Introduction
Osteosarcoma is one of the most fatal tumors
worldwide (1). The most common
treatments for osteosarcoma are radiotherapy, chemotherapy and
surgical resection (2,3). However, the side effects and cure
rate remain unsatisfactory. Understanding the molecular mechanisms
of oncogenes and tumor suppressor genes that are involved in cancer
progression may provide novel targets and biomarkers for diagnosis
and therapy of osteosarcoma.
Serpins are a group of proteins, which are able to
inhibit proteases (4). Certain
members of the Serpin family, including SerpinI1, SerpinB2,
SerpinE2 and SerpinD1, have been demonstrated to participate in
tumor progression, particularly in tumor metastasis (4–8).
These proteins are involved in sustaining cancer cells in brain
metastasis (5). Serpins also
exhibit a role as anti-plasminogen activators, which act as a
shield for cancer from deleterious signals of astrocytes (9,10).
Among these proteins, SerpinE2 is an extracellular plasminogen
activator inhibitor and regulates broad key factors of tumor
progression (11–13). In addition, SerpinE2 was found to
be increased in pancreatic tumors (14), breast tumors (15), colorectal tumors (16) and oral squamous carcinomas
(17). SerpinE2 also promotes
tumor metastatic activity (14,18).
Thus, it was hypothesized that SerpinE2 mediates cell growth and
invasion of osteosarcoma. However, the role of SerpinE2 in
osteosarcoma has not previously been reported and its molecular
mechanisms are poorly understood in osteosarcoma progression.
In the present study, the expression of SerpinE2 in
different osteosarcoma tissues was investigated. The cell
proliferation and drug resistance following SerpinE2 transfection
were determined. Moreover, the alteration of the cell cycle
following SerpinE2 over-expression and its mechanism was discussed.
Finally, the correlation between SerpinE2 expression and overall
survival rates were also investigated.
Materials and methods
Patients
Clinical osteosarcoma and tumor-adjacent normal
samples from 80 patients with osteosarcoma were obtained from the
Department of Orthopedics, The Second Xiangya Hospital, Central
South University (Changsha, China). Informed consent was obtained
from all patients, and the study was approved by the ethics
committee of the Second Xiangya Hospital. The patients had not
received any treatment prior to tissue collection. The tissue
histology classification was demonstrated using the World Health
Organization classification (19).
The characteristics of the patients are shown in Table I. After tissue collection, 20
patients with T1/T2 stage osteosarcoma were treated with
cis-platinum (30 mg once daily; Sigma-Aldrich, St. Louis, MO, USA)
and the samples of osteosarcoma and the tissues were collected at
all three stages prior to the second and fourth chemotherapeutic
treatment.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Variable | Number |
|---|
| Age (years) | |
| ≤50 | 43 |
| >50 | 37 |
| Gender | |
| Male | 48 |
| Female | 32 |
| T stage | |
| T1/T2 | 39 |
| T3/T4 | 41 |
| Histologic grade | |
| I | 30 |
| II | 36 |
| III | 14 |
| Metastasis | |
| No (M0) | 36 |
| Yes (M1) | 44 |
| Tumor size (cm) | |
| ≤5 | 29 |
| >5 | 51 |
Cell lines
MG-63 and SAOS-2 osteosarcoma cell lines (The Second
Xiangya Hospital, Central South University) were cultured at 37°C
in an incubator with 5% CO2 in Dulbecco's modified
Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco, Thermo
Fisher Scientific, Inc.) and penicillin/streptomycin (100
μg/ml, Sigma-Aldrich).
Reagents
Antibodies against SerpinE2 (rabbit polyclonal; cat.
no. ab75348), CyclinD1 (rabbit monoclonal; cat. no. ab134175), CDK4
(rabbit monoclonal; cat. no. 108357) and GAPDH (rabbit polyclonal;
cat. no. ab9485) were purchased from Abcam (Cambridge, UK), and
anti-Annexin V-APC was purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). Bortezomib was purchased from Toronto Research
Chemicals Inc. (North York, ON, Canada) and doxorubicin was
obtained from Sigma-Aldrich (St. Louis, MO, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen, Thermo Fisher
Scientific, Inc.) was used to extract the total RNAs (2 µg).
