was covered
with a coverslip and sealed with antifade solution. Cell morphology
was observed using an inverted fluorescence microscope (FV1200;
Olympus, Tokyo, Japan) and images were captured. Five visual fields
were randomly selected in each well and the cells were counted. The
rate of apoptosis was calculated as follows: Apoptotic index (%) =
Number of apoptotic cells/(number of normal cells + number of
apoptotic cells) × 100. The data were presented as the mean
±SD.
Statistical analysis
All data were analyzed using SPSS software, version
17.0 (SPSS, Inc., Chicago, IL, USA). Inter-group differences in
livin and survivin mRNA expression levels were compared by
univariate analysis of variance, and pairwise comparison was
performed with Fisher's least significant difference test.
Inter-group differences in livin and survivin protein expression
levels and the effect of RNA interference on cell survival and
apoptosis were compared using the Kruskal-Wallis test, and pairwise
comparison was conducted with the Bonferroni correction. P<0.05
was considered to indicate a statistically significant
difference.
Results
Morphologies of mPEG-CS, mPEG-CS-livin
shRNA and mPEG-CS-survivin shRNA NPs
Transmission electron micro-graphs of mPEG-CS-livin
shRNA and mPEG-CS-survivin shRNA NPs exhibit uniform approximate
spheres sized 100–200 nm that are distributed evenly, without
obvious aggregation (Fig. 1).
Inhibition of mRNA expression by RNA
interference measured using RT-PCR
RT-PCR products were resolved by 2% agarose gel
electrophoresis to analyze the effects of the different
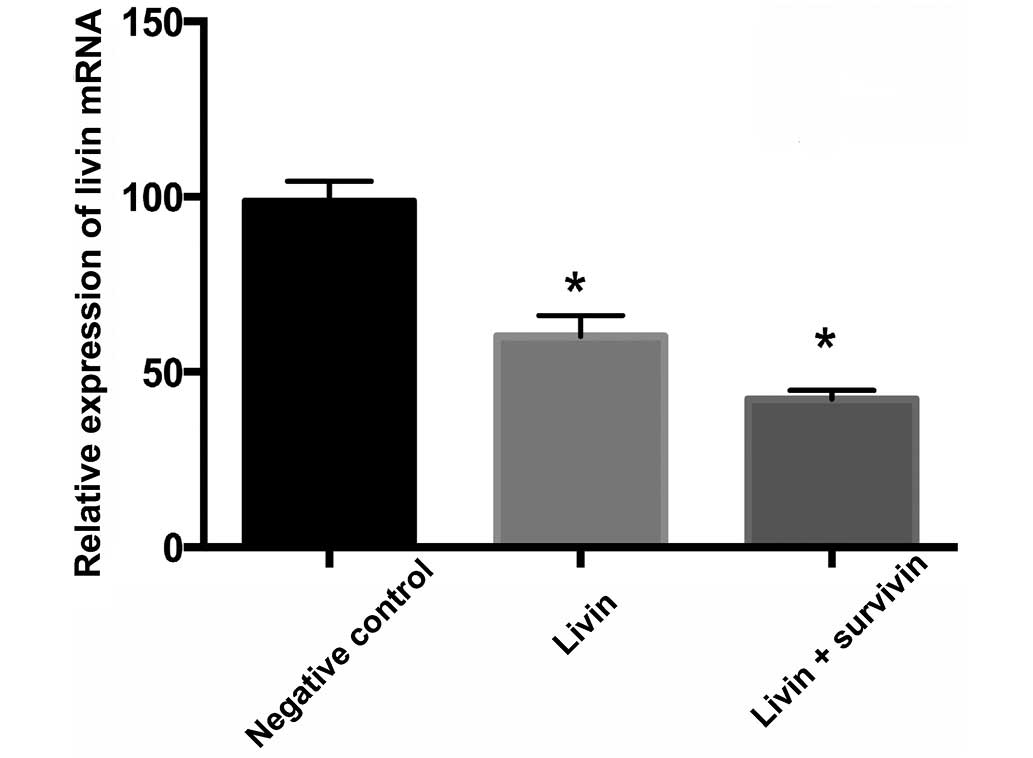
interference groups. Fig. 2
indicates that compared with the negative control group, the livin
and livin + survivin interference groups significantly reduced the
expression levels of livin mRNA (P<0.05). Fig. 3 indicates that compared with the
negative control group, the survivin and livin + survivin
interference groups significantly reduced the expression levels of
survivin mRNA (P<0.05).
