Introduction
Enterovirus 71 (EV71) is a major cause of hand, foot
and mouth disease (HFMD) in children (1). The pathogen was initially recognized
in l969 in California following extensive outbreaks in the United
States (1,2). Subsequently, outbreaks have been
recorded in Australia (3), Japan
(4,5), Brazil (6), Malaysia (7), Thailand (8), Singapore (9), Taiwan (10) and mainland China (11,12).
When infected by EV71, the majority of patients recover naturally
and generate neutralizing antibodies. However, there are certain
patients who experience severe clinical symptoms, including aseptic
meningitis, brainstem encephalitis, neurogenic pulmonary edema,
acute flaccid paralysis, myocarditis and even mortality (13–15).
The mechanisms underlying the difference in clinical severity
remain unknown. Numerous factors affect the outcome of the disease
during an EV71 infection epidemic, including genetic mutation of
the virus and the immune status of patient.
Polioviruses and EV71 are members of the
Enterovirus genus within the Picornaviridae family.
The EV71 genome is a positive-stranded RNA containing ~7,400
nucleotides. The genome includes an open reading frame, which
encodes one polyprotein that is further divided into P1, P2 and P3
by the self-coding proteinases, 2A and 3C. P1 is further divided
into four structural proteins, VP1-4. P2 and P3 contain the
non-structural proteins 2A, 2B, 2C, 3A, 3B, 3C and 3D (16,17).
The genome is flanked by 5′ and 3′ untranslated regions (UTRs)
(18). Studies investigating EV71
have previously demonstrated that amino acid mutations in VP2 and
VP1 (19,20), and a single nucleotide mutation in
the 5′-UTR (21) increase
infectivity and lethality. Furthermore, other reports demonstrated
that cluster of differentiation (CD)4 T cells function in a helper
role for B and CD8 T cell antiviral activities (22,23),
and induce immunopathogenesis elicited by viral infection (22). This suggests that CD4 T cells
exhibit opposing functions in viral-induced immunity or
immunopathogenesis.
The present study aimed to further characterize the
interactions between viral pathogens and the induced host
responses. The current study reports novel findings from two
representative isolated virus strains with severe or mild clinical
features, which were used to infect one-day-old mice. Results
demonstrated that during EV71 infection viral genomic mutation and
immunopathogenesis, potentially as a result of CD4 T cells, may
lead to histological damage to multiple tissues. Thus, the current
study has important implications for the identification of highly
and weakly pathogenic strains, and in aiding the treatment and/or
prevention of the EV71 infection.
Materials and methods
Cell culture and animals
All animal procedures were approved by the ethics
committee of Shandong Academy of Medical Sciences (Jinan, China),
and performed in compliance with the European Union regulations and
the National Institutes of Health standards (1996 Guide for the
Care and Use of Laboratory Animals) (24). BALB/c mice (n=56; weight, 1.2–1.5
g; age, 1 day) were purchased from Vital River Laboratories Co.,
Ltd. (Beijing, China). The animals were maintained according to the
guidelines of the National Science Council of the People's Republic
of China. Pups of the same experimental group were housed together
in an environment of 50% humidity at 22°C under a 12-h light/dark
cycle. They were kept with their mothers to provide food. The MA104
cell line (25) (Cell Bank of Type
Culture Collection of Chinese Academy of Sciences, Shanghai, China)
was maintained at 37°C in a humidified atmosphere with 5%
CO2 in modified Eagle's medium (MEM; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA).
Patient sample collection
Human sample collection was approved by the research
ethics committee of Jinan Infectious Diseases Hospital (Jinan,
China). Fecal samples were collected from a representative healthy
patient, and from 4 patients with mild and 2 with severe cases of
HFMD between 2008 and 2010. Prior to collecting fecal samples, all
parents or caregivers of the eligible children were asked to
provide written informed consent and given an explanation of the
study. Child assent was obtained from children ≥7 years of age.
Children were excluded if they and/or their caregivers refused
study participation. The four mild cases exhibited fever (<38°C)
with rash on the hands, feet and buttocks, and ulcers on the mouth.
The temperature of the patients returned to normal within 5 days
and other symptoms disappeared in a short time. The 2 patients with
severe cases experienced more severe symptoms. These included high
fever (>39°C) that lasted for 7 days, mild rash on the hands,
feet and buttocks, and frequent vomiting. The conditions of the two
severe cases developed rapidly with onset of pulmonary edema and
neurologic complications, in addition to dyspnea, acute flaccid
paralysis and coma.
Virus isolation and identification
The fecal samples were centrifuged at 10,000 × g for
10 min at 4°C and filtered to produce a sterile suspension. MA104
cells (1×106 cells) were plated into a culture flask,
incubated overnight to reach 65–70% confluence, and then infected
with 0.5 ml sterile suspension. Following adsorption for 2 h, the
virus suspension was replaced with MEM containing 2% FBS. The
culture medium was centrifuged at 10,000 × g for 10 min at 4°C. The
virus was collected from the supernatant by three freeze-thaw
cycles following the appearance of a 90% cytopathogenic effect
(CPE) determined by observation of cell shrinkage, rounding of the
cell, vacuolar changes and ecclasis using an ECLIPSE Ti-S
microscope (Nikon Corporation, Tokyo, Japan). Total RNA was
extracted from the virus supernatant using QIAamp Viral RNA Mini
kit (Qiagen, Inc., Valencia, CA, USA), according to the
manufacturer's protocols. Viral cDNA was obtained using Quantscript
RT kit (Tiangen Biotech Co., Ltd., Beijing, China) for 1 h at 37°C.
