Introduction
The incidence and mortality rates of primary liver
cancer are the fifth and second highest of all types of cancer in
the world, respectively, and hepatocellular carcinoma has a poor
prognosis with a 5-year survival rate of <5% (1). Current therapeutic methods are unable
to effectively improve the prognosis of patients with
hepatocellular carcinoma. Therefore, novel therapeutic strategies
are urgently required. The poly ADP-ribose polymerase (PARP) family
is involved in the regulation of several cellular functions,
including DNA damage and repair, RNA transcription, cell signaling,
cell cycle regulation and mitosis. It has been confirmed that PARP,
being involved in DNA damage and repair, is closely associated with
tumor therapies (2).
The PARP family includes PARP-1, PARP-2, PARP-3,
Vault-PARP tankyrase (TANK-1, TANK-2 and TANK-3) and other
subtypes. Several studies (3,4) have
focused on the role of PARP-1 in cancer, diabetes and inflammation.
PARP-1 is expressed at high levels in tumor cells with BRCA1/2
mutations, and PARP-1 inhibitors can be used as agents for the
treatment of tumor cells with breast cancer (BRCA)1/2 deficiency,
including breast cancer and ovarian cancer cells. PARP-1 inhibitors
have been used in clinical phase I and II trials for treating
BRCA-1/2 (−) breast cancer and ovarian cancer (3,4). In
addition, basic investigations have found that the PARP-1
inhibitor, olaparib, can increase the inhibition of radiation in
Ewing's sarcoma cell proliferation and induce the apoptosis of
sarcoma cells (5). Other studies
have shown that PARP-1 inhibitors can affect the proliferation,
apoptosis and invasion of tumor cells (6), and enhance the effects of
chemotherapy on tumor cells (3),
particularly in breast and ovarian cancer with BRCA1/2 mutations
(7). However the effect of PARP-1
inhibitors on liver cancer cells remains to be fully elucidated
(8).
It has been confirmed that phosphatase and tensin
homolog (PTEN) is a tumor suppressor gene involved in the
phosphoinositide 3-kinase/AKT/mammalian target of rapamycin
signaling pathways to maintain normal physiological activity of
cells, negatively regulates tumor cell cycle, induces tumor cell
apoptosis, and inhibits tumor cell invasion and metastasis
(9). Additionally, matrix
metalloproteinases (MMPs) are important in tumor invasion and
migration. The abnormal metabolism of extracellular matrix can lead
to tumor metastasis when the homeostasis between MMP and tissue
inhibitor of metalloproteinase (TIMP) is interrupted. MMP 3 is an
important member of the MMP family, and is capable of degrading
extracellular matrix involved in tissue morphogenesis,
physiological and pathological processes, and tumorigenesis
(10). In the present study, three
types of PARP-1 inhibitor were used, AG014699, BSI-201a and
AZD-2281, which have been used in previous clinical trials for the
treatment of breast cancer (11).
These were used to observe the effect of PARP-1 inhibitors on the
expression of PTEN, MMP and TIMP, examine their effects on the
proliferation, apoptosis and migration of human hepatoma HepG2
cells in vitro, and examine the possible underlying
mechanisms.
Materials and methods
Materials
The HepG2 cells were provided by the Institute of
Modern Physics, Chinese Academy of Science (Lanzhou, China). DMEM
was purchased from Hyclone; GE Healthcare Life Sciences (Logan, UT,
USA); fetal bovine serum (FBS) was purchased from Sijiqing Company
(Hangzhou, China). MTT was purchased from Amresco, LLC (Solon, OH,
USA), BSI-201 was purchased from Sigma; Merck KGaA (Darmstadt,
Germany). AZD2281 and AG014699 were purchased from Selleck
Chemicals (Shanghai, China). The primary antibodies (GAPDH, Caspase
3, Caspase 8, Bcl-2, Bax, PTEN, TIMP 3 and MMP3) were purchased
from Affinity Biologicals Inc. (Ancaster, ON, Canada). The
secondary antibody was purchased from Beyotime Institute of
Biotechnology (Haimen, China).
Cell culture
The HepG2 cells were cultured in DMEM medium
containing 10% FBS, and 100 U/ml antibiotics
(penicillin:streptomycin, 1:1) was added. The cells were incubated
at 37°C in an incubator with 5% CO2 and 95–98% relative
humidity. Cells at the logarithmic phase were treated with 0.25%
trypsin and passaged.
MTT assay
The HepG2 cells were cultured at a density of
5×103 /ml in 96-well plates, and divided into the
following control and drug groups: Blank group (complete medium);
control group (complete medium containing <1% DMSO); AG014699
group (5, 10, 20, 40 and 80 µmol/l); AZD2281 group (50, 100, 200,
400 and 800 µmol/l); BSI-201 group (10, 20, 40, 60 and 80 µmol/l).
Each group included six wells. Subsequently, 20 µl MTT was added to
each well following 24, 48, 72 and 96 h of culture. After 4 h, the
culture medium was aspirated and 150 µl DMSO was added to each
well. The optical density (OD) was measured on an absorbance
microplate reader with a wavelength of 570 nm. The inhibition rate
was calculated according to the following formula: Inhibition
ratio=1-(ODdrug group-ODcontrol
group)/(ODcontrol group-ODblank group)
×100%.
