Introduction
Pain is one of the most common health issues and
produces substantial personal, community and global burdens
(1–7). The majority of individuals have
experienced or are experiencing pain. However, there remains no
effective therapeutic agents for preventing pain. The majority of
drugs used for treating pain have been known to have obvious side
effects (8,9). Therefore, it is necessary to examine
the underlying mechanisms of pain and investigate novel therapeutic
strategies for the treatment of pain.
Recent studies have demonstrated that inactivated
ion channels and altered channel function results in
hypersensitivity to painful stimuli (10–14).
Peripheral neuropathic pain is a disorder caused by nerve injury,
and is characterized by the dysregulation of voltage-gated ion
channels, which are expressed in dorsal root ganglion (DRG) sensory
neurons (15).
Previous studies have revealed that microRNAs
(miRNAs/miRs) are involved in regulation of ion channels and affect
sensitivity to painful stimuli (16,17).
miRNAs may be important small molecules associated with pain
pathogenesis. miRNAs are a type of small, non-coding RNA, which act
as post-transcriptional regulators of gene expression (18–21).
Recent reports have identified that neural miRNAs serve critical
roles at different stages of neuronal development, including
neuritis outgrowth (22),
dendritogenesis (23) and spine
formation (24). Kusuda et
al reported that miRNAs are involved in regulating the microRNA
associated with the pathogenesis of chronic pain (25). Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analyses have demonstrated that
miRNA (miR)-1, -16 and -206 are expressed differentially in DRG
neurons and the dorsal horn of the spinal cord in different painful
situations. Furthermore, complete Freuds adjuvant-induced
inflammation significantly reduced miR-1, -16 and -206 levels in a
time-dependent manner. By contrast, miRNAs were upregulated in the
spinal dorsal horn (25). The
association between miRNA expression levels and numerous
pathophysiological processes implicates their role in neuropathic
pain.
With the development of genome libraries of miRNA
mimics, it is possible to identify target miRNA using a high
throughput screening method (26,27).
Therefore, the present study used this technique to investigate
miRNAs associated with neuropathic pain, and investigated
underlying molecular mechanisms.
Materials and methods
Animal model
All experimental procedures were approved by the
Committee on Animal Experimentation of the China-Japan Union
Hospital of Jilin University (Changchun, China). Adult male Kunming
mice (weight, 18–22 g; n=8) were purchased from the Animal Center
of Jilin University. Animals were housed in a 12 h light/dark cycle
and had free access to food and water at 25°C and 50+/−10% relative
humidity. Spared-nerve injury (SNI) was established according to a
previous report, with slight modifications (28). SNI was established by the
unilateral ligation and cutting of the peroneal and tibial nerve
branches, while the sural nerve was intact on the left (SNI-L, n=2)
or right (SNI-R, n=2) hind paws. Notably, only the lateral side of
the paw was innervated by SNI; therefore, only this area developed
neuropathy. In sham controls, all nerves and their branches were
exposed but no branch was ligated or transected on the left
(Sham-L, n=2) or right (Sham-R, n=2) hind paws. The wound was
sutured and the mice were allowed to recover. The Von Frey test for
mechanical allodynia was conducted as previously described by
Richner et al (28) one day
prior to surgery, and every two days following.
DRG culture
A total of 8 male and 8 female Kunming mice (age, 8
weeks; weight, 18–22 g) were maintained by the China-Japan Union
Hospital of Jilin University. Animals were housed in a 12 h
light/dark cycle and had free access to food and water at 25°C and
50 +/−10% relative humidity. Breeding and maintenance of the mouse
colony were performed until embryos were formed. Embryos were
sacrificed using ice-cold PBS followed by decapitation. DRGs from
vertebral levels were isolated and immersed in ice-cold PBS. DRGs
were digested in a solution of 0.6 mg/ml collagenase type IX
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and 1 mg/ml
dispase II (Sigma-Aldrich; Merck Millipore) at 37°C in 5%
CO2 and 95% air for 40 min. Following titration, the
DRGs were cultured in Dulbeccos modified Eagles medium/nutrient
mixture F-12 (Biofluids, Inc., Rockville, MD, USA) supplemented
with 2 mM glutamine and 10% fetal bovine serum (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA). The medium was
supplemented with 100 U/ml interferon gamma (R&D Systems, Inc.,
Minneapolis, MN, USA) to promote the formation of cell lines. The
cells were cultured at 37°C for 10 passages prior to cloning.
Establishment of DRG cell lines
Individual clones were isolated via cloning rings
and diluted in 24-well plates. The cell lines were routinely
cultured in a proliferating condition and passaged every week. The
clones were able to grow to >50 passages.