Taq DNA polymerase (Tiangen Biotech Co., Ltd.) was used in the PCR,
double distilled water served as a negative control and known EV71
cDNA served as a positive control. For identification of EV71, the
standard primer sequences were adopted, as follows: Sense,
5′-GCAGCCCAAAAGAACTTCAC-3′; and anti-sense,
5′-ATTTCAGCAGGTTGGAGTGC-3′. The reaction was conducted using an MJ
Mini Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at the following conditions: 94°C for 3 min; 30 cycles of 94°C for
30 sec, 55°C for 30 sec and 72°C for 1 min; and 72°C for 10 min.
The PCR products were separated using 2% agarose electrophoresis at
80 V for 40 min, with a D2000 DNA ladder (Tiangen Biotech Co.,
Ltd.) and double distilled water as a negative control. The
ethidium bromide was supplied by Sigma-Aldrich and the products
were extracted with TIANgel Midi Purification kit (Tiangen Biotech
Co., Ltd.), then sequenced by Beijing BioSune Biotechnology Co.,
Ltd. (Beijing, China).
Plaque purification
MA104 cells were seeded in a 24-well plate,
incubated overnight and then infected with 200 µl serially
diluted virus suspension (diluted 1:10, 1:102,
1:103 and so on). Following adsorption for 1 h, the
virus suspension was replaced with MEM supplemented with 2% FBS and
0.5% low melting agarose (Invitrogen; Thermo Fisher Scientific,
Inc.). An independent plaque was collected with a crooked pipet at
48 h post-infection until a monolayer of MA104 cells formed.
Virus titration
Infectivity titer, described as tissue culture
infectious dose 50 (TCID50), of the virus in MA104 cells
(1×105 cells/well) was performed following the
previously described method of Reed and Muench (26). Briefly, MA104 cells were seeded in
96-well plates at 1×104 cells/well. Following overnight
incubation, the virus was serially diluted 10-fold with RPMI-1640
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 2% FBS and
added to the monolayer MA104 cells. The plates were then incubated
at 37°C in a 5% CO2 atmosphere for 96 h and the presence
of CPE was observed (27).
Viral infection of mice
Seven groups of eight 1-day-old BALB/c pups were
inoculated with 1×104 TCID50 of the different
virus strains by intraperitoneal injection. One group was
administered MEM containing 2% FBS as a control. The mice were
monitored every 3–6 h for clinical symptoms and were sacrificed by
decapitation if mice met the following criteria: Body weight loss;
lethargy; hypoxemia; and hypothermia, <33.5°C.
Electromyography determination
The function of nerves and quadriceps of the
inoculated mice were examined using electromyography
(Keypoint® G4 Workstation; Natus Medical Incorporated,
Pleasanton, CA, USA). The responses of hind leg muscles were
measured using Ag-AgCl electrodes (Natus Medical Incorporated).
Thirty motor unit potentials were selected on the same muscles.
Histological analysis
At day 4 post-inoculation of the virus isolates,
tissues were obtained from the different groups of mice and
processed for histological analysis. Various tissue samples,
including the cerebrum, spinal cord, limb muscle, thymus gland,
spleen, liver, lungs, kidney, stomach and intestines were removed
from infected mice. The tissues were fixed with 4% formaldehyde
(Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) for 48 h
and embedded in paraffin (Sinopharm Chemical Reagent Co., Ltd.).
The 5-µm sections were stained with hematoxylin and eosin Y
(Sigma-Aldrich, St. Louis, MO, USA) for morphological examination.
The sections were observed under an ECLIPSE Ti-S microscope.
Immunohistochemical analysis
The 5-µm sections were placed on
poly-L-lysine-coated (Sigma-Aldrich) glass slides and fixed with 4%
paraformaldehyde (Sinopharm Chemical Reagent Co., Ltd.) in
phosphate-buffered saline (PBS; Invitrogen; Thermo Fisher
Scientific, Inc.). Endogenous peroxidase was inhibited by 3%
H2O2 (Sinopharm Chemical Reagent Co., Ltd.)
in PBS. The sections were blocked with 5% bovine serum albumin for
1 h at 37°C. EV71 was detected using mouse monoclonal anti-EV71
antibody (1:1,000; EMD Millipore, Billerica, MA, USA; cat. no.
MAB979) followed by incubation with goat anti-mouse immunoglobulin
G peroxidase-conjugated antibody (1:200; BIOSS, Beijing, China;
cat. no. bs-0296G). The slides were then stained with
3,3′-diaminobenzidine (Sigma-Aldrich) and counterstained with
hematoxylin for morphological examination using the ECLIPSE
Ti-S.
