Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney cancer in adults, accounting for ~90% of all renal tumors
and 3.9% of all cancers (1,2). In
the United States, renal cancer is the 6th and 8th leading
malignancy among men and women, respectively. In 2014, 63,920 novel
cases and 13,860-related fatalities from renal cancer are estimated
to have occurred (3). In China,
kidney cancer incidence has increased with an average annual growth
rate of 6.5% over the past 20 years, and kidney cancer-related
mortality has surpassed bladder cancer as the most common cancer of
the urinary system (4,5). RCC is characterized by a lack of
early-warning signs, protean clinical manifestations, and
resistance to radiotherapy and chemotherapy (6). As a result, 25% of patients present
with advanced disease when initially diagnosed with RCC, and 33.3%
of the patients who undergo resection of localized disease have a
recurrence (7). Survival of
patients with localized tumors who undergo radical nephrectomy is
significantly longer than patients with regional and distant
metastasis, thus underscoring the importance of early detection
(8). Therefore, it is critical to
identify novel molecular biomarkers for early detection, diagnosis
and targeted therapy.
MicroRNAs (miRNAs) are 21–25 nucleotide endogenously
produced non-coding RNAs, which regulate the expression of numerous
genes, and are significant role in a wide range of biological
processes, including animal and plant development, cell
proliferation, cell differentiation, apoptosis and metabolism.
Generally, miRNAs, as negative regulators, bind to a partially
complementary sequence usually located in the 3′-untranslated
region (3′-UTR) of their target mRNA and inhibit its translation
(9). Due to partial
complementation, a specific miRNA can regulate multiple genes and a
single gene can be modulated by hundreds of miRNAs at the
post-transcriptional level (10).
Recently, miR-20b-5p has been reported to be
dysregulated in malignancies of the breast (11–13),
stomach (14), cervix (15), blood (16), oropharynx (17) and colorectum (18). A recent miRNA microarray chip
analysis showed that miR-20b-5p was downregulated in ccRCC
(19). Thus, the expression and
function of miR-20b-5p in renal cancer requires further
investigation. The aim of the present study was to identify
miR-20b-5p as a tumor suppressor in RCC. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed to determine the downregulation of miR-20b-5p in RCC
tissues and cell lines compared with paired normal tissues and cell
lines and the effects of miR-20b-5p on cell migration,
proliferation and apoptosis were analyzed in RCC cell lines.
Materials and methods
Specimens
A total of 48 paired fresh RCC samples and adjacent
normal tissue samples (located 2.0 cm outside of visible RCC
lesions) were obtained from the Peking University Shenzhen Hospital
(Shenzhen, China). Written informed consent was obtained from all
patients. The study was approved by the ethics committee of the
Peking University Shenzhen Hospital (Shenzhen, China). The clinical
specimens were collected between September 2012 and November 2014.
All fresh tissue samples were immediately immersed in RNAlater® RNA
Stabilization agent (Qiagen, Inc., Hilden, Germany) following
surgical resection, snap-frozen in liquid nitrogen and stored in a
cryo freezer at −80°C for further use. The clinicopathological
information of the patients is shown in Table I. Stage classification was
performed according to the 2010 American Joint Committee on Cancer
staging system (20).
 | Table IClinicopathological features in RCC
patients. |
Table I
Clinicopathological features in RCC
patients.