Flow cytometric analysis
The HepG2 cells at the logarithmic phase were seeded
into 6-well plates at a density of 2.5×104 /ml. The
PARP-1 inhibitors AG014699 and BSI-201, which were sensitive to the
HepG2 cells, were selected for the apoptotic assay. After 48 h, the
cells were cultured with AG014699 at concentrations of 10, 30 and
50 µmol/l, or BSI-201 at concentrations of 20, 40 and 60 µmol/l for
48 h at 37°C, following which the cells were digested with 0.25%
trypsin and washed twice with PBS. To 100 µl of the solution
(1×104 cells), 5 µl of annexin V-fluorescein
isothiocyanate (BD GmbH, Heidelberg, Germany) and 10 µl propidium
iodide (20 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
were added. Analysis was performed using a FACScan and CELLQuest
software version 2.1.4.7 (BD Biosciences, Franklin Lakes, NJ,
USA).
Western blot analysis
The HepG2 cells at the logarithmic phase were seeded
into 6-well plates at a density of 5×104 /ml. After 48
h, AG014699 (10, 30 and 50 µmol/l) or BSI-201 (20, 40 and 60
µmol/l) were added then cultured for 48 h at 37°C. Cells were
digested with 0.25% trypsin containing 0.02% EDTA. After 1 min, the
cells were gently agitated with 1.5 ml of PBS buffer. After
centrifugation at 55 × g at 4°C for 5 min, the supernatant was
removed and washed three times with PBS. Then 200 µl of PMSF and
RIPA lysate (1:100) added for 30 min. The lysate was slowly
agitated with a 2 ml syringe and allowed to lyse sufficiently. The
centrifuge was preheated to 4°C and the lysate centrifuged at 8,051
× g for 15 min. The supernatant was dispensed into a 100 µl
centrifuge tube and the protein concentration was measured using a
Pierce BCA Protein Quantitative Assay kit (Pierce; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Sample proteins were separated
by electrophoresis on a 12% SDS-PAGE gel and transferred onto
polyvinylidene difluoride. Membranes were blocked with 5% non-fat
milk powder in TBST buffer (Tris-buffered saline, 0.05% Tween 20)
and incubated at 4°C for 2 h with primary antibodies against
Caspase 3 (AF835; 1:1,000), Caspase 8 (AF705; 1:1,000), Bax (AF820;
1:1,000), Bcl-2 (AF810; 1:1,000), PTEN (AF847; 1:1,000), TIMP3
(AF0265; 1:1,000) and MMP3 (AF548; 1:1,000; all from Affinity
Biologicals Inc.). The membranes were washed three times in TBST
and then incubated for 2 h at room temperature with the secondary
antibody (horseradish peroxidase-labeled goat anti-rabbit; s0001;
1:1,000; Beyotime Institute of Biotechnology, Haimen, China) and
then washed three times in TBST. The gray value of each band was
analyzed using ImageJ software, version 2.1.4.7 (National
Institutes of Health, Bethesda, MD, USA).
Cell migration analysis
The HepG2 cells at the logarithmic growth phase were
starved for 12 h, and then digested using trypsin. The cells, in
DMEM containing 5% FBS, were seeded into a Transwell chamber at a
density of 5×105 cells/ml. The chamber was placed into
24-well plates, and placed in an incubator at 37°C, 5%
CO2 overnight. Following incubation, the medium in the
chamber was removed, and AG014699 or BSI-201 were added with final
concentrations of 5 and 10 µmol/l, respectively. The control group
contained 300 µl serum-free medium with <1% DMSO. The lower
chamber contained 600 µl DMEM containing 25% FBS. The cells were
placed into the cell incubator and cultured for 48 h at 37°C. The
medium was then removed and the cells were washed twice in PBS
buffer, followed by fixing in 4% formaldehyde for 3–5 min at room
temperature and treatment with methanol for 20 min. Subsequently,
the cells were washed twice with PBS, and the dry Transwell chamber
was placed into the lower chamber for staining with crystal violet
for 15 min. The cells were washed twice in PBS and visualized using
an inverted microscope to examine cell migration.
Statistical analysis
The results are expressed as the mean ± standard
deviation. SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL,
USA) was used to analyze data. An independent t-test was used to
determine the differences between two groups. One-way analysis of
variance was used to analyze the differences between multiple
groups. P<0.05 was considered to indicate a statistically
significant difference.