Quantification of T and NK cells
At 0, 3, 7 and 12 days post-infection, spleens were
excised from two of the mice from each group, then homogenized and
the cells were harvested using 70-µm cell strainers (BD
Biosciences, Franklin Lakes, NJ, USA). Splenocyte suspensions were
centrifuged at 500 × g for 5 min at 4°C over Histopaque
(Sigma-Aldrich) and washed. Peripheral blood mononuclear cell
(PBMC) suspensions were prepared in PBS supplemented with 2% FBS
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). Staining
was performed according to the antibody manufacturer's protocols
(eBioscience, Inc., San Diego, CA, USA). Briefly, ~1 million PBMCs
were stained for the cell surface molecules, CD4, using fluorescein
isothiocyanate (FITC)-labeled rat monoclonal antibodies (1:200;
eBioscience, Inc., cat. no. 11-0042), and CD25, using phycoerythrin
(PE)-cyanine 5.5-labeled rat monoclonal antibodies (1:200;
eBioscience, Inc.; cat. no. 35-0251), for 30 min. They were then
fixed and permeated using Cytofix/Cytoperm buffer (BD Biosciences)
for the appropriate time according to the manufacturer's protocols.
Other PBMCs were stained in parallel using APC-labeled rat
monoclonal anti-CD16 (for NK cells; 1:200; eBioscience, Inc.; cat.
no. 17-0161-81), rat monoclonal FITC-labeled anti-CD3 (for total
lymphocytes; 1:200; eBioscience, Inc.; cat. no. 11-0032) and
PE-labeled rat monoclonal anti-CD8 antibody (1:200; eBioscience,
Inc.; cat. no. 12-0081). Cells were washed with 200 µl 1X
Perm/Wash buffer (BD Biosciences) and stained with PE-labeled rat
monoclonal forkhead box P3 (Foxp3) antibody (1:50; eBioscience,
Inc.; cat. no. 12-5773) for 1 h at room temperature. The cells were
washed again in 200 µl 1X Perm/Wash buffer and fixed in
paraformaldehyde-PBS. Cells were then analyzed using a BD
FACSCalibur™ flow cytometer (BD Biosciences) and FlowJo software
(version 7.6.1; Tree Star, Inc., Ashland, OR, USA).
Full-length gene sequencing and
analysis
The full-length sequencing of the virus genome was
performed by Beijing BioSune Biotechnology Co., Ltd. and analyzed
using Moleculat Evolutionary Genetics Analysis software (version
5.05) (28).
Statistical analysis
Student's t-test was used to determine statistical
significance. All analyses were performed using SPSS software
(version 13.5; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Six EV71 strains were isolated and a
weakly and highly pathogenic strain were selected for further
study
MA104 cells exhibited marked CPEs on day 1 following
inoculation with viral specimens. The CPEs included cell shrinkage,
rounding of the cell, cytoplasmic vacuolar changes and ecclasis.
Six specimens exhibited similar CPE and all were identified as EV71
following inoculation. The present study identified a weakly
(JN200803) and a highly pathogenic strain (JN200804; Fig. 1A).
No difference in virulence was determined
by TCID50 titration
The TCID50 of the six virus specimens
were calculated according to the method previously described by
Reed and Muench (26). The
TCID50 of all viruses were between 105.7 and
106.1/ml. No statistical significance was demonstrated
between the TCID50 titrations of different viral samples
(P>0.05; t-test).
Mice were infected with the weakly or
highly pathogenic strains
One-day-old BALB/c mice were inoculated with
1×104 TCID50 from each virus strain by
intraperitoneal injection. The data of two representative
highly/weakly pathogenic viruses are presented. By day 3
post-infection, 4 out of 7 JN200804-infected mice had developed
hind limb paralysis (Fig. 1B) and
by day 4 post-infection, all JN200804-infected mice had developed
hind limb paralysis and were sacrificed within 5–7 days
post-infection (Fig. 1C). By
contrast, the JN200803-infected and negative control mice remained
healthy and grew normally without any apparent neurological
symptoms.
Electromyography demonstrated an
increased time limit and decreased wave amplitude in
JN200804-infected mice
To further assess the function changes to the nerves
and muscles following viral infection, an electromyography machine
was employed to record the electric current time limit and wave
length. Electromyography of nerve and muscle function demonstrated
that the time limit was increased and the wave amplitude
significantly decreased in JN200804-infected mice compared with
control mice (P<0.05), whereas, the JN200803-infected mice were
normal with no statistical difference compared with the negative
control (P>0.05; Fig. 1D).
Numerous histological changes were
observed in JN200804-infected mice
Histological analysis was performed on a variety of
tissues and major organs from EV71-infected mice (Fig. 2). Compared with negative control
mice, numerous histopathological changes were observed in various
tissues following JN200804 inoculation. The number of Purkinje and
granular cells in the cerebellum were decreased compared with
non-infected mice (Fig. 2A).
Additionally, nerve fibers in the white matter of the spinal
midpiece appeared thick and swollen (Fig. 2C), and pneumorrhagia was observed
(Fig. 2E). Furthermore, lymphocyte
infiltration and acidophilic degeneration were present in the liver
(Fig. 2G). In the renal cortex,
the number of glomeruli was decreased and epithelial cells within
the kidney tubules appeared cloudy and were swollen in appearance
(Fig. 2I). The skeletal muscle
cells exhibited necrolysis with lymphocyte infiltration (Fig. 2K). However, the JN200803-infected
mice did not demonstrate apparent histopathological changes
(Fig. 2) when compared with
negative controls.