| Characteristic | Number of
cases |
|---|
| Mean age range
(years) | 52 (27–72) |
| Gender | |
| Male/female | 30/18 |
| Histological
type | |
| Clear
cell/papillary | 39/9 |
| pT-stage | |
| T1/T2/T3+T4 | 27/19/2 |
| Fuhrman grade | |
| I/II/III/IV | 15/22/8/3 |
| AJCC clinical
stages | |
| I/II/III+IV | 27/18/3 |
Cell culture and transfection
786-O and ACHN human RCC cell lines and the 293T
normal kidney human embryo kidney cell used in the present study
were obtained from the Guangdong and Shenzhen Key Laboratory of
Male Reproductive Medicine and Genetics (Shenzen, China). The human
RCC cell lines, including 786-O and ACHN, were originally obtained
from the American Type Culture Collection (Manassas, VA, USA). The
human embryo kidney cell line 293T (293T) was purchased from the
Type Culture Collection of the Chinese Academy of Medical Sciences
(Beijing, China). All cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific Inc.,
Waltham, MA, USA), with 10% fetal bovine serum (Gibco), 1%
antibiotics (100 µ/ml penicillin and 100 mg/ml streptomycin
sulfates) and 1% glutamate (Gibco), and then incubated at 37°C in a
humidified chamber containing 5% CO2. For the
upregulation of miR-20b-5p, synthesized miR-20b-5p mimics
(GenePharma, Shanghai, China) and Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific Inc.) were mixed into the Gibco™ Opti-MEM
I Reduced Serum medium (Thermo Fisher Scientific, Inc.) for
transfection, after the cells were seeded and cultured for 24 h.
Then, fluorescence microscopy (using a fluorescence microscope
obtained from Carl Zeiss Pte. Ltd., Oberkochen, Germany) and
RT-qPCR were used to verify transfection efficiency.
Extraction of total RNA and RT-qPCR
Total RNA was extracted from 48 paired RCC samples
and normal tissue using TRIzol reagent (Invitrogen) and were
purified using the RNeasy Maxi kit (Qiagen), according to the
manufacturer's instructions. 786-O, ACHN and 293T cells
(4×105 cells/well) were plated into 6-well plates (BD
Biosciences, USA) with three replicate wells, respectively. The
cells were trypsinized, using Gibco™ trypsin (Thermo Fisher
Scientific Inc.) to extract the total RNA using the miRNeasy Mini
kit (Qiagen, Valencia, CA, USA) 24 h later. The RNA samples with
260/280 ratios of 1.8–2.0 were used for further experiments. Total
RNA was converted into cDNA using the miScript II RT kit (Qiagen,
Valencia, CA, USA).
The expression level of miR-20b-5p was confirmed
with the miScriptSYBR green PCR kit (Qiagen, Valencia, CA, USA)
according to the manufacturer's instructions on the Roche
lightcycler 480 Real-Time PCR system (Roche). U6 was used as the
endogenous control to normalize the data. Their expression levels
were shown as fold differences relative to U6, which was based on
the equation relative quantification (RQ)=2−ΔΔCq
[ΔΔCq=(meanCqtumor-meanCqcontrol)−(meanCqnormal-meanCqcontrol)].
The miR-20b-5p forward primer was 5′-CAAAGTGCTCATAGTGCAGGTAG-3′ and
reverse primer was provided by the miScriptSYBR® green
PCR kit (Qiagen, Valencia, CA, USA). The forward primer of U6 was
5′-CTCGCTTCGGCAGCACA-3′ and reverse primer was
5′-ACGCTTCACGAATTTGCGT-3′. The reaction conditions for PCR were as
follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 sec,
55°C for 30 sec and 72°C for 30 sec.
Cell scratch assay
The migratory ability of 786-O and ACHN cells was
assessed by a wound scratch assay in vitro. Approximately
3×105 cells were seeded per 12-well dish and transfected
after 24 h with 100 pmol of either the miR-20b-5p mimics or a
negative control, using Lipofectamine 2000. After 6 h of
transfection, a sterile 200 µl pipette tip was used to
scrape a clear line through the cell layer. The cells were then
rinsed with phosphate-buffered saline (PBS) and cultured in
serum-free DMEM. Images of the scratches were acquired using a
digital camera system (Olympus Optical Co., Ltd., Tokyo, Japan) 0,
24 and 48 h after the scratches were made. The experiments were
performed in triplicate and repeated ≥3 times.