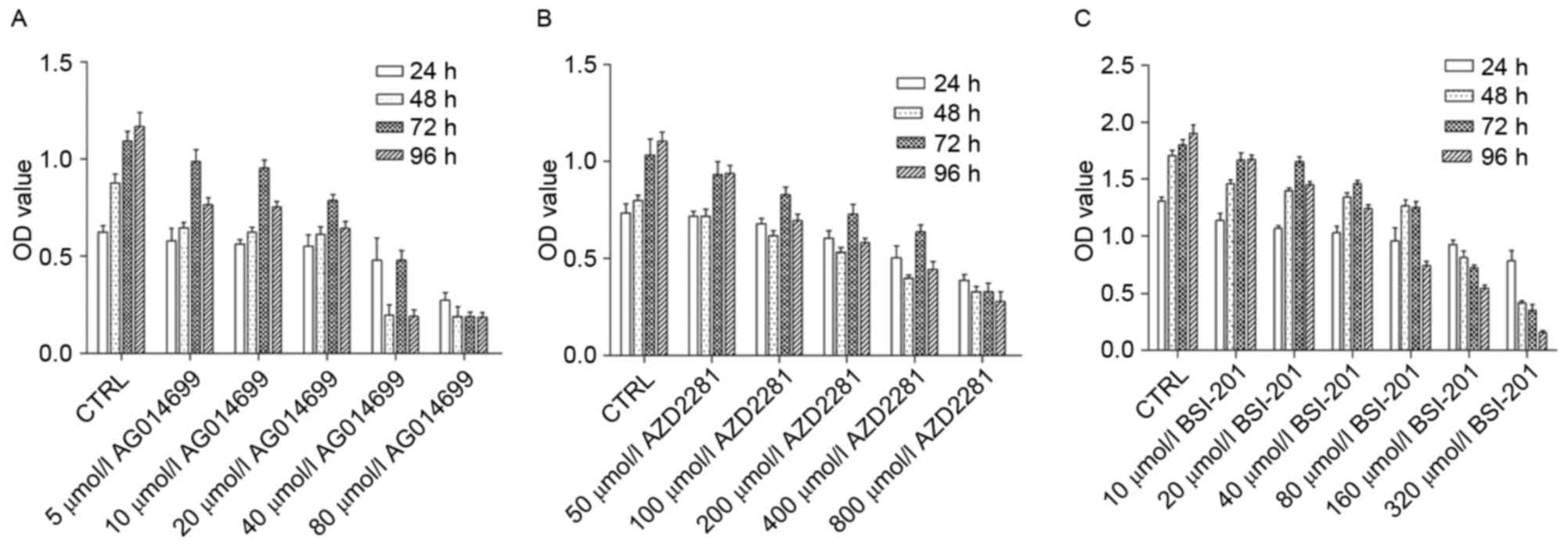
Results
Different concentrations of AG014699,
BSI-201 and AZD2281 exert different inhibitory effects on the
proliferation of HepG2 cells
The results showed that different concentrations of
the PARP-1 inhibitors inhibited the proliferation of HepG2 cells.
As concentrations increased, the inhibitory effect was enhanced
(Fig. 1). After 24 h, the half
maximal inhibitory concentrations of AG014699, BSI-201 and AZD2281
were 80, 160 and 800 µmol/l, respectively. After 48 h, the half
maximal inhibitory concentrations of AG014699, BSI-201, AZD2281
were 20, 30 and 400 µmol/l, and at 96 h, the half maximal
inhibitory concentrations were 10, 25 and 300 µmol/l at 72 h, and
5, 20 and 200 µmol/l.
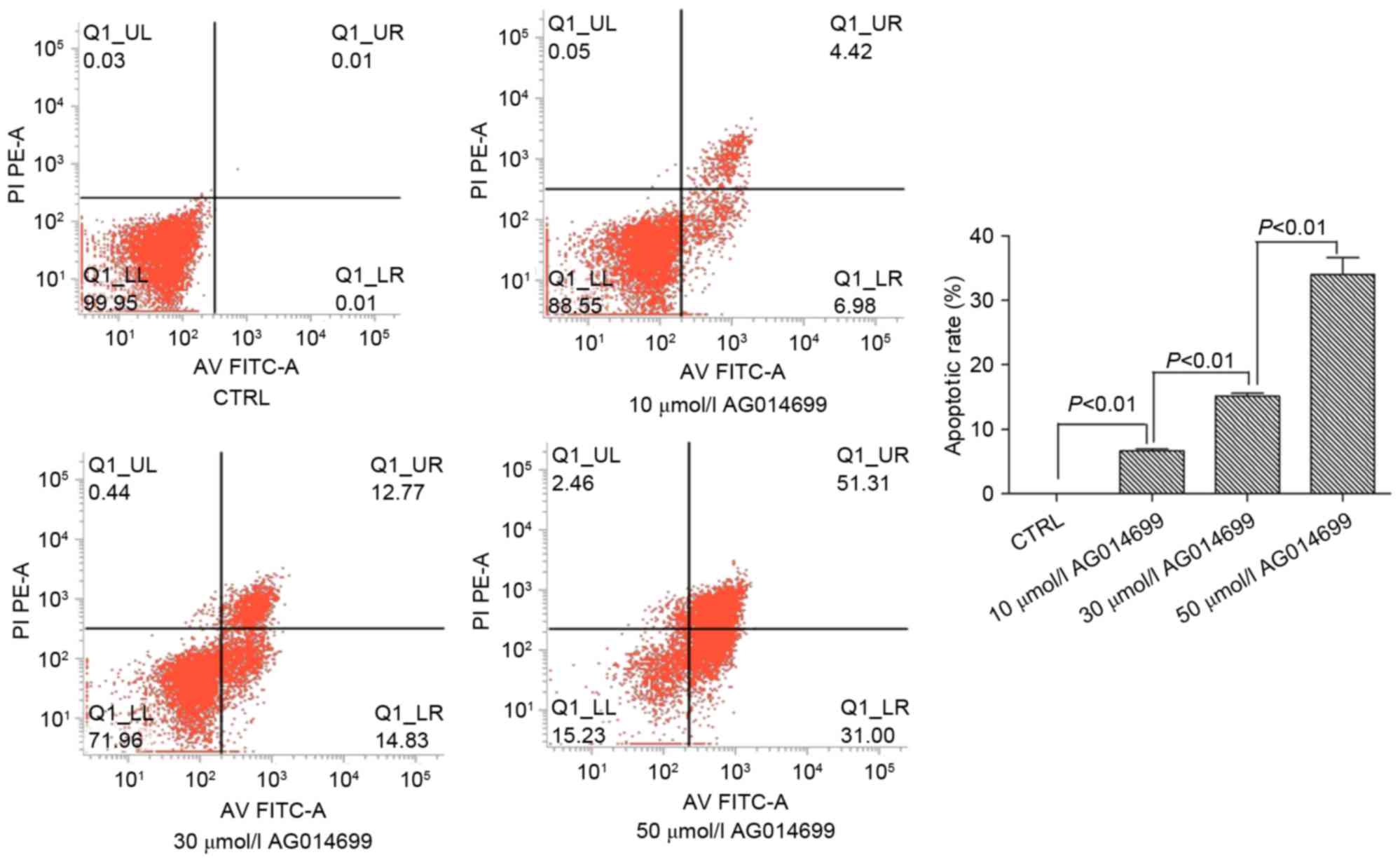
AG014699 and BSI-201 induce the
apoptosis of HepG2 cells
According to the results of the MTT assay, the two
PARP-1 inhibitors, AG014699 and BSI-201, were selected as they
exhibited sensitive inhibitory effects on the HepG2 cells. The
results showed that the apoptotic rates of the HepG2 cells
increased with increasing concentrations of these two inhibitors.