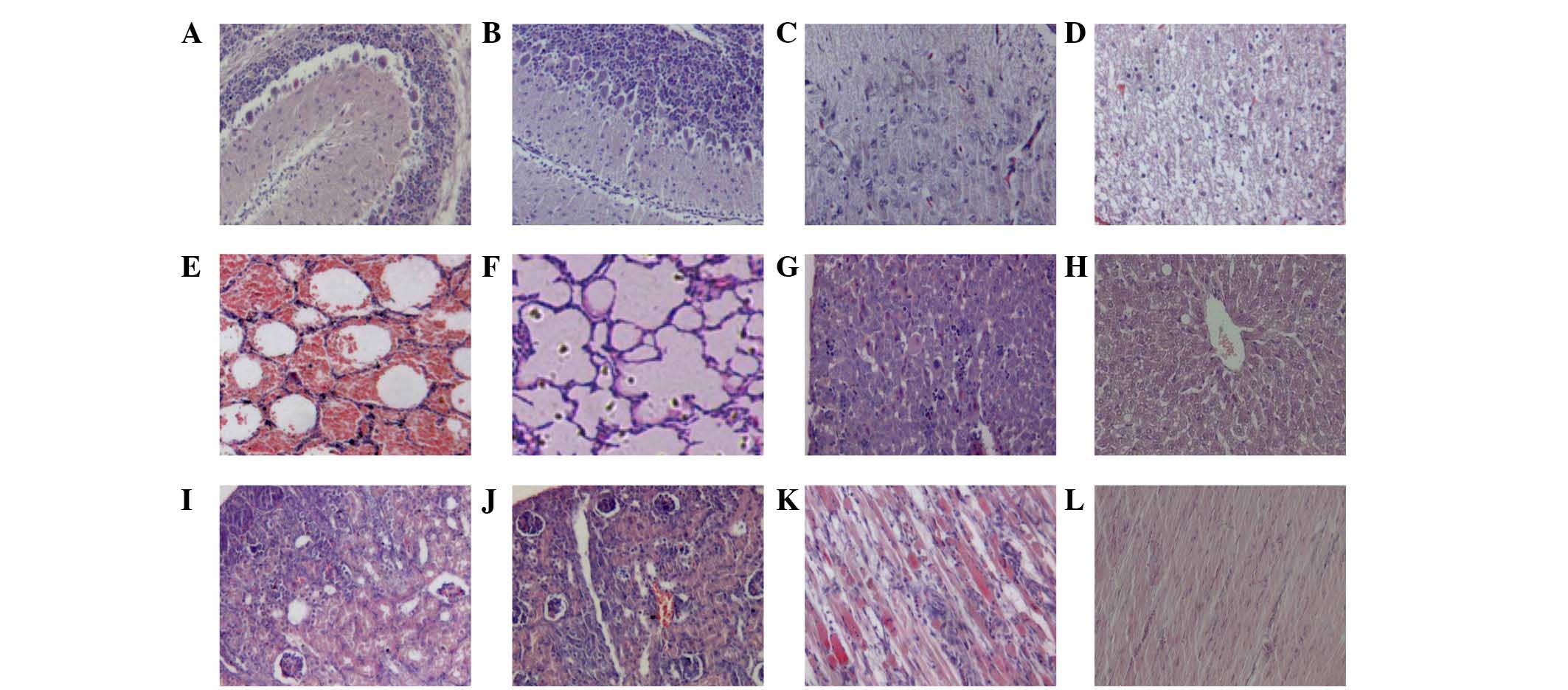 | Figure 2Histological examination of tissues
from enterovirus 71-infected mice using hematoxylin and eosin
staining. (A and B) Cerebellum, (C and D) spinal midpiece, (E and
F) lung, (G and H) liver, (I and J) renal cortex and (K and L)
skeletal muscle of JN200804-infected (A,C,E,G,I and K) and
JN200803-infected (B,D,F,H,J and L) mice. In JN200804-infected
mice, compared with negative controls, (A) the number of Purkinje
cells and granular cells of cerebellum were decreased, (C) nerve
fibers in white matter of spinal middle piece were thick, swollen
looking. (E) Pneumorrhagia was also observed under microscope. (G)
Lymphocyte infiltration and acidophilic degeneration was observed
in the liver. (I) In the renal cortex, the number of glomeruli was
decreased and epithelial cells within the kidney tubules appeared
cloudy and swollen. (K) Necrolysis and lymphocyte infiltration was
observed in skeletal muscle cells necrolysis with lymphocyte
infiltration There was no apparent histopathological changes to the
JN200803-infected mice compared with negative controls. The results
presented are representative of two experiments. |
Immunohistochemical analysis demonstrated
the presence of EV71 in brain tissue samples from JN200804-infected
mice
Having observed broad histological damages to
various tissues following infection with a highly pathogenic strain
of EV71, mice also exhibited apparent neurological damage. Thus,
the present study investigated whether the brain tissues were
directly infected with the virus. Evidence of EV71 infection was
confirmed by staining brain tissue sections from JN200804 and
JN200803-infected mice with anti-EV71-specific antibody (Fig. 3). EV71 staining was observed in the
brain tissue of JN200804-infected mice, but not in
JN200803-infected mice. Negative control tissues did not exhibit
any positive staining (data not shown).