Cell proliferation assay by MTT
The
3-(4,5-dimeth-ylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
(MTT, 5 mg/ml; Sigma-Aldrich) was used to analyze the cell
proliferation. 786-O and ACHN (5,000 cells/well) were plated into
96-well plates with 5 replicate wells of each condition. Each well
was transfected with 5 pmol miR-20b-5p mimics or a negative control
and assessed at 0, 24, 48 or 72 h post-transfection. The blank
control wells were just set up with DMEM. Before measurement, 20
µl MTT was added to the cells and incubated at 37°C in a
humidified chamber containing 5% CO2 for 6 h. Then, the
MTT medium mixtures were discarded and 120 µl dimethyl
sulphoxide (DMSO; Sigma-Aldrich, Shanghai, China) was added. After
agitating for 30 min at room temperature, the absorbance was
measured by the ELISA microplate reader (Bio-Rad, Hercules, CA,
USA) at a wavelength of 490 nm (with 630 nm as the reference wave
length).
Cell apoptosis assay
786-O or ACHN cells (3×105) were plated
in 6-well plates for the cell apoptosis assay. The cells were
transfected with 200 pmol of miR-20b-5p mimics or the negative
control (GenePharma) for 6 h. The sequence of the negative control
RNA was as follows: Forward, 5′-UCCAUAAAGUAGGAAACACUACA-3′; and
reverse, 5′-CAGUACUUUUGUGUAGUACAA-3′. After 48 h transfection, the
cells, including floating cells, were harvested, washed twice with
4°C PBS, resuspended in 100 µl of 1X binding buffer and
stained with 3 µl of propidium iodide (PI) and 5 µl
of Annexin V-fluorescein isothiocyanate (Invitrogen) for 15 min at
room temperature. Flow cytometry (EPICS, Xl-4, Beckman Coulter
Inc., Brea, CA, USA) was used to quantify the percentage of
apoptotic cells within 30 min of staining and 400 µl 1X
binding buffer was added to each sample prior to measurement. Each
experiment was conducted at least three times.
Bioinformatics
Predictions of potential targets of miR-20b-5p were
performed using computational algorithms based on 'seed regions'
between miRNAs and target genes. miRanda (http://mirdb.org/miRDB/index.html), TargetScan Release
6.2 (http://www.targetscan.org), microRNA
(http://www.microrna.org) and miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk)
were used to explore the association between miR-20b-5p and long
non-coding RNAs.
Statistical analysis
All data are presented as the mean ± standard
deviation from the three independent experiments. Statistical
analysis was conducted with SPSS 19.0 statistical software package
(IBM, Armonk, NY, USA). Statistical significance was determined
with Student's t-test. For the comparison of miR-20b-5p expression
levels between matched tumor and normal samples a paired t-test was
used. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-20b-5p is downregulated in human RCC
clinical specimens and RCC cell lines
A recent miRNA microarray chip analysis showed
downregulation of miR-20b-5p in RCC tissues (19). To confirm downregulation of
miR-20b-5p, RT-qPCR was performed in 48 paired RCC tissues and
adjacent normal tissues. Relative expression of miR-20b-5p
[Log2(T/N)] is shown in Fig. 1A.
The present study demonstrated that the expression of miR-20b-5p in
RCC tissues was significantly lower compared with adjacent normal
tissues (P<0.001), as shown in Fig.
1B.
The expression of miR-20b-5p in two RCC cell lines
(786-O and ACHN) and the 293T normal kidney human embryo kidney
cell line was analyzed. As shown in Fig. 2, miR-20b-5p expression was
significantly lower in 786-O and ACHN cells (P<0.001 and
P<0.001) than that in the 293T cells, which is in accordance
with the expression pattern of miR-20b-5p in tissues.
Validation of cell transfection
efficiency
As shown in Fig.
3A, the transfection efficiency was >90% when the cells were
transfected with fluorescein amidite-conjugated miRNA. RT-qPCR was
also used to quantify the transfection efficiency, and revealed
that expression of miR-20b-5p was 50.4 and 402.4 times that of
negative control after transfection in 786-O (P<0.001) and ACHN
(P<0.001), respectively (Fig.