Inhibition of protein expression by RNA
interference measured using western blotting
The effect of the different interference groups on
livin and survivin protein expression levels were measured by
western blotting. β-actin was used as the internal reference, and
the relative expression levels of livin protein in the negative
control, livin interference and livin + survivin interference
groups were 93.31±2.47, 50.22±3.42 and 6.43±2.16%, respectively
(Fig. 4). The expression levels of
livin in the livin and livin + survivin interference groups were
significantly downregulated compared with the negative control
group (P<0.05).
The relative expression levels of survivin protein
in the negative control, survivin interference and livin + survivin
interference groups were 86.29±4.24, 57.52±4.21 and 35.73±3.14%,
respectively (Fig. 5). Compared
with the negative control group, survivin protein expression levels
in the survivin and livin + survivin interference groups were
significantly downregulated (P<0.05).
Inhibition of MG-63 cell proliferation by
RNA interference measured using an MTT assay
The inhibitory effects of RNA interference on MG-63
cell proliferation were assessed using an MTT assay. The inhibitory
effects of livin, survivin and livin + survivin RNA interference
compared with the proliferation in the negative control group, were
significant (P<0.05) and, in the livin and livin + survivin
groups, increased in a time-dependent manner. In addition, livin +
survivin interference had a significantly greater effect compared
with livin or survivin interference alone (P<0.05; Table I).
 | Table IRate of proliferation inhibition (%)
in the livin and survivin RNA interference groups. |
Table I
Rate of proliferation inhibition (%)
in the livin and survivin RNA interference groups.
| Group | Rate of proliferation
inhibition over time (%)
|
|---|
| 24 h | 48 h | 72 h |
|---|
| Livin
interference | 31.12±1.02a | 38.72±0.68a | 41.29±1.51a |
| Survivin
interference | 34.23±0.97a | 39.92±1.67a | 44.71±0.96a |
| Livin + survivin
interference | 50.27±1.34a,b | 54.74±0.87a,b | 59.22±2.12a,b |
| Negative control | 2.36±1.54 | 3.05±1.14 | 5.54±1.44 |
Morphological analysis of apoptosis
Morphological alterations associated with apoptosis
were observed in MG-63 cells using a fluorescence microscope
following Hoechst staining. The nuclei of the normal cells were
blue, while those of apoptotic cells were fragmented, densely
stained and white. The apoptotic indices of the negative control,
livin, survivin and livin + survivin interference groups were
6.88±0.63, 31.76±2.45, 29.92±3.23 and 51.28±1.14%, respectively.
Compared with the negative control group, the apoptotic indices of
the three interference groups were significantly greater
(P<0.05). In addition, livin + survivin interference induced
significantly greater levels of apoptosis compared with the livin
and survivin interference groups alone (P<0.05; Fig. 6).
Discussion
Originating from bone mesenchymal cells,
osteosarcoma is a primary malignant bone cancer characterized by
the transformation of proliferating tumor cells into immature bone
or bone-like tissues. Upon diagnosis, 80% of patients with
osteosarcoma have suffered from micrometastases, with on average 8
months from surgical treatment to pulmonary metastasis (8). In addition, the 5-year survival rate
is low, with a high rate of cancer-associated mortality within one
year (9). In recent years,
amputation has been replaced with limb-salvage therapy due to the
development of chemotherapy, improvements in surgical techniques,
bone reconstruction, genotherapy, immunotherapy and targeted
molecular therapy (8).