Total RNA isolation from DRGs
A total of 2 weeks following SNI surgery, DRGs were
isolated according to a previous protocol (29). Each pool was added to 1 ml
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and homogenized with a homogenizer.
Following isolation with chloroform, RNA was precipitated using
isopropanol. The resultant pellets were re-suspended in Tris/EDTA
buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Following digestion
with DNase, the quality and purity of RNA was measured using a
Bioanalyzer system (Agilent Technologies, Inc., Santa Clara, CA,
USA). RNA integrity and contaminated genomes were identified by
agarose gel electrophoresis.
Microarray analysis
A TaqMan® Array Rodent MicroRNA Card was
used in a 7900HT Fast Real-Time PCR system (both purchased from
Applied Biosystems; Thermo Fisher Scientific, Inc.). The card
consisted of 384 TaqMan MicroRNA assays, which enables the
quantification of 226 miRs in mice. This work was conducted at
KangChen Bio-tech Inc. (Shanghai, China). Mus musculus miRNA
(mmu-miR)-449a and -185 DNA constructs demonstrated significant
alterations between the SNI and sham hind paws (P<0.05; Table I). Therefore, these two mmu-miRs
were selected for the subsequent experiments. mmu-miR-449a
(accession no. NR_029961) and mmu-miR-185 (accession no. NR_029706)
were cloned into the NdeI-EcoRI sites of a pcDNA3.1
vector (Invitrogen; Thermo Fisher Scientific, Inc.) using a
TOPO® Taq-Amplified Expression kit (Life Technologies;
Thermo Fisher Scientific, Inc.), and subsequently named
pcDNA3.1-miR-449a and pcDNA3.1-miR-185, respectively. Plasmids were
amplified in E. coli topoisomerase-10, purified using a
QIAprep® Miniprep kit (Qiagen, Inc., Valencia, CA, USA)
and identified by sequencing.
 | Table I.Significantly dysregulated microRNAs
in SNI and sham mice. |
Table I.
Significantly dysregulated microRNAs
in SNI and sham mice.
| miRs | logFC | P-value | Average Ct values
of miR levels (SNI) | Average Ct values
of miR levels (Sham) |
|---|
| mmu-miR-685 |
3.354 | 0.245957 | 8.268499971 | 6.167500019 |
| mmu-miR-337-3p |
1.7265 | 0.380265 | 9.698999882 | 8.222500086 |
| mmu-miR-139-3p |
1.4535 | 0.220282 | 4.266250015 | 3.332000137 |
| mmu-miR-687 |
1.357 | 0.370371 | 5.784999967 | 4.99000001 |
| mmu-miR-365 | −1.4085 | 0.225318 | 5.113250017 | 6.48149991 |
| mmu-miR-101a | −1.42 | 0.099638 | 6.218500018 | 7.390500069 |
| mmu-miR-339-5p | −1.5095 | 0.197606 | 5.355749965 | 6.210500002 |
| mmu-miR-130b | −2.002 | 0.144655 | 7.530249834 | 9.433000089 |
| mmu-miR-449a | −2.3225 | 0.039836 | 7.360249996 | 9.310999873 |
| mmu-miR-185 | −2.407 | 0.048612 | 6.308750033 | 8.404500151 |
Transfection of DRG cells
The DRG cell lines were transfected with 2 µg
pcDNA3.1-miR-449a or pcDNA3.1-miR-185 as treatment groups.
Transfection was conducted in >50% confluent cells in 6-well
plates by Lipofectamine® 2000 (Life Technologies; Thermo
Fisher Scientific, Inc.).
Channel gene selection
Microarray analyses revealed that FXYD
domain-containing ion transport regulator 3 (FXYD3), transient
receptor potential cation channel subfamily A member 1 (TRPA1) and
calcium-activated potassium channel subunit α-1 (KCNMA1) mRNA
expression levels were completely inhibited, whereas calcium
channel, voltage-dependent L-type calcium channel subunit α-1C
(CACNA1C), γ subunit of the epithelial sodium channel and
transmembrane phosphatase with tension homology (TPTE) mRNA
expression levels were significantly increased in DRG cells
transfected with mmu-miR-449a when compared with non-transfected
cells. TRPA1 has been widely reported to be associated with pain
pathogenesis (14,30). KCNMA1 regulates the potassium
channel (31), which has
additionally been demonstrated to be associated with pain
pathogenesis (32,33). TPTE is a transmembrane phosphatase,
which has tensin homology and reaches the highest level among all
channel proteins. Therefore, the most differentially expressed
mRNAs TRPA1, KCNMA1 and TPTE were selected for subsequent
experiments.