3B).
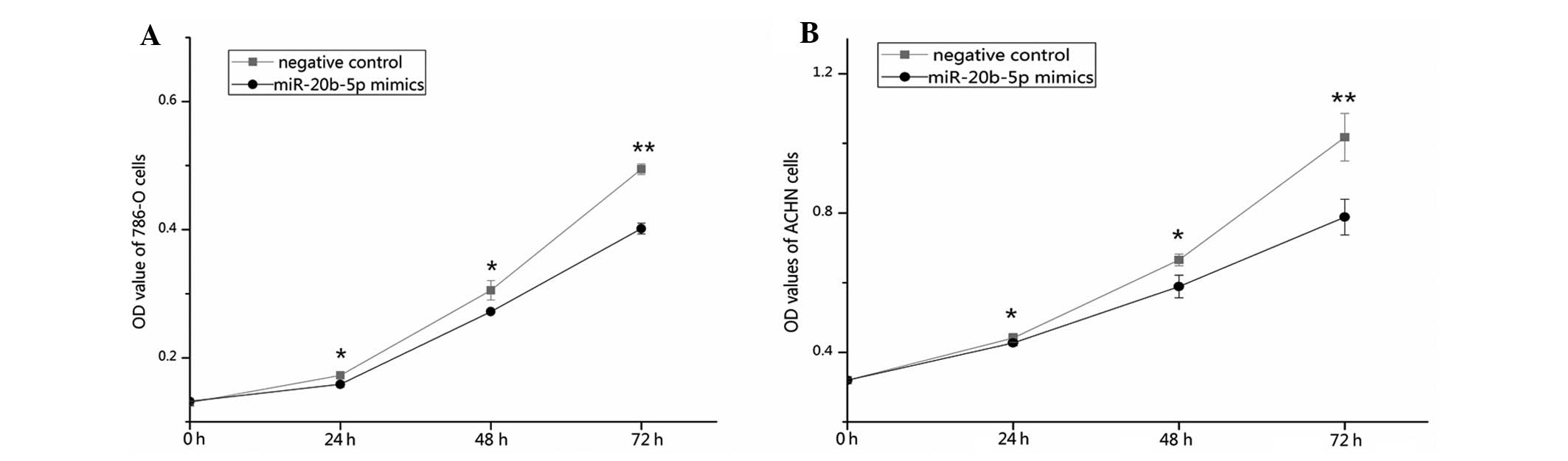
miR-20b-5p inhibits cell
proliferation
An MTT assay was used to determine whether
overexpression of miR-20b-5p affected the proliferation of RCC
cells. The outcomes revealed that the proliferation of 786-O cells
was decreased by 8.11 (P<0.05), 10.92 (P<0.05) and 18.74%
(P<0.01), and the proliferation of ACHN cells was decreased by
3.26 (P<0.05), 11.52 (P<0.05), and 22.53% (P<0.01) at 24,
48 and 72 h after transfection with miR-20b-5p mimics as compared
with the negative control. The results indicated that the
upregulation of miR-20b-5p expression significantly decreased the
proliferation of renal cancer cells (Fig. 4).