There were significant differences between the groups (P<0.01;
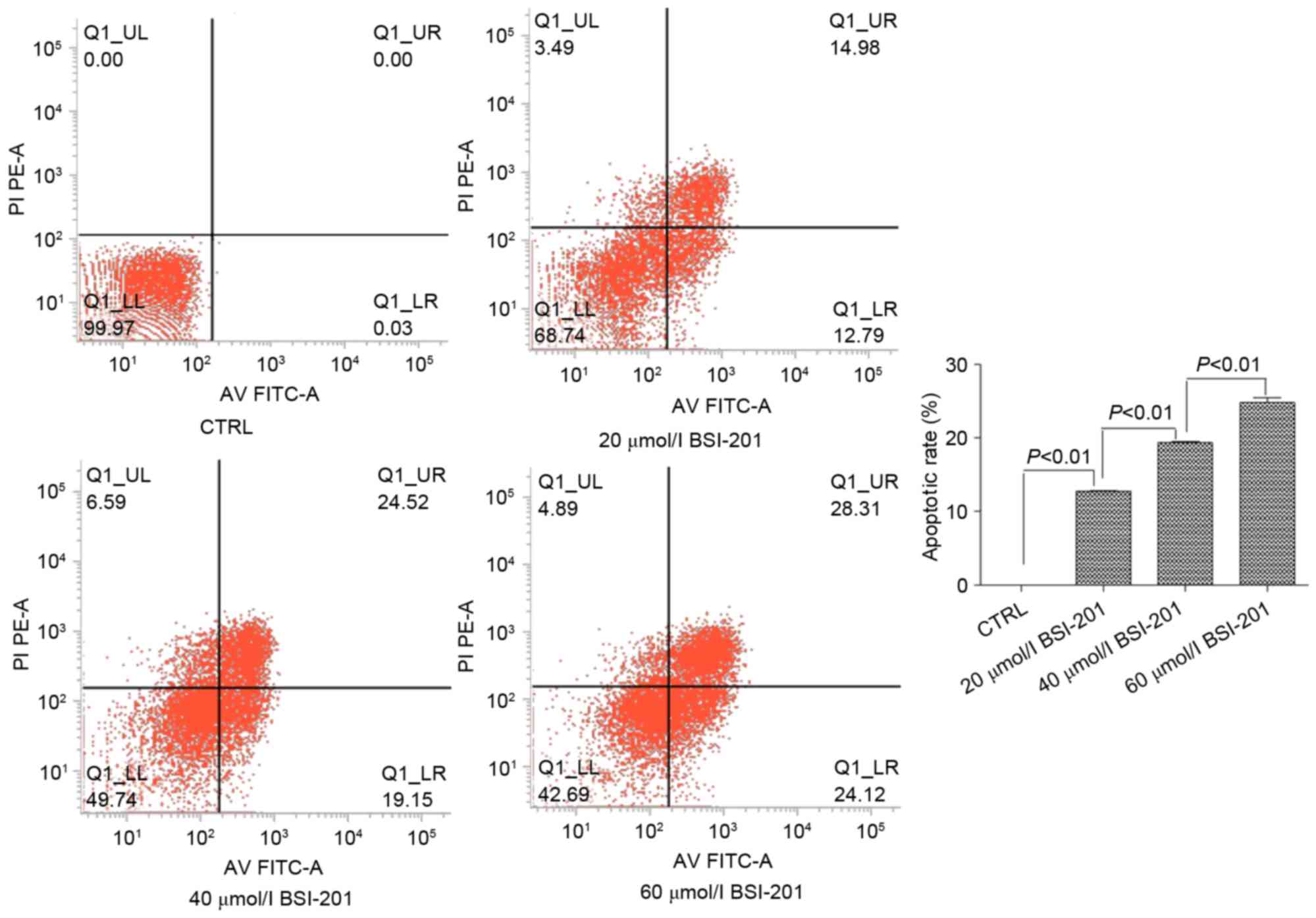
Fig. 2). Similarly, in the HepG2
cells treated with 20, 40 or 60 µmol/l BSI-201 for 48 h, the
apoptotic rates of the HepG2 cells also increased, and significant
differences were found between the groups (P<0.01; Fig. 3).