RT was performed using a PrimeScript RT Reagent Kit with a gDNA
Eraser (Takara Bio, Inc., Dailan, China) qPCR was performed on an
ABI 7500 Real Time PCR system (Thermo Fisher Scientific, Inc.)
using SYBR premix Ex Taq II (Takara Bio Inc.). The primers were
designed by Primer Express version 3.0 software (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and the sequences were
as follows: Forward: 5′-TCTCATTGCAAGATCATCGCC-3′ and reverse:
5′-CCCCATGAATAACACAGCACC-3′ for SerpinE2; forward:
5′-CCTAGTACTGCAATTCGGGAAATT-3′ and reverse:
5′-CCTGGAATCCTGCATAAGCAC-3′ for cyclin-dependent kinase (CDK)1;
forward: 5′-CCAGGAGTTACTTCTATGCCTGA-3′ and reverse:
5′-AATCCGCTTGTTAGGGTCGTA-3′ for CDK2; forward:
5′-CACAGTTCGTGAGGTGGCTTTA-3′ and reverse:
5′-TGTCCTTAGGTCCTGGTCTACATG-3′ for CDK4; forward:
5′-TGCACAGTGTCACGAACAGA-3′ and reverse: 5′-ACCTCGGAGAAGCTGAAACA-3′
for CDK6; forward: 5′-CTCTTAACCGCGATCCTCCAG-3′ and reverse:
5′-CAATAAAAGATCCAGGGTACATGATTG-3′ for Cyclin A; forward:
5′-AAAGGCGTAACTCGAATGGA-3′ and reverse: 5′-CCGACCTTTTATTGAAGAGCA-3′
for Cyclin B; forward: 5′-TCGCTGGAGCCCGTGAA-3′ and reverse:
5′-CCGCCTCTGGCATTTTGG-3′ for Cyclin D1; forward:
5′-ATACAGACCCACAGAGACAG-3′ and reverse: 5′-TGCCATCCACAGAAATACTT-3′
for Cyclin E; and forward: 5′-CGCTCTCTGCTCCTCCTGTT-3′ and reverse:
5′-CCATGGTGTCTGAGCGATGT-3′ for glyceraldehyde 3-phosphate
dehydrogenase (GAPDH). The reaction conditions were as follows: 1
cycle at 95°C for 10 sec, 40 cycles at 95°C for 10 sec and 60°C 30
sec. The relative expression levels were normalized to GAPDH
expression using the 2−ΔΔCq method (20).
Cell transfection
The DNA fragment of SerpinE2 was obtained from
Invitrogen, Thermo Fisher Scientific, Inc. and cloned into the
pEF-BOS-EX expression vector, as previously described (21). The vectors were then transfected
into MG-63 and SAOS-2 cells using Lipofectamine 2000 (Invitrogen,
Thermo Fisher Scientific, Inc.). The wild-type controls of MG-63
and SAOS-2 cells were not transfected with vectors. Briefly, 1
µg SerpinE2 over-expressing (OE) vector was added to cells
in the OE groups. There were five replicates for each group. After
24 h transfection according to the manufacturer's instructions, the
cells were used for the following experiments.
Cell proliferation
The cells were counted using a hemocytometer (cat.
no. Z359629; Sigma-Aldrich) under an inverted microscope (Axiovert
200 M; Zeiss, Thornwood, NY, USA). The dead cells were determined
by 0.4% Trypan Blue (Sigma-Aldrich, St. Louis, MO, USA) staining. A
minimum of 100 cells were counted.