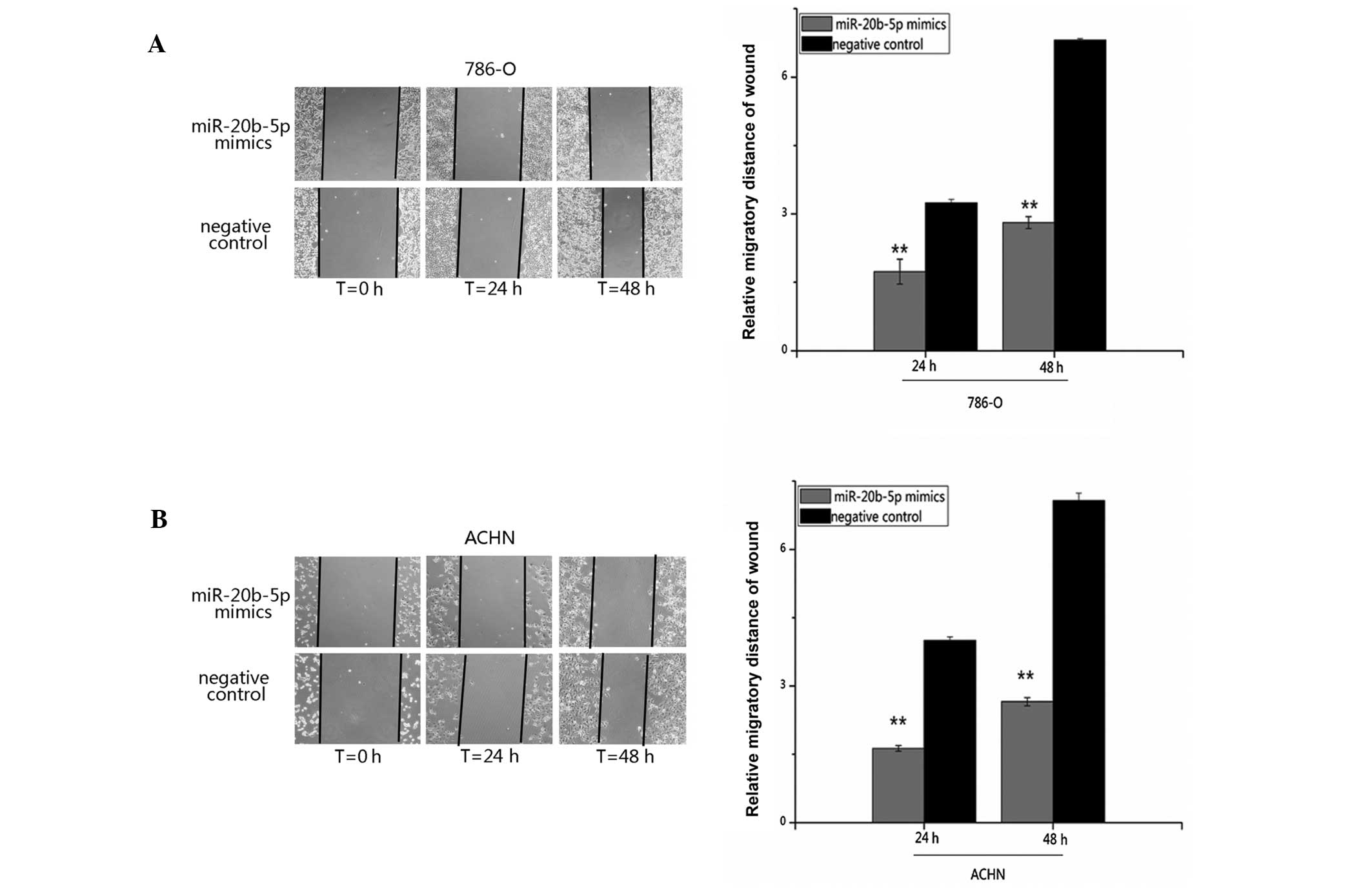
miR-20b-5p suppresses cell migration
Cell migration was examined by a wound scratch
assay. The results demonstrated that migratory distances of cells
transfected with miR-20b-5p mimics were markedly shorter than that
of the negative control group. The inhibition rates of migration of
miR-20b-5p were 46.45 and 58.74% for 786-O cells (P<0.001), and
59.25 and 62.49% for ACHN cells (P<0.001) 2 and 48 h after
transfection, compared with the negative control group. It is
suggested that upregulation of miR-20b-5p inhibited the migratory
ability in renal cancer cells (Fig.