Bulk CD4 T cell expansion was increased
in highly pathogenic EV71 JN200804 strain-infected mice
Viral pathogens often induce strong CD4 T cell
responses that are characterized for their ability to aid B cell
and CD8 T cell responses (20).
However, a previous study has elucidated alternative pathogenic
mechanisms by which vaccine-elicited CD4 T cells induce
immunopathogenesis following chronic lymphocytic choriomeningitis
virus (LCMV) infection (22).
Thus, the present study investigated the CD4 T cell expansion
levels during EV71 infection (Fig.
4). Notably, at day 3 post-JN200804 infection, the expansion of
spleen bulk CD4 T cells was increased by 6-fold compared with the
weakly pathogenic (JN200803) strain (P<0.01). This result
indicates that overexpansion of CD4 T cells may be an important
factor in tissue damage following infection with highly pathogenic
EV71 (Fig. 4). Furthermore,
regulatory T cells isolated from the spleen were also quantified by
intracellular staining with Foxp3. No significant difference was
observed in the level of Foxp3-positive cells between JN200804 and
JN200803-infected samples. Additionally, no significant difference
was observed in the levels of CD8 T or NK cells between JN200804
and JN200803-infected samples (data not shown).
 | Figure 4CD4 T cells expansion is increases in
highly pathogenic EV71-infected mice. Mice (n=5) were inoculated
with EV71 JN200803 and JN200804 strains (1×104 tissue
culture infectious dose50, intraperitoneal injection),
controls were injected with phosphate-buffered saline. At 0, 3, 7
and 12 days post-inoculation, spleens were isolated and
homogenized. Splenocytes were stained with anti-CD4 and CD25
antibodies, and intra-cellularly stained with anti-Foxp3
antibodies. Flow cytometry and FlowJo software were used to analyze
the CD4 T cells. (A) Kinetics of CD4 T cells expansion. A
significant increase of CD4 T cell expansion was observed at 3 days
post-inoculation with the highly pathogenic EV71 JN200804 strain.
Representative flow cytometry analysis of CD4 T cell population in
(B) uninfected mice, and mice 3 days post-inoculation with EV71 (C)
JN200804 and (D) JN200803 strains. The results presented are
representative of three experiments. Data are presented as the mean
± standard error. **P<0.01 vs. JN200803. CD, cluster
of differentiation; EV71, enterovirus 71; Foxp3, forkhead box
P3. |
Full-length gene sequencing and analysis
indicated differences between the two strains in the non-coding and
coding regions
Having demonstrated that the different virus strains
resulted in either weakly or highly pathogenic changes in patients
and animal models, the present study aimed to further investigate
factors that may be leading to the differences. As viral genomic
mutations can be a causative factor of various clinical
differences, the two viruses were sequenced and the nucleotide
sequences were analyzed. Sequence variation was detected at 106
nucleotide sequences, which included 4 nucleotide sequence
variances in non-coding regions and 16 changes to the resulting
amino acid sequence in the coding regions of the two strains
(Table I). These sequences were
deposited in GenBank (www.ncbi.nlm.nih.gov/genbank/) with the accession
numbers JF913464 and HQ825317 for JN200803 and JN200804,
respectively.
 | Table ISequence comparison of JN200803 and
JN200804 strains. |
Table I
Sequence comparison of JN200803 and
JN200804 strains.
| Region | Nucleotide sequence
(JN200803>JN200804) | Amino acid sequence
(JN200803>JN200804) |
|---|
| 5′-UTR | C115T | – |
| T132C | – |
| T811C | – |
| VP3 | A2196G | Q485R |
| C2253T | A504V |
| G2351A | V537I |
| VP1 | G2657A | G639S |
| 2A | G3551A | G937S |
| 2B | C3783T | A1014V |
| A3875G | I1045V |
| C3927T | T1062I |
| 2C | A4146G | K1135R |
| G4177T | E1145D |
| T4206C | V1155A |
| G4823A | V1361M |
| 3C | A5531G | I1597V |
| G5591A | D1617N |
| 3D | C5978T | L1746F |
| C7179T | A2146V |
| 3′-UTR | A7347G | – |
Discussion
EV71 infection has become a severe health problem
for young children in China, and worldwide, due to frequent
epidemics. The disease is typically mild and self-limiting in
children, however, the infection may also result in severe
neurological disease, which can be fatal. It is generally
understood that the level of clinical severity depends on the
patient's own health and immune status. Antibodies generated from a
previous infection may protect patients from a later EV71
infection. Epidemiological studies have also demonstrated that the
severity of HFMD is not directly associated with certain genotypes
of EV71 (29,30). Thus, different genotypes may
present with mild clinical symptoms and also severe complications
at the same time. This finding is supported by research on the
infections of cynomolgus monkeys. Previous results have
demonstrated that monkeys inoculated with five EV71 strains
isolated from individual patients with fatal HFMD encephalitis and
meningitis all became infected and developed neurological
manifestations within 1–6 days post-inoculation, irrespective of
the inoculated strain used (31).