Clonogenicity assay
Clonogenic growth was assayed by seeding
1×103 cells in 0.5 ml DMEM containing 10% FBS and 0.33%
agar. The cells were cultured for 1 week, the medium was removed
and the cells were fixed in 75% methanol and 25% acetic acid for 15
min. The cells were then stained using 0.5% crystal violet
(Shanghai Sangon Biological Engineering Technology Co., Ltd.,
Shanghai, China) in methanol for 20 min. Then the colonies were
counted. The clonogenicity was indicated as % colonies relative to
MG-63 WT cells. The clonogenic analysis was performed in
triplicate.
Flow cytometry
To analyze cell differentiation, the cells
(1×105 cells) were collected using TrypLE (Invitrogen,
Thermo Fisher Scientific, Inc.) with 100 U/ml DNase (Invitrogen,
Thermo Fisher Scientific, Inc.). The cells were fixed in cold
methanol, blocked using 4% FBS and washed with phosphate-buffered
saline with Tween 20 (EMD Millipore, Billerica, MA, USA) and then
incubated with Annexin V-APC antibody (cat. no. A35110; Invitrogen,
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Finally, the cells were stained with propidium iodide
(Invitrogen; Thermo Fisher Scientific, Inc.) and were analyzed
using an FC500 machine and CXP version 2.1 software (both purchased
from Beckman Coulter, Pasadena, CA, USA).
Effects of SerpinE2 on drug resistance in
osteosarcoma
The cells were treated with bortezomib (5 nM) and
doxorubicin (50 nM) as pervious reported (21). After 48 h treatment, the cells were
analyzed by a clonogenicity assay.
Western blotting
The proteins were extracted using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Wuhan, China). After determining the protein
concentration using a Bradford assay method (Beyotime Institute of
Biotechnology), 15 µg protein for each sample was
electrophoresed using 15% sodium dodecyl sulfate-polyacrylamide gel
and transferred to a polyvinylidene fluoride membrane (Millipore,
Billerica, MA, USA). The membranes were incubated with primary
antibodies (dilution SerpinE2 1:1,000; Cyclin D1 1:500; CDK4
1:1,000; GAPDH 1:2,000) overnight at 4°C. After three (5 min/time)
washes in phosphate-buffered saline with Tween-20, the secondary
antibodies (dilution 1:200) were incubated with the membrane for 30
min at room temperature. Finally, the protein signals were
visualized with an enhanced chemiluminescent detection system
(EMD).
Statistical analysis
The survival data of osteosarcoma patients were
indicated using a Kaplan-Meier curve and analyzed using log-rank
test by SPSS v 17.0 (SPSS Inc, Chicago, IL, USA) using the data of
the 80 patients. The difference between paired groups was analyzed
by paired t-test and the differences among multiple groups (>2)
was analyzed by one-way analysis of variance followed by Tukey's
test. Gene expression correlations were analyzed using the
Spearman's rank correlation coefficient test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Increased expression of SerpinE2 in
osteosarcoma
Significantly higher expression of SerpinE2 was
found in osteosarcoma tissues compared with the tumor-adjacent
normal tissues from 80 patients (Fig.
1A). In addition, higher SerpinE2 expression in tissues that
had metastasized compared with non-metastatic tissues (Fig. 1B). Notably, SerpinE2 expression in
tumor-node-metastasis stage II–III was markedly higher than
expression in stage I (Fig. 1C).
In addition, the levels of SerpinE2 increased substantially at the
three serial stages at diagnosis, prior to the second and fourth
chemotherapeutic treatment (Fig.
1D).