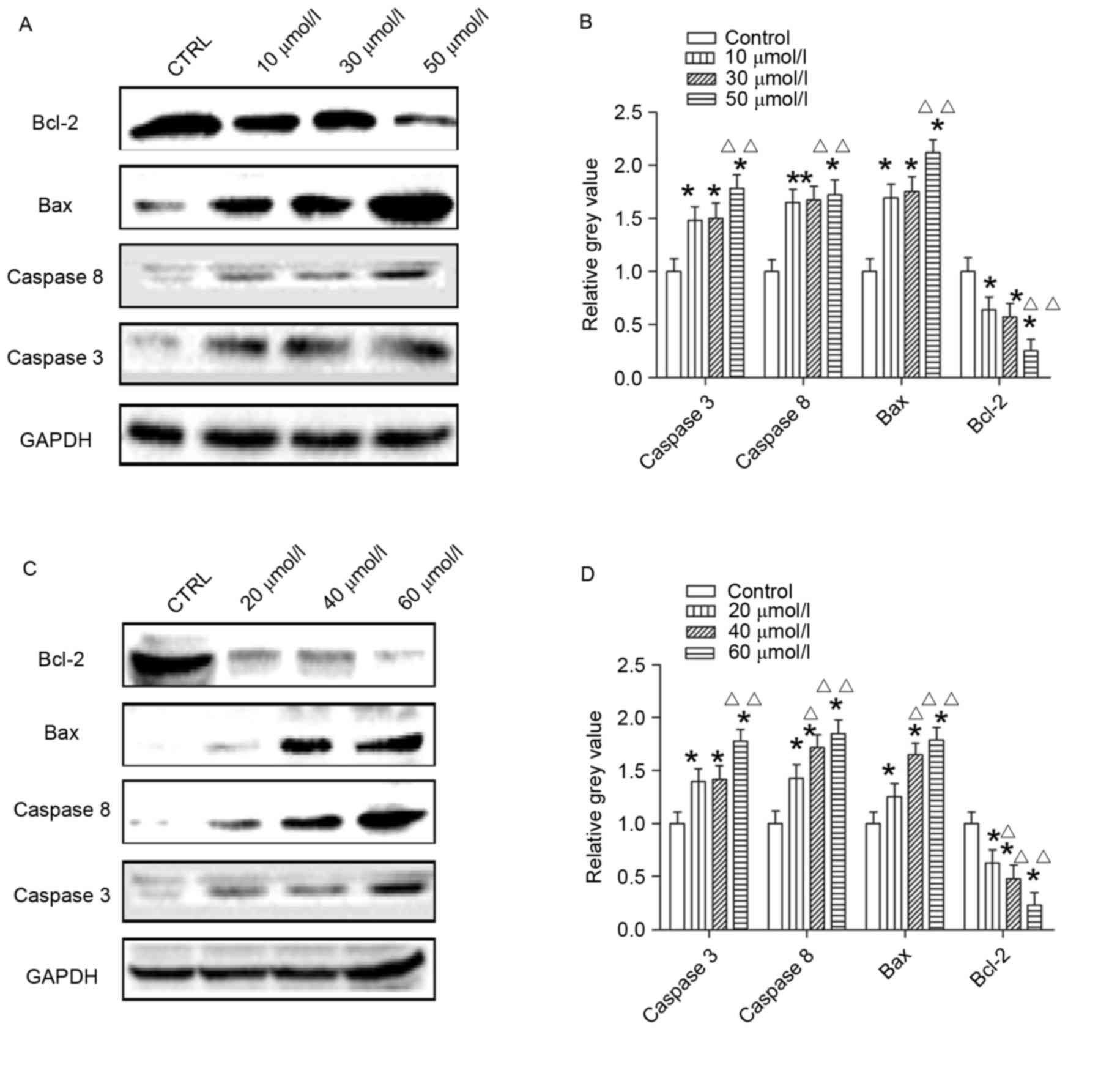
Expression levels of
apoptosis-associated proteins are induced by AG014699 and
BSI-201
Following treatment of cells for 48 h with AG014699
at concentrations of 10, 30 and 50 µmol/l, the protein levels of
Caspase 3, Caspase 8 and Bax were higher, compared with those in
the control group (Fig. 4A and B),
which was also the case in the cells treated with BSI-201
concentrations of 20, 40 and 60 µmol/l (Fig. 4C and D). The protein levels of
Bcl-2 in the two treatment groups were reduced, compared with those
in the control groups (Fig.
4).