5).
miR-20b-5p promotes cell apoptosis
The effects of miR-20b-5p on apoptosis were
determined by flow cytometric analysis. After transfection with
miR-20b-5p mimics or negative control for 48 h, 786-O and ACHN
cells were harvested, and stained and the number of cells was
quantified. The results demonstrated that the early apoptosis rate
of 786-O cells was 11.77 and 7.21% (P<0.001) and of ACHN cells
was 17.66 vs. 6.52% (P<0.05) when each were transfected with
miR-20b-5p mimics or negative control, respectively. These data
suggest that upregulation of miR-20b-5p promoted RCC cell apoptosis
(Fig. 6).
Target gene prediction
miRanda, TargetScan Release 6.2, microRNA and
miRWalk all predicted vascular endothelial growth factor A (VEGFA),
proteinase-activated receptor 1 (PAR-1; also known as F2R), MAP3K8
and CAMP responsive element binding protein 1 (CREB1) to be
putative targets of miR-20b-5p. VEGFA and PAR-1 have been
demonstrated to be modulated by miR-20b in cancer (21,22),
while MAP3K8 and CREB1 have not been investigated. microRNA also
predicted that MALAT1 (lncRNA) is a target of miR-20b-5p.
Discussion
Cancer, a complex multistep process that involves
the accumulation of sequential alterations of numerous genes,
including the activation of oncogenes and dysfunction of
anti-oncogenes, is characterized by unrestricted proliferation,
invasion and metastasis (23). By
inducing either mRNA degradation or translational repression,
miRNAs regulate a large proportion of the transcriptome (~50% in
humans) (10,24). Therefore, mRNAs function as
potential oncogenes and tumor repressors and have the ability to
affect all cellular processes. Thus, an aberrant miRNA expression
signature is a hallmark of cancer, including kidney cancer
(23). miRNA profiling in kidney
cancers revealed that a number of miRNAs were up- or downregulated,
including miR-20b, and specific miRNA profiles could serve as a
valuable tool for diagnosis (19,25).
miR-20b has been found to be upregulated in several
types of human cancer, such as breast cancer (13,26),
gastric cancer (14) and cervical
neoplasms (15). miR-20b could
function as an oncogene through downregulating tumor suppressor
genes, such as PTEN, BRCA1 and p21 (13,27).
Studies also demonstrated that upregulation of miR-20b was found in
gastric and cervical cancer tissues and may be associated with the
prognosis of patients and could serve as a diagnostic tool
(14,15,26,28).
However, certain studies have demonstrated that miR-20b is
significantly downregulated in early T-cell precursor acute
lymphoblastic leukemia, oropharyngeal carcinoma and colorectal
tumors, and the downregulation of miR-20b-5p is important in
metastasis and early recurrence of breast cancer (11,12,16–18).
Perez-Rivas et al, identified a set of recurrence-related
microRNAs, including miR-20b, with potential to identify patients
that are likely to develop metastasis early after primary breast
surgery (11). The results of
another study (12) also confirmed
that miR-20b-5p is important in metastasis and early recurrence of
breast cancer. According to lymph node metastases, pathology and
immunohistochemistry, patients in the study by Li et al
(12) were divided into three
groups: The high invasive and metastatic group (HIMG), the low
invasive and metastatic group (LIMG) and the normal group. The
authors detected the expression of miR-20b in the centre and at the
edge of breast cancer tissues and normal tissues, and their results
showed that the relative expression of miR-20a and miR-20b was
lower in the center of the tumor than at the edge in the LIMG,
lower at the edge of the tumor than in the center in the HIMG, and
lower in breast cancer tissues than in normal tissues (12)
Moreover, miR-20b-5p was reported to be
downregulated in RCC tissues by next-generation small
RNA-sequencing (19). However, the
function and clinical significance of miR-20b-5p in RCC has yet to
be explored in renal cancer. In this study, RT-qPCR was performed
to quantify the relative miR-20b-5p expression in 48 paired RCC
tissues and cell lines compared with adjacent normal tissues and
293T cells. The result, which showed that miR-20b-5p was
downregulated significantly in RCC tissues and RCC cell lines, was
in accordance with the previous sequencing. Furthermore, the role
of miR-20b-5p in cellular process such as proliferation, migration
and apoptosis were determined by an MTT assay, a scratch assay and
flow cytometry after transfection with synthetic miR-20b-5p mimics
or negative controls. In the present study, upregulation of
miR-20b-5p decreased the ability of the cells to proliferate and
migrate, and induced the apoptosis of RCC cells, signifiantly. The
results of the present study demonstrated that the effects of
miR-20b-5p on the migration, proliferation and apoptosis of the
cells were more marked in ACHN cells than in 786-O cells. This
phenomenon may have two alternative possible explanations: first,
since the expression of miR-20b-5p is lower in ACHN than in 786-O
cells, the same quantity of exogenous miR-20b-5p may lead to a more
notable effect on the ACHN cells; second, the transfection
efficiency experiments revealed that, comparatively, a much greater
quantity of miR-20b-5p mimics could be transfected into the ACHN
cells than into the 786-O cells. The function assay suggested that
miR-20b-5p may be a tumor suppressor gene in RCC by inhibiting cell
proliferation and migration, and promoting cell apoptosis.