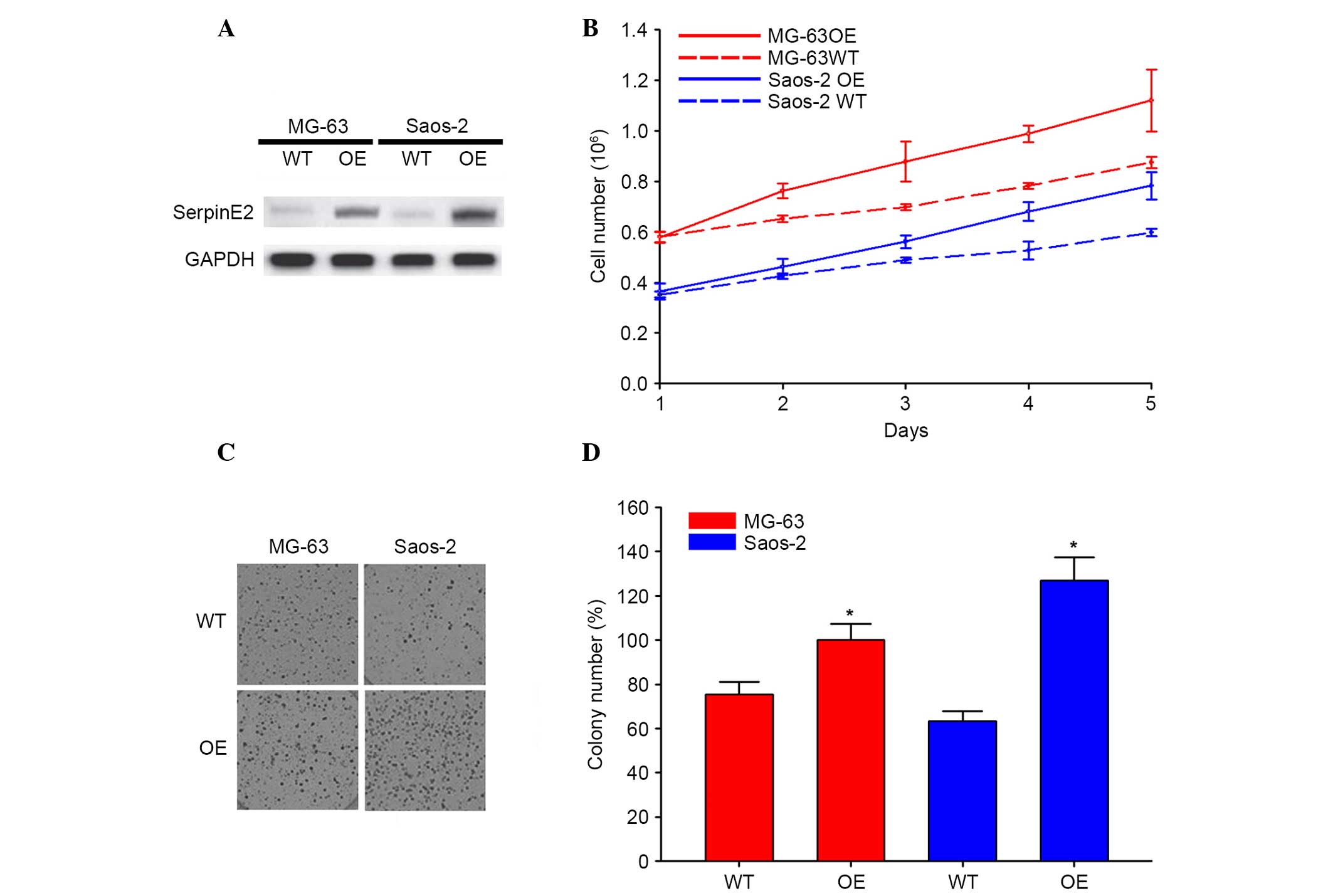
SerpinE2 increases cell proliferation of
osteosarcoma
To demonstrate the effects of SerpinE2 on
osteosarcoma cells, MG-63 and SAOS-2 cell lines were transfected
with SerpinE2 OE vectors. The increased expression of SerpinE2 in
the OE cells compared with untransfected control cells (WT) were
verified by western blotting (Fig.
2A). The growth of WT and OE cells was evaluated for 5 days and
the results showed significantly increased cell growth of OE cells
compared with WT cells (Fig. 2B).
The colony formation of MG-63 and SAOS-2 cells was also
significantly promoted by over-expression of SerpinE2 (Fig. 2C and D).
SerpinE2 promotes drug resistance in
osteosarcoma
Colony formation assays were used to investigate the
possibility that SerpinE2 promotes drug resistance in osteosarcoma.