The pathogenesis of osteosarcoma has been attributed
to a complicated process involving the activation of numerous
anti-apoptotic genes and the deactivation of pro-apoptotic genes.
Alterations in apoptosis serve an essential role in tumor onset,
with the inhibition of the expression of pro-apoptotic genes and
excess upregulation of anti-apoptotic genes contributing to tumor
onset, progression and metastasis (10).
Livin and survivin, as important members of the
inhibitor of apoptosis family (11), are overexpressed in numerous types
of malignant tumor tissues (12,13)
and are novel targets for genotherapy, due to their effect on
invasion and metastasis in addition to prognosis. Through various
pathways, livin is able to suppress apoptosis by blocking apoptotic
receptors and by inhibiting the mitochondria-based intrinsic
apoptotic pathways (11).
Additionally, downregulated livin expression is able to promote
apoptosis and inhibit tumor growth (14). Livin expression is upregulated in
human osteosarcoma tissue, with a rate of positivity for livin
expression of 58.7%, which is significantly correlated with
microvessel density (15).
Survivin has a similar molecular structure to that of livin.
Besides suppressing apoptosis, survivin also facilitates cell
transformation by participating in angiogenesis of tumor tissues
and generation of drug resistance (16). Survivin is expressed in malignant
tumors, however, not in normal tissue (17); thus, it is possible that tumor
cells may be killed selectively by targeted survivin immunotherapy
and genotherapy. Therefore, survivin is a tumor antigen that is
widely applicable to the clinical genotherapy of osteosarcoma.
Compared with traditional carriers such as plasmids,
NPs are superior due to their large surface area, high adsorption
capacity, facile surface modification and passive targeting
(18). In the current study CS, a
marine organism-derived polysaccharide, was selected due to its
biocompatibility, safety, non-toxicity and degradability (18). Carrying a positive charge, CS is
able to react with negatively charged DNA to form a polyelectrolyte
complex that effectively protects DNA from degradation by DNA
enzymes (19). However, ordinary
CS is insoluble in water or common organic solvents, so it alone is
not applicable in the biomedical field (20). Therefore, the current study altered
the characteristics of CS through chemical modification. mPEG-CS
NPs had low cytotoxicity, a suitable size for gene delivery,
protective effects on genes and long circulating ability, in
addition to high drug loading and encapsulation efficiency.
Together, this indicates that this derivative is promising in gene
transfection and genotherapy.
To verify the biological effects of livin and
survivin on osteosarcoma cells and the applicability of CS NPs in
tumor treatment, the current study generated mPEG-CS, mPEG-CS-livin
shRNA, mPEG-CS-survivin shRNA and mPEG-CS-(livin shRNA + survivin
shRNA) NPs to transfect MG-63 cells, and to reduce livin and
survivin mRNA and protein expression levels. The present study
indicates that mPEG-CS NPs are suitable carrier surrogates for
traditional plasmids to mediate transfection.
Furthermore, the current study demonstrated that
downregulated expression of livin and survivin in MG-63 cells
significantly suppressed proliferation and enhanced apoptosis in
MG-63 cells. Notably, simultaneous livin and survivin RNA
interference exerted greater inhibitory effects on the cells
compared with livin or survivin interference alone. In conclusion,
the anti-apoptotic genes livin and survivin participate in the
onset and progression of osteosarcoma, and simultaneous inhibition
is potentially eligible for future genotherapy.
References
|
1
|
Salah S, Ahmad R, Sultan I, Yaser S and
Shehadeh A: Osteosarcoma with metastasis at initial diagnosis:
Current outcomes and prognostic factors in the context of a
comprehensive cancer center. Mol Clin Oncol. 2:811–816.
2014.PubMed/NCBI
|
|
2
|
Sarela AI, Macadam RC, Farmery SM, Markham
AF and Guillou PJ: Expression of the antiapoptosis gene, survivin,
predicts death from recurrent colorectal carcinoma. Gut.