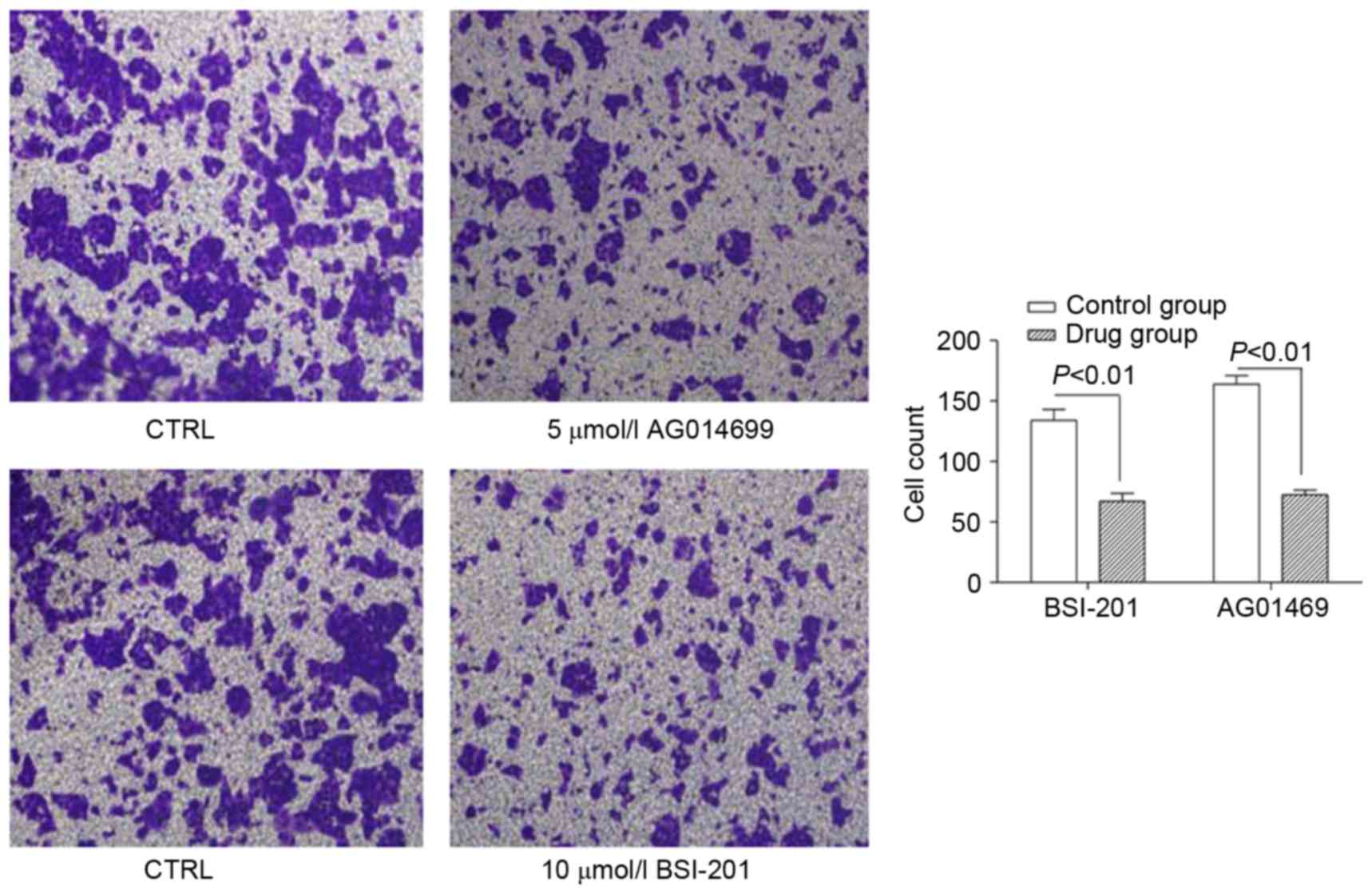
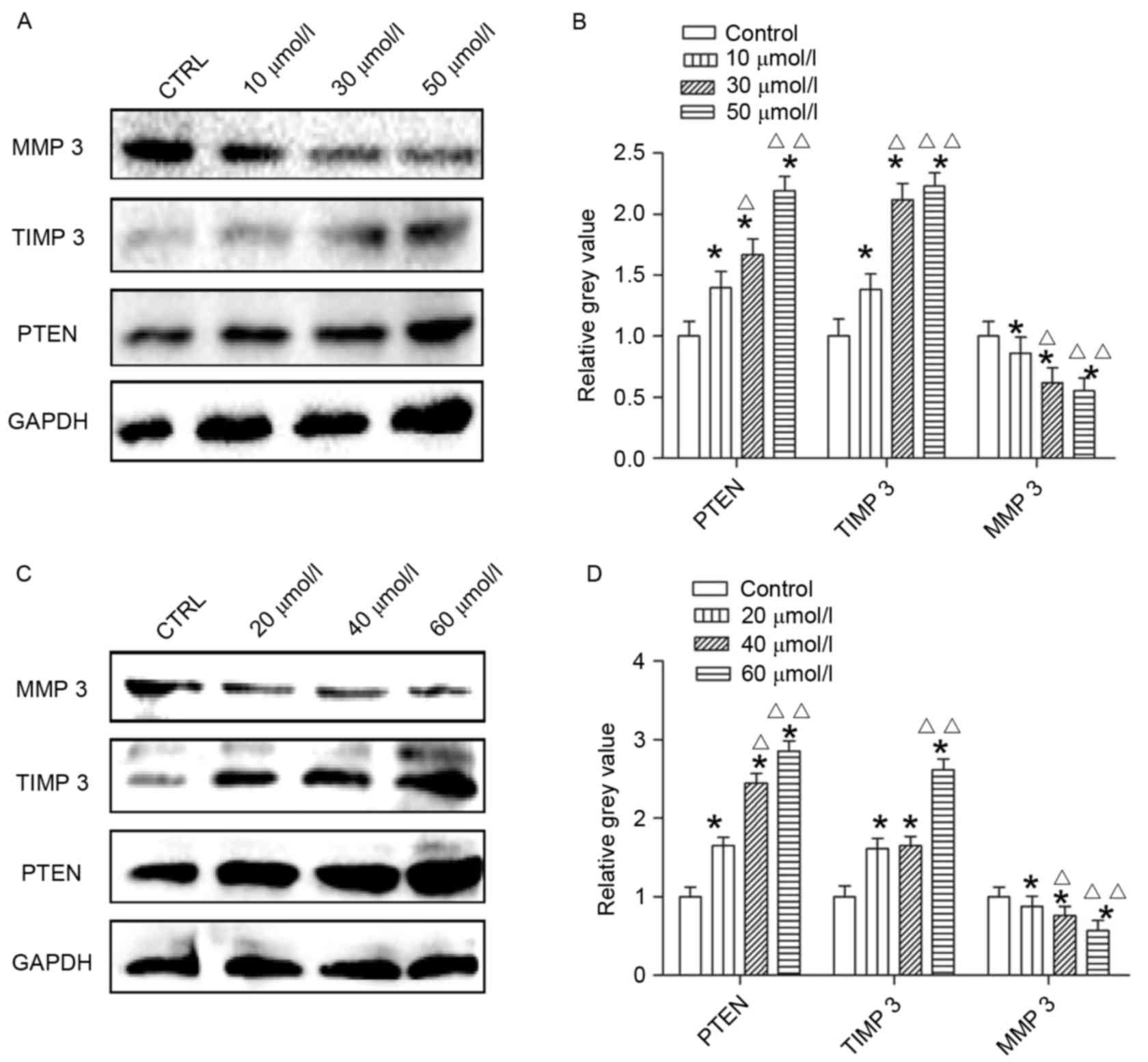
Effects of AG014699 and BSI-201 on
HepG2 cell migration
Following treatment of the HepG2 cells for 48 h with
10 µmol/l of BSI-201 or 5 µmol/l AG01469, the results showed that
the numbers of cells, which migrated into the lower chamber were
lower, compared with those in the control groups, and this
difference was significant (P<0.01; Fig. 5). When the HepG2 cells were treated
with AG014699 at concentrations of 10, 30 and 50 µmol/l (Fig. 6A and B) or with BSI-201 at
concentrations of 20, 40 and 60 µmol/l (Fig. 6C and D) for 48 h, the protein
levels of PTEN and TIMP3 were higher, compared with those in the
control group, whereas the levels of MMP3 were lower, compared with
those in the control group.