miR-20b-5p is a negative regulator of numerous
important genes in cancer progression. miR-20b has been
demonstrated to cause downregulation of vascular endothelial growth
factor (VEGF) at the mRNA and protein level under hypoxia-mimicking
conditions, through reducing the levels of nuclear hypoxia
inducible factor-1α subunit in breast cancer cells (21). VEGF is the key gene of angiogenic
factors that induce angiogenesis, thereby ensuring the delivery of
enough blood to support the growth of the tumor (29). RCC is a highly vascularized tumor
that originates in the renal cortex. VEGF-targeted agents, such as
sorafenib and axitinib have recently been tested in a phase III
clinical trial and have been shown to improve prognosis in patients
with metastatic RCC (30).
Therefore, miR-20b-5p may be identified as an anti-VEGF agent for
RCC patients. PAR-1 was also confirmed as a target gene of miR-20b
by a luciferase reporter assay in melanoma (22). PAR-1 mediates angiogenesis and
impacts the process of tumor growth and disease progression, and de
Martino M et al reported that the AA genotype of the PAR-1
variation IVSn-14 A>T is associated with an increased risk of
metastasis and poorer prognosis in patients with RCC (31).
To the best of our knowledge, this is the first
study to identify that miR-20b-5p was downregulated in RCC tissue
and cell lines and that it functioned as a tumor suppressor gene in
RCC via inducing cellular migration reduction, proliferation
inhibition and cell apoptosis. Further studies are required to
explore the role of the miR-20b-5p-mediated molecular pathway in
tumor suppression in RCC. The potential clinical significance of
miR-20b-5p in early detection, prognosis prediction and therapeutic
target is worthy of researching.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (no. 81101922), the Science and
Technology Development Fund Project of Shenzhen (nos.
JCYJ20130402114702124 and JCYJ20150403091443329) and the fund of
Guangdong Key medical subject.
References
|
1
|
Tavani A and La Vecchia C: Epidemiology of
renal-cell carcinoma. J Nephrol. 10:93–106. 1997.PubMed/NCBI
|
|
2
|
National Cancer Institute: Surveillance,
epidemiology and end results program. SEER stat fact sheets: Kidney
and renal pelvis cancer. http://seer.cancer.gov/statfacts/html/kidrp.html.
Accessed March 30, 2014.
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 20142014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhejiang Channel XNA: World Kidney Day:
Kidney Cancer incidence increased year by year become 'invisible
killer'. Available at: http://www.zj.xinhuanet.com/news-center/science/2014-03/13/c_119759340.htm.