RT-qPCR
A total of one month following SNI surgery, DRGs
were isolated according to a previous protocol (29). RNA was isolated using a Takara
MiniBEST Universal RNA Extraction kit (code no. 9767; Takara
Biotechnology Co., Ltd., Dalian, China). RNA (5 µg) was
reverse-transcribed using an RT kit (Takara Biotechnology Co.,
Ltd.). The mRNA expression levels of TPA1, KCNMA1 and TPTE were
further measured in the mmu-miR-449a transfected and
non-transfected DRG cell lines, SNI models and sham controls.
Platinum® TaqDNA polymerase was purchased from Applied
Biosystems; Thermo Fisher Scientific, Inc. qPCR with
SYBR® Green (Invitrogen; Thermo Fisher Scientific, Inc.)
detection was performed using Mouse Neuroscience Ion Channels and
Transporters RT2 Profiler™ PCR Array containing 84
primer sets for ion channel and transporter genes (Qiagen, Inc.).
qPCR was performed in triplicate using a SYBR Green PCR Master Mix
with the following primers: Forward, 5-gaa ctg atc atc aat ggt tc-3
and reverse, 5-agg ttt gga ttt gct cct tg-3 for TRPA1; forward,
5-aaa acc ctt gag cgc aac ag-3 and reverse, 5-gcc taa ctc tca ggt
gct cc-3 for KCNMA1; forward, 5-cct gct gaa cga ggg ttc ag-3 and
reverse, 5-tgt gct tgg ctc ttt cca gc-3 for TPTE; forward, 5-ttc
ccc tcc atc gtg ggc cg-3 and reverse, 5-gtc cca gtt ggt aac aat
gc-3 for β-actin. Reactions were carried out using a 7500 Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the following conditions: Initial denaturation at 95°C
for 10 min, followed by 40 cycles of denaturation at 95°C for 10
sec, annealing at 60°C for 34 sec and extension at 60°C for 60 sec.
The relative expression value was calculated by the
2∆∆Cq method (34).
Due to the small sample size, the experiment was
repeated on 8 further Kunming male mice (age, 8 weeks; weight,
18–22 g). Mice were purchased from the Animal Center of Jilin
University and maintained by the China-Japan Union Hospital of
Jilin University, as aforementioned. SNI and sham surgeries were
performed as already described.
Statistical analysis
To compare groups, one-way analysis of variance was
performed, followed by Tukeys post hoc test. Statistical analyses
were performed using GraphPad Prism software version 5.0.4
(GraphPad Software Inc., La Jolla, CA, USA). Data are expressed as
the mean ± standard deviation. *P<0.05 was considered to
indicate a statistically significant difference.
Results
Evaluation of the SNI mouse model
In the SNI mouse model, the onset of mechanical
allodynia was assessed 1 day prior to and 2 days following surgery.
Von Frey testing of SNI was performed on the SNI-L, SNI-R, Sham-R
and Sham-L groups. The unilateral SNI hind paws maintained strong
mechanical allodynia during the whole observation period of two
weeks. The SNI hind paws developed obvious hypersensitivity on the
operated paw (P<0.05), particularly at days 3 and 5, whereas the
sham hind paws were not significantly affected. The sham animals
exhibited no significant alterations in the threshold for
mechanical allodynia over time (Fig.
1). These results implied that a SNI model was successfully
established.
Hierarchical cluster analysis of
differentially dysregulated microRNAs in the SNI model and sham
controls
The spectrum of the differentially dysregulated
microRNAs was analyzed in the DRG cells of SNI models and sham
animals within the context of the four mRNA-expression subtypes.
Numerous differentially dysregulated microRNAs exhibited
mRNA-subtype-specific and clinical-subtype-specific patterns. The
differentially dysregulated microRNAs were considerably more
diverse and recurrent within the SNI and sham groups. However, the
overall upregulated microRNAs were increased in the sham groups and
reduced in the SNI groups (Fig.
2).
Hierarchical cluster analysis of differentially
dysregulated microRNAs revealed 154 dysregulated microRNAs in the
sham groups and reduced levels in the SNI groups. The levels of
mmu-miR-685, -337-3p, -139-3p, -687, -365, -101a, -339-5p, -130b,
-449a and -185 were markedly increased in sham animals. Among these
10 microRNAs, only mmu-miR-449a and -185 had statistically
significant alterations in SNI models compared with sham groups
(P<0.05). No significant differences were observed in the
expression levels of the additional 8 miRs assessed between the SNI
and sham groups (P>0.05; Table
I).