There were more colonies of SerpinE2 MG-63-OE cells generated
following bortezomib (5 nM) and doxorubicin (50 nM) treatment
compared with those of MG-63-WT cells (Fig. 3A). Annexin V as a marker of
apoptotic cell death was detected by flow cytometry, which revealed
that treatment of cells for 48 h with bortezomib (5 nM) and
doxorubicin (50 nM) contributed to fewer apoptotic MG-63-OE cells
compared with MG-63-WT cells (Fig.
3B).
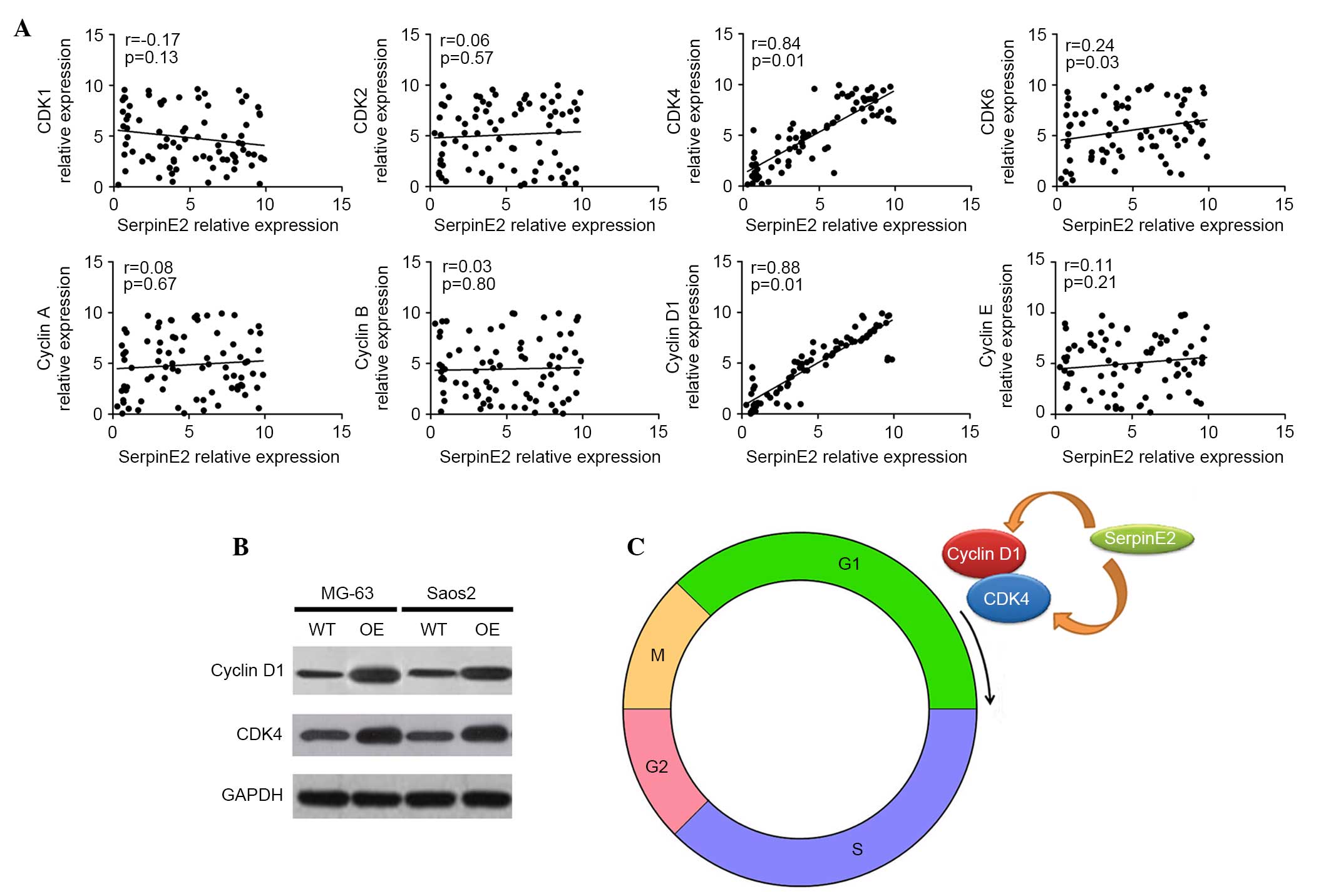
SerpinE2 is positively correlated with
CDK4 and cyclin D1
To detect the mechanism of SerpinE2 promoting cell
proliferation in osteosarcoma, the cell cycle-related genes, CDK1,
CDK2, CDK4, CDK6, cyclin A, cyclin B, cyclin D1 and cyclin E were
analyzed after transfection with SerpinE2 (Fig. 4A). Only CDK4, CDK6 and cyclin D1
were significantly correlated with SerpinE2 expression (P<0.05).