Discussion
The present study is the first, to the best of our
knowledge, to use the AG014699, BSI-201 AZD-2281 PARP-1 inhibitors
in vitro to treat hepatoma cell lines, and to show that the
three PARP-1 inhibitors were able to inhibit the proliferation of
HepG2 cells. In addition, AG014699 and BSI-201 showed superior
sensitivity, and were able to induce apoptosis and inhibit the
migration of hepatoma cells, the mechanisms of which may be
associated with altered apoptosis and migration signaling
pathways.
Primary liver cancer is a complex pathological
process and its detailed mechanism remains to be fully elucidated.
DNA damage caused by a variety of factors are important in the
process (12). There have been
increasing reports on DNA damage repair and tumor
occurrence/development, particularly those investigating the
association between PARP and the development of tumors (13). PARP-1 may also be involved in the
biological function of tumor cells, including tumor cell
proliferation, apoptosis, migration and invasion. Previously, Yang
et al (14) found that the
PARP-1 inhibitor, olaparib, inhibits the cloning of JF-305
pancreatic cancer cells, and inhibits the cell cycle of cells in
the S phase and G2/M phase of cell formation in vivo. In
liver cancer, Munoz-Gamez et al (15) found that the PARP-1 inhibitor,
ABT-888, combined with acetazolamide inhibited the proliferation of
liver cancer cells and induced cell apoptosis. However, there have
been no reports of a sensitive PARP-1 inhibitor of liver cancer
cells.
The present study demonstrated that three types of
PARP-1 inhibitor, AG014699, BSI-201 and AZD2281, showed inhibitory
effects on the proliferation of human hepatoma cells, however,
their sensitivities differed. The most sensitive was AG014699,
followed by BSI-201 and AZD2281. Chuang et al (16) found that the sensitivities to
PARP-1 inhibitors in breast cancer were
AG014699>AZD2281>BSI-201, whereas the present study
demonstrated that the sensitivities to the PARP-1 inhibitors on
HepG2 cells were AG014699>BSI-201>AZD2281. Therefore,
different tumor cells may have different sensitivities to different
inhibitors. The present study also detected the apoptosis of HepG2
cells treated with AG014699 and BSI-201, which cells were shown to
be more sensitive to, and found that AG014699 and BSI-201 induced
apoptosis of the HepG2 cells. The highest rates of apoptosis were
31 and 24.82%, respectively. In addition, the protein expression
levels of Caspase 3, Caspase 8 and Bax increased, whereas that of
Bcl-2 decreased following treatment with the two types of PARP-1
inhibitor. Cell apoptosis includes the mitochondrial pathway,
endoplasmic reticulum and death receptor pathway (17). The Caspase enzyme system is core to
apoptosis, and a variety of apoptotic pathways and apoptotic
factors can ultimately activate Caspase enzymes to cause apoptosis
(17). The results of the present
study showed that Caspase 3, Caspase 8, Bax and Bcl-2 were key
molecules in the mitochondrial apoptotic pathway, indicating that
PARP-1 inhibitors induced the apoptosis of HepG2 cells through the
mitochondrial pathway.
Preventing metastasis in liver cancer is a challenge
requiring urgent solutions in the treatment of liver cancer. The
present study found that fewer HepG2 cells migrated to the lower
Transwell chamber in the inhibitor-treated group, compared with
those in the control group. This suggested that AG014699 and
BSI-201 inhibited the migration of HepG2 cells. Forster et
al (18) found that patients
with endometrial cancer, which was sensitive to cisplatin, had
prolonged survival rates following treatment with iniparib, and
metastases of brain tissue reduced. Biopsy showed that patients
were deficient in the PTEN gene, therefore, it was suggested that
iniparib may be a novel method for the treatment of tumors with
PTEN gene deletion. The present study found that AG014699 and
BSI-201 upregulated the expression of PTEN in HepG2 cells, and
suggested that AG014699 and BSI-201 may increase the expression of
PTEN in HepG2 cells, thereby reducing the migration of the
cells.