PCR array
PCR array analysis indicated that mmu-miR-449a
transfection of DRG cells affected the mRNA expression levels of
the majority of ion channel genes, compared with non-transfected
cells (Fig. 3). For convenience,
the three most differentially expressed ion channel genes were
selected for subsequent experiments: TRPA1, KCNMA1 and TPTE
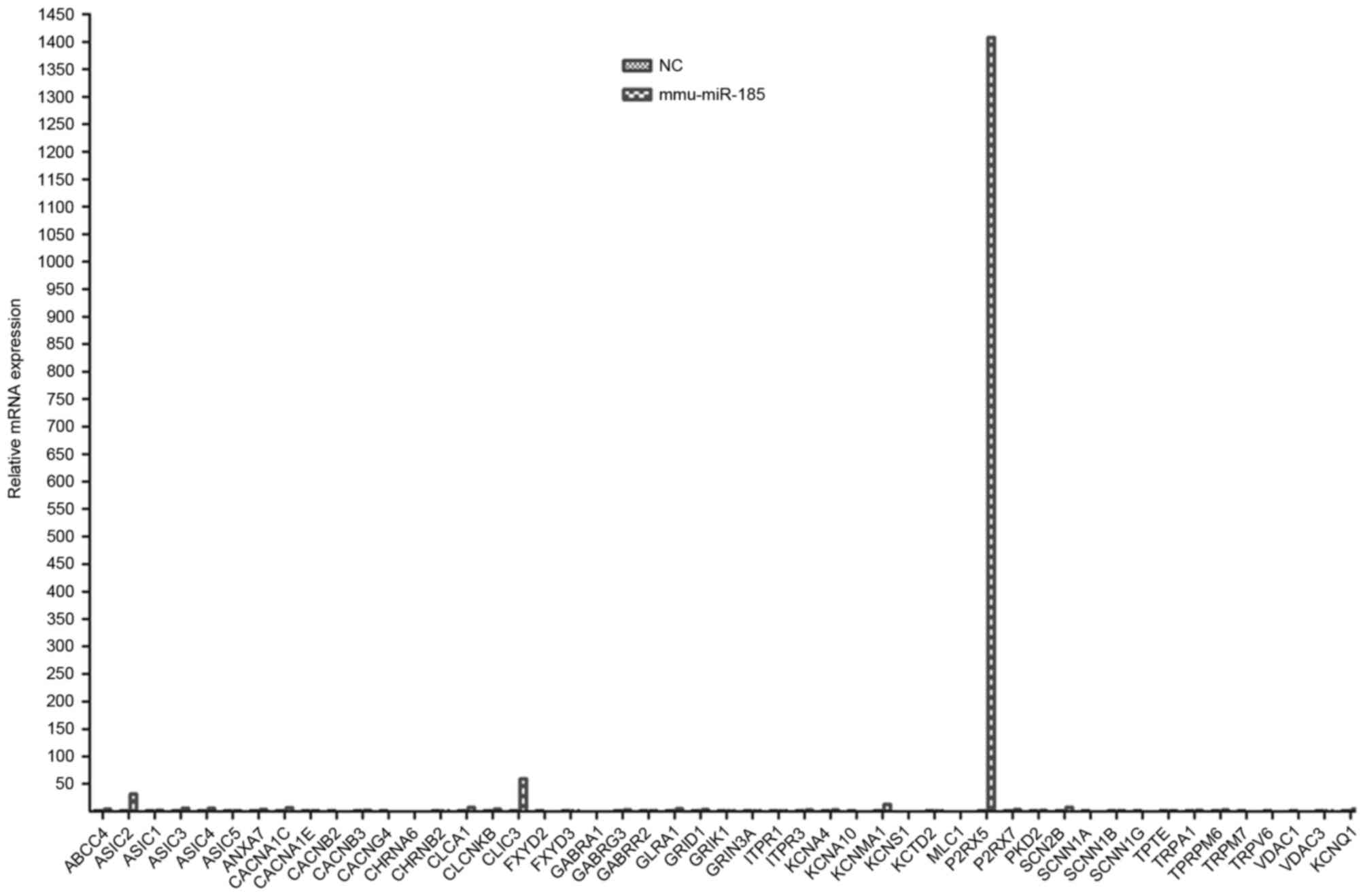
(P<0.05). Unlike mmu-miR-449a, PCR array analysis demonstrated
that mmu-miR-185 transfection did not alter the majority of ion
channel encoding genes, except for the members of the P2X family of
ligand-gated cation channels, including P2X purinoceptor 5 (P2RX5),
in DRG cells compared with non-transfected cells (Fig. 4). These results suggested that
mmu-miR-185 does not serve an important role in regulating the
levels of the majority of ion channel proteins.