This is in accordance with the results of the present study, as
despite the TCID50 of the two virus strains (JN200804
and JN200803) being similar in vitro, they induced starkly
contrasting clinical features and pathogenicity when used to
inoculate animals. The present study demonstrated that JN200804
infection resulted in a vast array of histological changes in major
tissues and organs, and immunohistochemical analysis additionally
demonstrated damage to brain tissues (Fig. 3). The current results are
consistent with the findings discussed in a recent review (32); viral tropisms are completely
distinct in the two viral strains investigated in the present
study.
Viral infections are the result of coefficient
interaction between a virus and host. One of the common features of
RNA virus infection is genetic mutation, particularly for HIV and
influenza viruses. This type of mutation is also a causative factor
that leads to various clinical outcomes in HIV and viral influenza
infections. Regarding EV71 infection, a previous report has
demonstrated that mutations in the VP1 capsid proteins are
responsible for the viral infectivity in vitro and virulence
in vivo (19). Likewise,
virulence loci of coxsachievirus and poliovirus have also been
reported within the VP1 region (33). The full-length sequencing results
of the current study demonstrated that one amino acid mutation
existed between the two strains in the VP1 region (Table I). Although non-structural proteins
are not essential in forming the basic frame of a viral particle,
they are understood to be crucial for infection and replication.
Currently, the virulence and infectivity as a result of genomic
changes remain largely unknown. However, in EV71 infection, an
increasing number of reports, including the present study (Table I), have demonstrated that mutations
in different regions, including the 3′-UTR (18) and 5′-UTR (34,35),
in structural (19,36) and non-structural (37–42)
protein coding regions are responsible for virulence and tissue
tropisms. Further study will elucidate the genetic virulent points
important for therapeutic and preventive applications. As such, the
ongoing aim of the present research is to investigate
mutation-associated virulence in an EV71-infected mouse model.
Viral infection also induces immunopathogenesis,
particularly in CD4 T cells. A previous report by the
Penaloza-MacMaster et al (22) has demonstrated that
vaccine-elicited CD4 T cells increased tissue immunopathogenesis in
response to LCMV (16). In support
of this previous finding, the present study demonstrated that the
level of CD4 T cells was significantly increased at day 3 following
injection of mice with JN200804 (Fig.
4B), the highly pathogenic EV71 strain. This indicates an
important function for CD4 T cell response-induced pathogenesis
during EV71 infection. Thus, further research will focus on CD4 T
cell function in order to explore the mechanisms by which CD4 T
cells result in immunopathogenesis during EV71 infection.
In conclusion, the results of the current study
demonstrated the genomic variation, and the different clinical,
histological CD4 T cell responses as a result of infection with the
two EV71 strains. The results of the present study suggest that
genomic and immunological factors may mediate the tissue damage
caused by highly pathogenic EV71 infection. Thus, by characterizing
highly and weakly pathogenic EV71 strains, the results of the
present study have important implications regarding the treatment
and prevention of EV71.
Acknowledgments
The present study was supported by the National
Science and Technology Major Special Project (grant no.
2014ZX09509-001001008); the Shandong Provincial Natural Science
Foundation (grant no. ZR2013HQ039); the Institute of Basic Medicine
grant (grant no. 2014-2); Shandong Academy of Medical Sciences
grant (grant no. 2014-11); the Department of Health and Family-plan
Bureau (grant no. 2014WS0068); and Shandong Provincial College of
Traditional Chinese Medicine Antiviral Collaborative Innovation
Center (grant no. XTCX2014).
References
|
1
|
Schmidt NJ, Lennette EH and Ho HH: An
apparently new enterovirus isolated from patients with disease of
the central nervous system. J Infect Dis. 129:304–309. 1974.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alexander JP Jr, Baden L, Pallansch MA and
Anderson LJ: Enterovirus 71 infections and neurologic disease -
United States, 1977–1991. J Infect Dis. 169:905–908. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gilbert GL, Dickson KE, Waters MJ, Kennett
ML, Land SA and Sneddon M: Outbreak of enterovirus 71 infection in
Victoria, Australia, with a high incidence of neurologic
involvement. Pediatr Infect Dis J. 7:484–488. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mizuta K, Aoki Y, Suto A, Ootani K,
Katsushima N, Itagaki T, Ohmi A, Okamoto M, Nishimura H, Matsuzaki
Y, et al: Cross-antigenicity among EV71 strains from different
genogroups isolated in Yamagata, Japan, between 1990 and 2007.
Vaccine. 27. pp. 3153–3158. 2009, View Article : Google Scholar
|
|
5
|
Iwai M, Masaki A, Hasegawa S, Obara M,
Horimoto E, Nakamura K, Tanaka Y, Endo K, Tanaka K, Ueda J, et al:
Genetic changes of coxsackievirus A16 and enterovirus 71 isolated
from hand, foot, and mouth disease patients in Toyama, Japan
between 1981 and 2007. Jpn J Infect Dis. 62:254–259.