In the present study, it was found that the AG014699
and BSI-201 inhibitors of PARP-1 regulated the protein expression
of TIMP3 in HepG2 cells and downregulated the expression of MMP3.
These results suggested that PARP-1 inhibitors upregulated the
TIMP-3/MMP-3 ratio to reduce migration of the HepG2 cells.
In conclusion, the present study showed that the
three PARP-1 inhibitors inhibited the proliferation of human
hepatoma cells in vitro, however, the sensitivity of the
three PARP-1 inhibitors were different. AG014699 and BSI-201 may
induce the apoptosis of HepG2 cells through the mitochondrial
pathway, and reduce the migration of HepG2 cells by upregulating
the protein expression of PTEN and increasing the TIMP-3/MMP-3
ratio. However, further investigations are required to elucidate
the detailed mechanism for the treatment of liver cancer.
Acknowledgements
This study was supported by the Technical Research
and Development Project of Gansu Province (grant no.
1305TCYA023).
References
|
1
|
Lafaro KJ, Demirjian AN and Pawlik TM:
Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am.
24:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan Y and Zong WX: PARP Activation and
Necrotic Cell Death. Necrotic Cell Death. Shen HM and Vandenabeele
P: New York, NY: Springer; pp. 163–175. 2014, View Article : Google Scholar
|
|
3
|
Speers C, Feng FY and Pierce LJ: PARP-1
inhibitors and radiotherapy sensitivity: Future prospects for
therapy? Breast Cancer Management. 3:281–296. 2014. View Article : Google Scholar
|
|
4
|
Somlo G, Frankel P, Luu T, Ma C, Arun B,
Garcia A, Cigler T, Harvey HA, Sparano JA, et al: Phase II trial of
single agent PARP inhibitor ABT-888 (veliparib [vel]) followed by
postprogression therapy of vel with carboplatin (carb) in patients
(pts) with stage BRCA-associated metastatic breast caner (MBC):
California Cancer Consortium trial PHII-96. J Clin Oncol.
32:(suppl). S10212014.
|
|
5
|
Stewart E, Goshorn R, Bradley C, Griffiths
LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB III,
Bahrami A, et al: Targeting the DNA repair pathway in ewing
sarcoma. Cell Rep. 9:829–841. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cipak L and Jantova S: PARP-1 inhibitors:
A novel genetically specific agents for cancer therapy. Neoplasma.
57:401–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ibrahim YH, Garcia-Garcia C, Serra V, He
L, Torres-Lockhart K, Prat A, Anton P, Cozar P, Guzmán M, Grueso J,
et al: PI3K inhibition impairs BRCA1/2 expression and sensitizes
BRCA-proficient triple-negative breast cancer to PARP inhibition.
Cancer Discov. 2:1036–1047. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou X, Huang ZY, Chen XP and Huang SH:
Suppressive effect of poly (ADP-ribose)polymerase-1 inhibitor PJ34
on human hepatoma cell line HepG2. World Chinese Journal of
Digestology. 15:1806–1809. 2007.(In Chinese).
|
|
9
|
Milella M, Falcone I, Conciatori F, Incani
Cesta U, Del Curatolo A, Inzerilli N, Nuzzo CM, Vaccaro V, Vari S,
Cognetti F and Ciuffreda L: PTEN: Multiple functions in human
malignant tumors. Front Oncol. 5:242015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Banik D, Netherby CS, Bogner PN and Abrams
SI: MMP3-mediated tumor progression is controlled transcriptionally
by a novel IRF8-MMP3 interaction. Oncotarget. 6:15164–15179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Domagala P, Huzarski T, Lubinski J, Gugala
K and Domagala W: PARP-1 expression in breast cancer including
BRCA1-associated, triple negative and basal-like tumors: Possible
implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat.
127:861–869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Lope CR, Tremosini S, Forner A, Reig M
and Bruix J: Management of HCC. J Hepatol. 56:(Suppl 1). S75–S87.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Golia B, Singh HR and Timinszky G:
Poly-ADP-ribosylation signaling during DNA damage repair. Front
Biosci (Landmark Ed). 20:440–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang X, Ndawula C Jr, Zhou H, Gong X and
Jin J: JF-305, a pancreatic cancer cell line is highly sensitive to
the PARP inhibitor olaparib. Oncol Lett. 9:757–761. 2015.PubMed/NCBI
|
|
15
|
Muñoz-Gámez JA, Viota López J, Barrientos
A, Carazo Á, Sanjuán-Nuñez L, Quiles-Perez R, Muñoz-de-Rueda P,
Delgado Á, Ruiz-Extremera Á and Salmerón J: Synergistic
cytotoxicity of the poly (ADP-ribose) polymerase inhibitor ABT-888
and temozolomide in dual-drug targeted magnetic nanoparticles.
Liver Int. 35:1430–1441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chuang HC, Kapuriya N, Kulp SK, Chen CS
and Shapiro CL: Differential anti-proliferative activities of
poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative
breast cancer cells. Breast Cancer Res Treat. 134:649–659. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fiandalo MV and Kyprianou N: Caspase
control: Protagonists of cancer cell apoptosis. Exp Oncol.
34:165–175. 2012.PubMed/NCBI
|
|
18
|
Forster MD, Dedes KJ, Sandhu S, Frentzas
S, Kristeleit R, Ashworth A, Poole CJ, Weigelt B, Kaye SB and
Molife LR: Treatment with olaparib in a patient with PTEN-deficient
endometrioid endometrial cancer. Nat Rev Clin Oncol. 8:302–306.
2011. View Article : Google Scholar : PubMed/NCBI
|