Mmu-miR-449a affects the mRNA
expression levels of TRPA1, KCNMA1 and TPTE
The effects of mmu-miRNA-449a on the ion channel
protein levels of TRPA1, KCNMA1 and TPTE were assessed in DRG cells
by RT-qPCR. Of the three proteins examined, TRPA1 demonstrated the
most significantly altered mRNA expression levels between
mmu-miRNA-449a-transfected cells and non-transfected cells, and
between SNI and sham cells. TRPA1 mRNA expression levels
significantly decreased in mmu-miRNA-449a-transfected cells and
increased in the SNI models (P<0.05). KCNMA1 mRNA levels were
additionally differentially expressed between
mmu-miRNA-449a-transfected cells and non-transfected cells, and
between SNI and sham cells. Its mRNA expression levels were
additionally significantly increased in the SNI models (P<0.01;
Fig. 5). TPTE mRNA levels were
differentially expressed between mmu-miRNA-449a-transfected and
non-transfected cells (P<0.05), and were increased in
mmu-miRNA-449a-transfected cells and decreased in SNI models
(P<0.05).
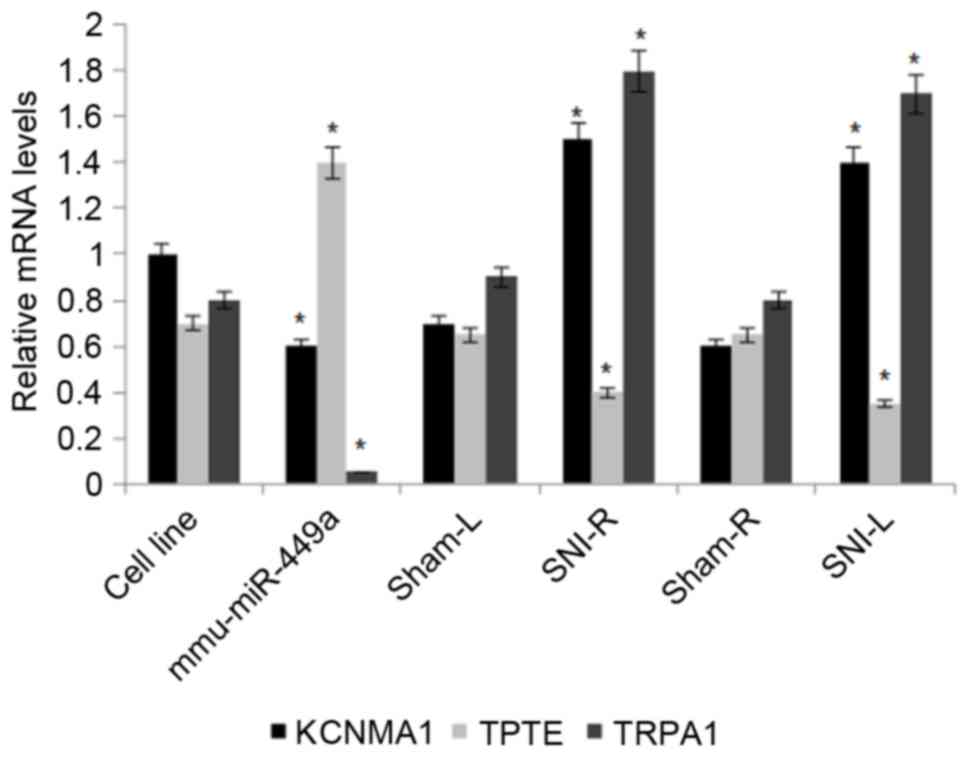 | Figure 5.Reverse transcription-quantitative
polymerase chain reaction analysis of the mRNA expression levels of
TRPA1, KCNMA1 and TPTE in non-transfected and
mmu-miR-449a-transfected cell line, SNI models and sham controls.
β-actin served as an internal control. Data are presented are
presented as the mean ± standard deviation (n=5). *P<0.05 vs.
the respective sham group. mmu, Mus musculus; miR, microRNA;
TRPA1, transient receptor potential cation channel subfamily A
member 1; KCNMA1, calcium-activated potassium channel subunit α-1;
TPTE, transmembrane phosphatase with tension homology; miR,
microRNA; L, left; R, right. |
Discussion
The results of the present study demonstrated that
two microRNAs in the SNI models and sham mice were significantly
downregulated: mmu-miR-449a and -185. mmu-miR-185 overexpression
did not affect the levels of most ion channel proteins, except
P2RX5. P2RX5 is expressed in neurons, smooth and cardiac muscle
cells, and leukocytes (35). In
contrast, the overexpression of mmu-miR-449a affected the levels of
most ion channel proteins. This result suggested that the
peripheral deletion of mmu-miR-449a enhances novice inputs in DRG
cells. Therefore, downregulation of mmu-miR-449a may be sufficient
to affect the global pain mechanisms in SNI mouse models.