According to the correlation coefficients, CDK4 (r=0.84) and cyclin
D1 (r=0.88) were positively correlated with SerpinE2 expression,
while no correlation was observed between CDK6 and SerpinE2
(r=0.24). In the cell lines, the protein levels also suggested the
correlation between CDK4 and SerpinE2 as well as cyclin D1 and
SerpinE2 expression (Fig. 4B).
Notably, CDK4 and cyclin D1 mediate the development of tumors by
driving cell-cycle progression through G1, which leads to cell
proliferation (Fig. 4C). Thus,
SerpinE2 can promote cell proliferation of osteosarcoma cells via
promoting CDK4 and cyclin D1 expression (Fig. 4C).
Correlation between SerpinE2 expression
and survival rates
The expression of SerpinE2 was determined by
RT-qPCR. To confirm the correlation between SerpinE2 expression and
survival rates, it was demonstrated that patients with osteosarcoma
with high SerpinE2 expression [cut off: 30%, high (>30%) vs. low
(<30%)] had the worst outcome compared with the patients with
low SerpinE2 expression and medium SerpinE2 expression (P<0.05),
indicating that patients with high SerpinE2 expression had poor
overall survival (Fig. 5).
Discussion
Osteosarcoma, as one of the most dangerous malignant
bone tumors, shows high propensity for invasion and proliferation.
Recent studies have reported that SerpinE2 is highly expressed in
different tumor tissues (12–14,16).
The over-expression of SerpinE2 can promote tumor progression. A
previous study suggested that SerpinE2 is upregulated by RAS, BRAF
and MEK1, which results in oncogenesis of intestinal epithelial
cells (16). Thus, SerpinE2 may be
a potential target for cancer treatment. The present study
demonstrated that SerpinE2 increased the proliferation and invasion
of osteosarcoma cells and discussed the mechanism underlying these
effects.
The expression levels of SerpinE2 were upregulated
in high-grade osteosarcoma cells, which exhibited a similar
expression profile to other cancer types. This evidence strongly
suggested that SerpinE2 could be regarded as an oncogene as
previously suggested (22). In
addition, by tracking 20 patients following chemotherapy, SerpinE2
expression profiles during the therapy were analyzed. Prior to the
second and fourth chemotherapeutic treatment, the expression of
SerpinE2 increased significantly compared with the samples at the
beginning of treatment. The higher expression of SerpinE2 was
observed in the present study, and this is in accordance with
previous studies (23–25). These results showed that SerpinE2
may lead to drug resistance and promote cell proliferation.
Next, the effects of SerpinE2 on osteosarcoma cell
lines were assayed. Over-expression of SerpinE2 induced
proliferation and increased clonogenic formation of MG-63 and
SAOS-2 cells confirming the hypothesis that SerpinE2 promotes the
proliferation of osteosarcoma cells. Similar results were found in
pancreatic cancer (14),
testicular cancer (11) and
medulloblastoma (26). In
addition, the association between the drug resistance and the
expression of SerpinE2 was investigated. Following treatment with
bortezomib and doxorubicin, drug resistance was shown. MG-63 cells
were shown to be less sensitive to bortezomib and doxorubicin
following transfection with SerpinE2. To the best of our knowledge,
this is the first study to demonstrate that SerpinE2 contributes to
drug resistance in osteosarcoma. Vaillant et al (27) indicated that Serpine2 is required
for drug resistance.