Accessed 13 March 2014.
|
|
5
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
6
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perez-Rivas LG, Jerez JM, Carmona R, de
Luque V, Vicioso L, Claros MG, Viguera E, Pajares B, Sánchez A,
Ribelles N, et al: A microRNA signature associated with early
recurrence in breast cancer. PLoS One. 9:e918842014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li JY, Zhang Y, Zhang WH, Jia S, Kang Y
and Zhu XY: Differential distribution of miR-20a and miR-20b may
underly metastatic heterogeneity of breast cancers. Asian Pac J
Cancer Prev. 13:1901–1906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li D, Ilnytskyy Y, Kovalchuk A, Khachigian
LM, Bronson RT, Wang B and Kovalchuk O: Crucial role for early
growth response-1 in the transcriptional regulation of miR-20b in
breast cancer. Oncotarget. 4:1373–1387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo J, Miao Y, Xiao B, Huan R, Jiang Z,
Meng D and Wang Y: Differential expression of microRNA species in
human gastric cancer versus non-tumorous tissues. J Gastroenterol
Hepatol. 24:652–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Dysregulated
microRNAs in the pathogenesis and progression of cervical neoplasm.
Cell Cycle. 11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coskun E, Neumann M, Schlee C, Liebertz F,
Heesch S, Goekbuget N, Hoelzer D and Baldus CD: MicroRNA profiling
reveals aberrant microRNA expression in adult ETP-ALL and
functional studies implicate a role for miR-222 in acute leukemia.
Leuk Res. 37:647–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hui AB, Lin A, Xu W, Waldron L,
Perez-Ordonez B, Weinreb I, Shi W, Bruce J, Huang SH, O'Sullivan B,
et al: Potentially prognostic miRNAs in HPV-associated
oropharyngeal carcinoma. Clin Cancer Res. 19:2154–2162. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamaguchi T, Iijima T, Wakaume R,
Takahashi K, Matsumoto H, Nakano D, Nakayama Y, Mori T, Horiguchi S
and Miyaki M: Underexpression of miR-126 and miR-20b in hereditary
and nonhereditary colorectal tumors. Oncology. 87:58–66. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Müller S and Nowak K: Exploring the
miRNA-mRNA regulatory network in clear cell renal cell carcinomas
by next-generation sequencing expression profiles. Biomed Res Int.
2014:9484082014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer Verlag; New York, NY: pp. 479–489. 2010
|
|
21
|
Cascio S, D'Andrea A, Ferla R, Surmacz E,
Gulotta E, Amodeo V, Bazan V, Gebbia N and Russo A: miR-20b
modulates VEGF expression by targeting HIF-1 alpha and STAT3 in
MCF-7 breast cancer cells. J Cell Physiol. 224:242–249.
2010.PubMed/NCBI
|
|
22
|
Saleiban A, Faxälv L, Claesson K, Jönsson
JI and Osman A: miR-20b regulates expression of
proteinase-activated receptor-1 (PAR-1) thrombin receptor in
melanoma cells. Pigment Cell Melanoma Res. 27:431–441. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Visone R and Croce CM: MiRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Voinnet O: Origin, biogenesis and activity
of plant microRNAs. Cell. 136:669–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Junker K, Ficarra V, Kwon ED, Leibovich
BC, Thompson RH and Oosterwijk E: Potential role of genetic markers
in the management of kidney cancer. Eur Urol. 63:333–340. 2013.
View Article : Google Scholar
|
|
26
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
27
|
Wu S, Huang S, Ding J, Zhao Y, Liang L,
Liu T, Zhan R and He X: Multiple microRNAs modulate p21Cip1/Waf1
expression by directly targeting its 3′ untranslated region.
Oncogene. 29:2302–2308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
George DJ and Kaelin WG Jr: The von
Hippel-Lindau protein, vascular endothelial growth factor and
kidney cancer. N Engl J Med. 349:419–421. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Motzer RJ and Basch E: Targeted drugs for
metastatic renal cell carcinoma. Lancet. 370:2071–2073. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Martino M, Haitel A, Schatzl G and
Klatte T: The protease activated receptor 1 gene variation IVSn-14
A>T is associated with distant metastasis and cancer specific
survival in renal cell carcinoma. J Urol. 190:1392–1397. 2013.
View Article : Google Scholar : PubMed/NCBI
|