2009.PubMed/NCBI
|
|
6
|
da Silva EE, Winkler MT and Pallansch MA:
Role of enterovirus 71 in acute flaccid paralysis after the
eradication of poliovirus in Brazil. Emerg Infect Dis. 2:231–233.
1991. View Article : Google Scholar
|
|
7
|
Chan LG, Parashar UD, Lye MS, Ong FG, Zaki
SR, Alexander JP, Ho KK, Han LL, Pallansch MA, Suleiman AB, et al:
For the Outbreak Study Group: Deaths of children during an outbreak
of hand, foot, and mouth disease in sarawak, malaysia: Clinical and
pathological characteristics of the disease. Clin Infect Dis.
31:678–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chatproedprai S, Theanboonlers A, Korkong
S, Thongmee C, Wananukul S and Poovorawan Y: Clinical and molecular
characterization of hand-foot-and-mouth disease in Thailand,
2008–2009. Jpn J Infect Dis. 63:229–233. 2010.PubMed/NCBI
|
|
9
|
Shah VA, Chong CY, Chan KP, Ng W and Ling
AE: Clinical characteristics of an outbreak of hand, foot and mouth
disease in Singapore. Ann Acad Med Singapore. 32:381–387.
2003.PubMed/NCBI
|
|
10
|
Liu CC, Tseng HW, Wang SM, Wang JR and Su
IJ: An outbreak of enterovirus 71 infection in Taiwan, 1998:
Epidemiologic and clinical manifestations. J Clin Virol. 17:23–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun LM, Zheng HY, Zheng HZ, Guo X, He JF,
Guan DW, Kang M, Liu Z, Ke CW, Li JS, et al: An enterovirus 71
epidemic in Guangdong Province of China, 2008: Epidemiological,
clinical, and virogenic manifestations. Jpn J Infect Dis. 64:13–18.
2011.PubMed/NCBI
|
|
12
|
Tan X, Huang X, Zhu S, Chen H, Yu Q, Wang
H, Huo X, Zhou J, Wu Y, Yan D, et al: The persistent circulation of
enterovirus 71 in People's Republic of China: Causing emerging
nationwide epidemics since 2008. PLoS One. 6:e256622011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang LY, King CC, Hsu KH, Ning HC, Tsao
KC, Li CC, Huang YC, Shih SR, Chiou ST, Chen PY, et al: Risk
factors of enterovirus 71 infection and associated hand, foot, and
mouth disease/herpangina in children during an epidemic in Taiwan.
Pediatrics. 109:e882002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li CC, Yang MY, Chen RF, Lin TY, Tsao KC,
Ning HC, Liu HC, Lin SF, Yeh WT, Chu YT and Yang KD: Clinical
manifestations and laboratory assessment in an enterovirus 71
outbreak in southern Taiwan. Scand J Infect Dis. 34:104–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen KT, Chang HL, Wang ST, Cheng YT and
Yang JY: Epidemiologic features of hand-foot-mouth disease and
herpangina caused by enterovirus 71 in Taiwan, 1998–2005.
Pediatrics. 120:e244–e252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen C, Wang Y, Shan C, Sun Y, Xu P, Zhou
H, Yang C, Shi PY, Rao Z, Zhang B and Lou Z: Crystal structure of
enterovirus 71 RNA-dependent RNA polymerase complexed with its
protein primer VPg: Implication for a trans mechanism of VPg
uridylylation. J Virol. 87:5755–5768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu G, Qi J, Chen Z, Xu X, Gao F, Lin D,
Qian W, Liu H, Jiang H, Yan J and Gao GF: Enterovirus 71 and
coxsackievirus A16 3C proteases: Binding to rupintrivir and their
substrates and anti-hand, foot, and mouth disease virus drug
design. J Virol. 85:10319–10331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kok CC and Au GG: Novel marker for
recombination in the 3′-untranslated region of members of the
species Human enterovirus A. Arch Virol. 158:765–773. 2013.
View Article : Google Scholar
|
|
19
|
Huang SW, Wang YF, Yu CK, Su IJ and Wang
JR: Mutations in VP2 and VP1 capsid proteins increase infectivity
and mouse lethality of enterovirus 71 by virus binding and RNA
accumulation enhancement. Virology. 422:132–143. 2012. View Article : Google Scholar
|
|
20
|
Cordey S, Petty TJ, Schibler M, Martinez
Y, Gerlach D, van Belle S, Turin L, Zdobnov E, Kaiser L and
Tapparel C: Identification of site-specific adaptations conferring
increased neural cell tropism during human enterovirus 71
infection. PLoS Pathog. 8:e10028262012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yeh MT, Wang SW, Yu CK, Lin KH, Lei HY, Su
IJ and Wang JR: A single nucleotide in stem loop II of
5′-untranslated region contributes to virulence of enterovirus 71
in mice. PLoS One. 6:e270822011. View Article : Google Scholar
|
|
22
|
Penaloza-MacMaster P, Barber DL, Wherry
EJ, Provine NM, Teigler JE, Parenteau L, Blackmore S, Borducchi EN,
Larocca RA, Yates KB, et al: Vaccine-elicited CD4T cells induce
immunopathology after chronic LCMV infection. Science. 347:278–282.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manzke N, Akhmetzyanova I, Hasenkrug KJ,
Trilling M, Zelinskyy G and Dittmer U: CD4+ T cells
develop antiretroviral cytotoxic activity in the absence of
regulatory T cells and CD8+ T cells. J Virol.