Currently there is no information regarding the
association between SerpinE2 and the cell cycle. Herein, by
screening 8 cell cycle-related genes from the 80 patients, it was
demonstrated that SerpinE2 expression was positively correlated
with cyclin D1 (R=0.88) and CDK4 (R=0.84), respectively. Notably,
CDK4 and cyclin D1 mediate the development of tumors by driving
cell-cycle progression through G1, which leads to cell
proliferation. Therefore, it was proposed that SerpinE2 promotes
osteosarcoma by inducing the expression of cyclin D1-CDK4 kinase
complexes. These results were further demonstrated by western blot
analysis, in which increased SerpinE2 expression upregulated CDK4
and cyclin D1 in cells. Thus, it was suggested that increased
SerpinE2 expression in osteosarcoma induces cell proliferation by
activating cyclin D1/CDK4 expression resulting in poor survival of
patients with osteosarcoma.
In conclusion, the present study demonstrated that
increased SerpinE2 expression demonstrates drug-resistance,
promotes osteosarcoma cell proliferation and contributes to poor
survival in osteosarcoma patients. Thus, targeting SerpinE2 may be
a potential therapeutic strategy for patients with
osteosarcoma.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Janeway KA and Grier HE: Sequelae of
osteosarcoma medical therapy: A review of rare acute toxicities and
late effects. Lancet Oncol. 11:670–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silverman GA, Bird PI, Carrell RW, Church
FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer
RW, et al: The serpins are an expanding superfamily of structurally
similar but funtionally diverse proteins: Evolution, mechanism of
inhibition, novel functions, and a revised nomenclature. J Biol
Chem. 276:33293–33296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valiente M, Obenauf AC, Jin X, Chen Q,
Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E and Massagué
J: Serpins promote cancer cell survival and vascular co-option in
brain metastasis. Cell. 156:1002–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paik PK, Shen R, Won H, Rekhtman N, Wang
L, Sima CS, Arora A, Seshan V, Ladanyi M, Berger MF and Kris MG:
Next-Generation sequencing of stage IV squamous cell lung cancers
reveals an association of PI3K aberrations and evidence of clonal
heterogeneity in patients with brain metastases. Cancer Discov.
5:610–621. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shiiba M, Nomura H, Shinozuka K, Saito K,
Kouzu Y, Kasamatsu A, Sakamoto Y, Murano A, Ono K, Ogawara K, et
al: Down-regulated expression of SERPIN genes located on chromosome
18q21 in oral squamous cell carcinomas. Oncol Rep. 24:241–249.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Z, Liu Q, Song M, Zheng Y, Nan P, Cao
Y, Chen G, Li Y and Zhong Y: Detecting correlation between sequence
and expression divergences in a comparative analysis of human
serpin genes. Biosystems. 82:226–234. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Erler JT: Cancer: Disabling defences in
the brain. Nature. 508:46–47. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Medress Z and Hayden Gephart M: Molecular
and genetic predictors of breast-to-brain metastasis: Review and
case presentation. Cureus. 7:e2462015.PubMed/NCBI
|
|
11
|
Nagahara A, Nakayama M, Oka D, Tsuchiya M,
Kawashima A, Mukai M, Nakai Y, Takayama H, Nishimura K, Jo Y, et
al: SERPINE2 is a possible candidate promotor for lymph node
metastasis in testicular cancer. Biochem Biophys Res Commun.