MiR-449a has been demonstrated to serve as a tumor
suppressor in various cancers by regulating cell differentiation,
particularly in neuroblastoma (36). miR-449a has additionally been
revealed to be involved in the executive functioning of the brain,
as demonstrated by the Wisconsin Card Sorting Test (37). However, the association between
miR-449A and neuropathic pain remains to be elucidated. Therefore,
the present study selected to use mmu-miR-449a. The present
research provided evidence that mmu-miR-449a has a strong effect on
neuronal excitability in DRG neurons, and that downregulation of
the regulator contributes to pain hyposensitivity. This was
primarily due to the upregulation of the channel protein TRAP1 in
DRG cells.
Peripheral neuropathic pain in different disorders
may be caused by various mechanisms and multiple etiological
elements. In the present study, a unilateral SNI model was
successfully established by a common traumatic nerve injury, which
induces neuropathic pain. Although the model mimics the mechanisms
of other types of neuropathic pain, its long-lasting behavioral
mechanical hypersensitivity renders it useful for research on the
underlying molecular mechanisms.
In the neuropathic pain model, mmu-miR-449a
expression was observed to be significantly decreased in DRG
neurons, with a dysregulation in levels of the ion channels
proteins TRPA1, KCNMA1 and TPTE. Among these ion channel proteins,
TRPA1 and KCNMA1 were markedly increased, whereas TPTE was
significantly downregulated. Furthermore, these results
demonstrated that increased expression of mmu-miR-449A caused the
reverse changing trends of TRPA1, KCNMA1 and TPTE. Ionic channels
serve important functions in sensory neurons, including transducing
specific stimuli via electrical signals.
The results of the present study demonstrated that
downregulation of mmu-miR-449a caused upregulation of TRPA1 protein
expression levels in the SNI model, and may result in
hypersensitivity to neuropathic pain. TRPA1 is an important member
of transient receptor potential of cation channels family, and
primarily exists in the sensory neurons of the trigeminal nerve and
DRG cells (38). Furthermore,
TRPA1 serves an important role in mechanical and norepinephrine
hypersensitivity to neuropathic pain following nerve injury and may
attenuate hyperalgesia (39,40).
Therefore, mmu-miR-449a may relieve neuropathic pain by
down-regulating the levels of TRPA1.
KCNMA1 may be associated with neuropathic pain as
increasing studies on potassium channel physiology have
demonstrated the potential for the development of analgesics
(33). Potassium channels have
been reported to be associated with severe persistent breast pain
following surgery (41). Similar
to TRPA1, the results of the present study additionally revealed
that KCNMA1 levels increased in SNI models and decreased in sham
controls, and mmu-miR-449a effectively decreased the levels of
KCNMA1.
There are certain limitations to the present study.
Despite the effective establishment of the SNI mouse model, the
precise molecular mechanisms underlying the downregulation of
mmu-miR-449a and neuropathic pain remains unclear. mmu-miR-449a
silencing or overexpressing animal models should be established to
confirm the functional roles of mmu-miR449. In addition, two
animals in each group are a small population and the results may be
unstable for microarray analysis. To reduce the bias caused by the
limited sample size, RT-qPCR analysis was repeated on eight further
mice; the SNI and sham procedures were performed as aforementioned
(data not shown), and this demonstrated similar results.
Furthermore, assessing a broader range of ion channels may have
revealed additional differential expression levels. The present
work was limited to a mouse model; a rat model may be more
representative due to its larger size.
In conclusion, in the present study, an SNI-mediated
decrease in mmu-miR-449a levels suggested the functional importance
of the microRNAs in trauma-induced neuropathic pain. Lower levels
of mmu-miR-449a led to an increase in the ion channel proteins
TRPA1 and KCNMA1, and a decrease in TPTE in DRG neurons, which was
associated with the development of mechanical allodynia. In
contrast, overexpression of mmu-miR-449a led to a decrease in
expression levels of TRPA1 and KCNMA1, and an increase in TPTE in
DRG neurons. From these results, mmu-miR-449a may be a potential
therapeutic molecule for the alleviation of neuropathic pain.
References
|
1
|
Trost Z, Zielke M, Guck A, Nowlin L,
Zakhidov D, France CR and Keefe F: The promise and challenge of
virtual gaming technologies for chronic pain: The case of graded
exposure for low back pain. Pain Manag. 5:197–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gerges FJ, Manchanda C, Novak G, Al-Kimawi
M, Semenovski M and Williams S: Occult spinal dysraphism: A
challenge in pain management. Pain Physician. 18:E225–E228.