87:6306–6313. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
National Research Council: Guide for the
Care and Use of Laboratory Animals. 7th edition. National Academy
Press; Washington, DC: 1996
|
|
25
|
Cuadras MA, Arias CF and López S:
Rotaviruses induce an early membrane permeabilization of MA104
cells and do not require a low intracellular Ca2+
concentration to initiate their replication cycle. J Virol.
71:9065–9074. 1997.PubMed/NCBI
|
|
26
|
Reed LJ and Muench H: A simple method of
estimating fifty per cent endpoints. Am J Hyg. 27:493–497.
1938.
|
|
27
|
Chua BH, Phuektes P, Sanders SA, Nicholls
PK and McMinn PC: The molecular basis of mouse adaptation by human
enterovirus 71. J Gen Virol. 89:1622–1632. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tamura K, Peterson D, Peterson N, Stecher
G, Nei M and Kumar S: MEGA5: Molecular evolutionary genetics
analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods. Mol Biol Evol. 28:2731–2739. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimizu H, Utama A, Yoshii K, Yoshida H,
Yoneyama T, Sinniah M, Yusof MA, Okuno Y, Okabe N, Shih SR, et al:
Enterovirus 71 from fatal and nonfatal cases of hand, foot and
mouth disease epidemics in Malaysia, Japan and Taiwan in 1997–1998.
Jpn J Infect Dis. 52:12–15. 1999.
|
|
30
|
Shih SR, Ho MS, Lin KH, Wu SL, Chen YT, Wu
CN, Lin TY, Chang LY, Tsao KC, Ning HC, et al: Genetic analysis of
enterovirus 71 isolated from fatal and non-fatal cases of hand,
foot and mouth disease during an epidemic in Taiwan, 1998. Virus
Res. 68:127–136. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagata N, Shimizu H, Ami Y, Tano Y,
Harashima A, Suzaki Y, Sato Y, Miyamura T, Sata T and Iwasaki T:
Pyramidal and extrapyramidal involvement in experimental infection
of cynomolgus monkeys with enterovirus 71. J Med Virol. 67:207–216.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Muehlenbachs A, Bhatnagar J and Zaki SR:
Tissue tropism, pathology and pathogenesis of enterovirus
infection. J Pathol. 235:217–228. 2015. View Article : Google Scholar
|
|
33
|
McMinn P, Lindsay K, Perera D, Chan HM,
Chan KP and Cardosa MJ: Phylogenetic analysis of enterovirus 71
strains isolated during linked epidemics in Malaysia, Singapore,
and Western Australia. J Virol. 75:7732–7738. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thompson SR and Sarnow P: Enterovirus 71
contains a type I IRES element that functions when eukaryotic
initiation factor eIF4G is cleaved. Virology. 315:259–266. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Klinck R, Sprules T and Gehring K:
Structural characterization of three RNA hexanucleotide loops from
the internal ribosome entry site of polioviruses. Nucleic Acids
Res. 25:2129–2137. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang PN and Shih SR: Update on
enterovirus 71 infection. Curr Opin Virol. 5:98–104. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang CH, Li HC, Jiang JG, Hsu CF, Wang YJ,
Lai MJ, Juang YL and Lo SY: Enterovirus type 71 2A protease
functions as a transcriptional activator in yeast. J Biomed Sci.
17:652010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cui S, Wang J, Fan T, Qin B, Guo L, Lei X,
Wang J, Wang M and Jin Q: Crystal structure of human enterovirus 71
3C protease. J Mol Biol. 408:449–461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shih SR, Chiang C, Chen TC, Wu CN, Hsu JT,
Lee JC, Hwang MJ, Li ML, Chen GW and Ho MS: Mutations at KFRDI and
VGK domains of enterovirus 71 3C protease affect its RNA binding
and proteolytic activities. J Biomed Sci. 11:239–248. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
de Jong AS, de Mattia F, Van Dommelen MM,
Lanke K, Melchers WJ, Willems PH and van Kuppeveld FJ: Functional
analysis of picornavirus 2B proteins: Effects on calcium
homeostasis and intracellular protein trafficking. J Virol.
82:3782–3790. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang WF, Yang SY, Wu BW, Jheng JR, Chen
YL, Shih CH, Lin KH, Lai HC, Tang P and Horng JT: Reticulon 3 binds
the 2C protein of enterovirus 71 and is required for viral
replication. J Biol Chem. 282:5888–5898. 2007. View Article : Google Scholar
|
|
42
|
Choe SS, Dodd DA and Kirkegaard K:
Inhibition of cellular protein secretion by picornaviral 3A
proteins. Virology. 337:18–29. 2005. View Article : Google Scholar : PubMed/NCBI
|


