46:645–650. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang L, Zhang Q, Liu B, Han M and Shan B:
Challenge and promise: Roles for Livin in progression and therapy
of cancer. Mol Cancer Ther. 7:3661–3669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zaffaroni N and Daidone MG: Survivin
expression and resistance to anticancer treatments: Perspectives
for new therapeutic interventions. Drug Resist Updat. 5:65–72.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kulkarni AR, Lin YH, Liang HF, Chang WC,
Hsiao WW and Sung HW: A novel method for the preparation of
nanoaggregates of methoxy polyethyleneglycol linked chitosan. J
Nanosci Nanotechnol. 6:2867–2873. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Tian B, Yin X, Zhang Y, Hu D, Hu
Z, Liu M, Pan Y, Zhao J, Li H, et al: Preparation, characterization
and transfection efficiency of cationic PEGylated PLA nanoparticles
as gene delivery systems. J Biotechnol. 130:107–113. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosen G, Tan C, Sanmaneechai A, Beattie EJ
Jr, Marcove R and Murphy ML: The rationale for multiple drug
chemotherapy in the treatment of osteogenic sarcoma. Cancer.
35(suppl): 936–945. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu X, Ding J, Zhang Z, Li Q, Zhuang X and
Chen X: Polymeric nanocarriers for drug delivery in osteosarcoma
treatment. Curr Pharm Des. Sep 22–2015.Epub ahead of print.
View Article : Google Scholar
|
|
10
|
Fang J, Ding M, Yang L, Liu LZ and Jiang
BH: PI3K/PTEN/AKT signaling regulates prostate tumor angiogenesis.
Cell Signal. 19:2487–2497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salvesen GS and Duckett CS: IAP proteins:
Blocking the road to death's door. Nat Rev Mol Cell Biol.
3:401–410. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lopes RB, Gangeswaran R, McNeish IA, Wang
Y and Lemoine NR: Expression of the IAP protein family is
dysregulated in pancreatic cancer cells and is important for
resistance to chemotherapy. Int J Cancer. 120:2344–2352. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tanabe H, Yagihashi A, Tsuji N, Shijubo Y,
Abe S and Watanabe N: Expression of survivin mRNA and livin mRNA in
non-small-cell lung cancer. Lung Cancer. 46:299–304. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu B, Han M, Wen JK and Wang L:
Livin/ML-IAP as a new target for cancer treatment. Cancer Lett.
250:168–176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie CH, Wu BY, Zhao Y and Hu HL:
Association between Livin expression and microvascular density in
osteosarcoma. J Pract Med. 27:1529–1532. 2011.In Chinese.
|
|
16
|
Church DN and Talbot DC: Survivin in solid
tumors: Rationale for development of inhibitors. Curr Oncol Rep.
14:120–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dallaglio K, Marconi A and Pincelli C:
Survivin: A dual player in healthy and diseased skin. J Invest
Dermatol. 132:18–27. 2012. View Article : Google Scholar
|
|
18
|
Erbacher P, Zou S, Bettinger T, Steffan AM
and Remy JS: Chitosan-based vector/DNA complexes for gene delivery:
Biophysical characteristics and transfection ability. Pharm Res.
15:1332–1339. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Richardson SC, Kolbe HV and Duncan R:
Potential of low molecular mass chitosan as a DNA delivery system:
Biocompatibility, body distribution and ability to complex and
protect DNA. Int J Pharm. 178:231–243. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kievit FM, Veiseh O, Bhattarai N, Fang C,
Gunn JW, Lee D, Ellenbogen RG, Olson JM and Zhang M:
PEI-PEG-chitosan copolymer coated iron oxide nanoparticles for safe
gene delivery: Synthesis, complexation, and transfection. Adv Funct
Mater. 19:2244–2251. 2009. View Article : Google Scholar :
|