2015.PubMed/NCBI
|
|
3
|
Rogachov A, Cheng JC and DeSouza DD:
Discriminating neural representations of physical and social pains:
How multivariate statistics challenge the ‘shared representation’
theory of pain. J Neurophysiol. 114:2558–2560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baquie P, Fooks L, Pope J and Tymms G: The
challenge of managing mid-foot pain. Aust Fam Physician.
44:106–111. 2015.PubMed/NCBI
|
|
5
|
Hayes K and Gordon DB: Delivering quality
pain management: The challenge for nurses. AORN J. 101:328–334;
quiz 335–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Foster DC, Falsetta ML, Woeller CF,
Pollock SJ, Song K, Bonham A, Haidaris CG, Stodgell CJ, Messing SP,
Iadarola M and Phipps RP: Site-specific mesenchymal control of
inflammatory pain to yeast challenge in vulvodynia-afflicted and
pain-free women. Pain. 156:386–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Frisch S: Perceptions of pain. Cultural
differences add to the challenge of treating patients' pain. Minn
Med. 97:14–16. 2014.
|
|
8
|
Dale O, Moksnes K and Kaasa S: European
palliative care research collaborative pain guidelines: Opioid
switching to improve analgesia or reduce side effects. A systematic
review. Palliat Med. 25:494–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosenbaum D, Dallongeville J, Sabouret P
and Bruckert E: Discontinuation of statin therapy due to muscular
side effects: A survey in real life. Nutr Metab Cardiovasc Dis.
23:871–875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Caires R, Luis E, Taberner FJ,
Fernandez-Ballester G, Ferrer-Montiel A, Balazs EA, Gomis A,
Belmonte C and de la Peña E: Hyaluronan modulates TRPV1 channel
opening, reducing peripheral nociceptor activity and pain. Nat
Commun. 6:80952015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dib-Hajj SD, Black JA and Waxman SG:
NaV1.9: A sodium channel linked to human pain. Nat Rev Neurosci.
16:511–519. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng CF, Wang WC, Huang CY, Du PH, Yang
JH and Tsaur ML: Coexpression of auxiliary subunits KChIP and DPPL
in potassium channel Kv4-positive nociceptors and pain-modulating
spinal interneurons. J Comp Neurol. 524:846–873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Skerratt SE and West CW: Ion channel
therapeutics for pain. Channels (Austin). 9:344–351. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sherkheli MA, Schreiner B, Haq R, Werner M
and Hatt H: Borneol inhibits TRPA1, a proinflammatory and noxious
pain-sensing cation channel. Pak J Pharm Sci. 28:1357–1363.
2015.PubMed/NCBI
|
|
15
|
Laedermann CJ, Cachemaille M, Kirschmann
G, Pertin M, Gosselin RD, Chang I, Albesa M, Towne C, Schneider BL,
Kellenberger S, et al: Dysregulation of voltage-gated sodium
channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J Clin
Invest. 123:3002–3013. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakai A and Suzuki H: microRNA and Pain.
Adv Exp Med Biol. 888:17–39. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Linnstaedt SD, Walker MG, Parker JS, Yeh
E, Sons RL, Zimny E, Lewandowski C, Hendry PL, Damiron K, Pearson
C, et al: MicroRNA circulating in the early aftermath of motor
vehicle collision predict persistent pain development and suggest a
role for microRNA in sex-specific pain differences. Mol Pain.
11:662015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mallik S and Maulik U: MiRNA-TF-gene
network analysis through ranking of biomolecules for
multi-informative uterine leiomyoma dataset. J Biomed Inform.
57:308–319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Galicia-Vázquez G, Chu J and Pelletier J:
eIF4AII is dispensable for miRNA-mediated gene silencing. RNA.
21:1826–1833. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding M, Li J, Yu Y, Liu H, Yan Z, Wang J
and Qian Q: Integrated analysis of miRNA, gene and pathway
regulatory networks in hepatic cancer stem cells. J Transl Med.
13:2592015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan L, Lee S, Lazzaro DR, Aranda J, Grant
MB and Chaqour B: Single and Compound Knockouts of MicroRNA
(miRNA)-155 and its Angiogenic Gene Target CCN1 in Mice Alter
Vascular and Neovascular Growth in the Retina via Resident
Microglia. J Biol Chem. 290:23264–23581. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van Spronsen M, van Battum EY, Kuijpers M,
Vangoor VR, Rietman ML, Pothof J, Gumy LF, van Ijcken WF, Akhmanova
A, Pasterkamp RJ and Hoogenraad CC: Developmental and
activity-dependent miRNA expression profiling in primary
hippocampal neuron cultures. PLoS One. 8:e749072013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Ueno Y, Liu XS, Buller B, Wang X,
Chopp M and Zhang ZG: The microRNA-17-92 cluster enhances axonal
outgrowth in embryonic cortical neurons. J Neurosci. 33:6885–6894.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Impey S, Davare M, Lesiak A, Fortin D,
Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH and
Wayman GA: An activity-induced microRNA controls dendritic spine
formation by regulating Rac1-PAK signaling. Mol Cell Neurosci.