391:1641–1646. 2010. View Article : Google Scholar
|
|
12
|
Lee RK, Fan CC, Hwu YM, Lu CH, Lin MH,
Chen YJ and Li SH: SERPINE2, an inhibitor of plasminogen
activators, is highly expressed in the human endometrium during the
secretory phase. Reprod Biol Endocrinol. 9:382011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Demeo DL, Mariani TJ, Lange C, Srisuma S,
Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH,
et al: The SERPINE2 gene is associated with chronic obstructive
pulmonary disease. Am J Hum Genet. 78:253–264. 2006. View Article : Google Scholar :
|
|
14
|
Buchholz M, Biebl A, Neesse A, Wagner M,
Iwamura T, Leder G, Adler G and Gress TM: SERPINE2 (protease nexin
I) promotes extracellular matrix production and local invasion of
pancreatic tumors in vivo. Cancer Res. 63:4945–4951.
2003.PubMed/NCBI
|
|
15
|
Sarrió D, Rodriguez-Pinilla SM, Hardisson
D, Cano A, Moreno-Bueno G and Palacios J: Epithelial-mesenchymal
transition in breast cancer relates to the basal-like phenotype.
Cancer Res. 68:989–997. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bergeron S, Lemieux E, Durand V, Cagnol S,
Carrier JC, Lussier JG, Boucher MJ and Rivard N: The serine
protease inhibitor serpinE2 is a novel target of ERK signaling
involved in human colorectal tumorigenesis. Mol Cancer. 9:2712010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao S, Krogdahl A, Sørensen JA, Kousted
TM, Dabelsteen E and Andreasen PA: Overexpression of protease
nexin-1 mRNA and protein in oral squamous cell carcinomas. Oral
Oncol. 44:309–313. 2008. View Article : Google Scholar
|
|
18
|
Neesse A, Wagner M, Ellenrieder V, Bachem
M, Gress TM and Buchholz M: Pancreatic stellate cells potentiate
proinvasive effects of SERPINE2 expression in pancreatic cancer
xenograft tumors. Pancreatology. 7:380–385. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schajowicz F, Sissons HA and Sobin LH: The
World Health Organization's histologic classification of bone
tumors. Cancer. 75:1208–1214. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Watanabe R, Iida S, Shimizu Y, Nagata S
and Fukunaga R: SEI family of nuclear factors regulates
p53-dependent transcriptional activation. Genes Cells. 10:851–860.
2005. View Article : Google Scholar
|
|
22
|
Kashkar H, Deggerich A, Seeger JM,
Yazdanpanah B, Wiegmann K, Haubert D, Pongratz C and Krönke M:
NF-kappaB-independent down-regulation of XIAP by bortezomib
sensitizes HL B cells against cytotoxic drugs. Blood.
109:3982–3988. 2007. View Article : Google Scholar
|
|
23
|
Wang K, Wang B, Xing AY, Xu KS, Li GX and
Yu ZH: Prognostic significance of SERPINE2 in gastric cancer and
its biological function in SGC7901 cells. J Cancer Res Clin Oncol.
141:805–812. 2015. View Article : Google Scholar
|
|
24
|
Hannemann J, Oosterkamp HM, Bosch CA,
Velds A, Wessels LF, Loo C, Rutgers EJ, Rodenhuis S and van de
Vijver MJ: Changes in gene expression associated with response to
neoadjuvant chemotherapy in breast cancer. J Clin Oncol.
23:3331–3342. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van't Veer LJ, Dai H, Van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar
|
|
26
|
Holleman A, Cheok MH, den Boer ML, Yang W,
Veerman AJ, Kazemier KM, Pei D, Cheng C, Pui CH, Relling MV, et al:
Gene-expression patterns in drug-resistant acute lymphoblastic
leukemia cells and response to treatment. N Engl J Med.
351:533–542. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vaillant C, Valdivieso P, Nuciforo S, Kool
M, Schwarzentruber-Schauerte A, Méreau H, Cabuy E, Lobrinus JA,
Pfister S, Zuniga A, et al: Serpine2/PN-1 is required for
proliferative expansion of pre-neoplastic lesions and malignant
progression to medulloblastoma. PLoS One. 10:e01248702015.
View Article : Google Scholar : PubMed/NCBI
|