43:146–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kusuda R, Cadetti F, Ravanelli MI, Sousa
TA, Zanon S, De Lucca FL and Lucas G: Differential expression of
microRNAs in mouse pain models. Mol Pain. 7:172011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang W, Shi Q, Mattes WB, Mendrick DL and
Yang X: Translating extracellular microRNA into clinical biomarkers
for drug-induced toxicity: From high-throughput profiling to
validation. Biomark Med. 9:1177–1188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Asha S, Sreekumar S and Soniya EV:
Unravelling the complexity of microRNA-mediated gene regulation in
black pepper (Piper nigrum L.) using high-throughput small RNA
profiling. Plant Cell Rep. 35:53–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Richner M, Bjerrum OJ, Nykjaer A and
Vaegter CB: The spared nerve injury (SNI) model of induced
mechanical allodynia in mice. J Vis Exp pii. 30922011.
|
|
29
|
Scroggs RS, Todorovic SM, Anderson EG and
Fox AP: Variation in IH, IIR and ILEAK between acutely isolated
adult rat dorsal root ganglion neurons of different size. J
Neurophysiol. 71:271–279. 1994.PubMed/NCBI
|
|
30
|
Zima V, Witschas K, Hynkova A, Zimová L,
Barvík I and Vlachova V: Structural modeling and patch-clamp
analysis of pain-related mutation TRPA1-N855S reveal inter-subunit
salt bridges stabilizing the channel open state. Neuropharmacology.
93:294–307. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanthesh BM, Sandle GI and Rajendran VM:
Enhanced K(+) secretion in dextran sulfate-induced colitis reflects
upregulation of large conductance apical K(+) channels (BK;
Kcnma1). Am J Physiol Cell Physiol. 305:C972–C980. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsantoulas C: Emerging potassium channel
targets for the treatment of pain. Curr Opin Support Palliat Care.
9:147–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pereira V, Busserolles J, Christin M,
Devilliers M, Poupon L, Legha W, Alloui A, Aissouni Y, Bourinet E,
Lesage F, et al: Role of the TREK2 potassium channel in cold and
warm thermosensation and in pain perception. Pain. 155:2534–2544.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
gene expression data using real-time quantitative PCR and the
2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abramowski P, Ogrodowczyk C, Martin R and
Pongs O: A truncation variant of the cation channel P2RX5 is
upregulated during T cell activation. PLoS One. 9:e1046922014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Z, Ma X, Sung D, Li M, Kosti A, Lin
G, Chen Y, Pertsemlidis A, Hsiao TH and Du L: microRNA-449a
functions as a tumor suppressor in neuroblastoma through inducing
cell differentiation and cell cycle arrest. RNA Biol. 12:538–554.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY,
Wen CC, Huang YH, Hsiao PC, Hsiao CK, Liu CM, et al: MicroRNA
expression aberration as potential peripheral blood biomarkers for
schizophrenia. PLoS One. 6:e216352011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nassini R, Materazzi S, Benemei S and
Geppetti P: The TRPA1 channel in inflammatory and neuropathic pain
and migraine. Rev Physiol Biochem Pharmacol. 167:1–43. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kogure W, Wang S, Tanaka K, Hao Y,
Yamamoto S, Nishiyama N, Noguchi K and Dai Y: Elevated H2 O2 levels
in trinitrobenzene sulfate-induced colitis rats contributes to
visceral hyperalgesia through interaction with the ransient
receptor potential ankyrin 1 cation channel. J Gastroenterol
Hepatol. 31:1147–1453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
DeBerry JJ, Saloman JL, Dragoo BK, Albers
KM and Davis BM: Artemin immunotherapy is effective in preventing
and reversing cystitis-induced bladder hyperalgesia via TRPA1
regulation. J Pain. 16:628–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Langford DJ, Paul SM, West CM, Dunn LB,
Levine JD, Kober KM, Dodd MJ, Miaskowski C and Aouizerat BE:
Variations in potassium channel genes are associated with distinct
trajectories of persistent breast pain after breast cancer surgery.
Pain. 156:371–380. 2015. View Article : Google Scholar : PubMed/NCBI
|