Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney cancer and accounts for ~2–3% of all cases of cancer in
adults (1). RCC may be resistant
to chemotherapy and conventional radiation therapy, although
surgical resection is an effective treatment (2–4).
However, 30% of patients with RCC who undergo surgical resection
suffer from recurrence (3,5). The prognosis of patients with RCC
remains unfavorable, particularly for those with advanced tumors
(6). It is essential to diagnose
RCC as early as possible and to identify novel biomarkers for
RCC.
Long non-coding RNAs (lncRNAs) are a group of
non-protein coding transcripts that have a length of >200
nucleotides (7,8). lncRNAs were regarded as
‘transcriptional noise’ when identified initially. However,
accumulating evidence has demonstrated that lncRNAs may influence
gene expression via transcriptional, post-transcriptional or
epigenetic regulation (9). With
the development of whole-genome sequencing technologies, it is now
clear that non-coding RNAs account for ≥90% of the human genome and
serve important roles in different biological processes (10,11),
including genome rearrangement (12), chromatin modifications (13), gene imprinting (14), cell cycle control (15) and X-chromosome inactivation
(16). The lncRNA eosinophil
granule ontogeny transcript (EGOT) is located at 3p26.1. A previous
study revealed that lncRNA EGOT was downregulated in breast cancer,
and its expression was associated with malignant status and poor
prognosis (17). In addition,
next-generation deep sequencing revealed that lncRNA EGOT may be
functional in clear cell carcinoma (18); however, there appears to be no
associated studies about lncRNA EGOT in RCC. In the present study,
the expression and function of lncRNA EGOT in RCC was examined.
Materials and methods
Sample collection
A total of 24 paired RCC tissues (RCC and adjacent
normal tissues) were collected from Peking University Shenzhen
Hospital (Shenzhen, China) from December 2012 to December 2014. The
adjacent normal tissues were 2 cm away from visible RCC lesions and
negative for tumor tissues, as determined by microscopy. Tissues
were immersed in RNAlater (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) for ≥30 min while being dissected, and subsequently stored
at −80°C until further use. Written informed consent was obtained
from all patients. Collection and usage of the samples were
reviewed and approved by the Ethics Committees of Peking University
Shenzhen Hospital (Shenzhen, China). The tissues collected were
reviewed and classified by hematoxylin and eosin staining. The
clinical and pathological characteristics of the patients are
presented in Table I.
 | Table I.Clinicopathological features of renal
cell carcinoma patients. |
Table I.
Clinicopathological features of renal
cell carcinoma patients.
| Clinical
characteristic | No. patients |
|---|
| Age (years) |
|
| Mean
(range) | 51 (25–70) |
| Sex |
|
|
Male | 15 |
|
Female | 9 |
| Histological
type |
|
| Clear
cell | 21 |
|
Papillary | 3 |
| TNM stage |
|
| T1 | 15 |
| T2 | 7 |
| T3 +
4 | 2 |
| AJCC clinical
stages |
|
| I | 14 |
| II | 7 |
| III +
IV | 3 |
Cell culture
The 2 RCC cell lines, 786-O and ACHN, were purchased
from the American Type Culture Collection (Manassas, VA, USA).
Cells were cultured in a humidified incubator containing 5%
CO2 at a temperature of 37°C in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.), 1% glutamine (Gibco; Thermo Fisher Scientific, Inc.), 1
µl/ml penicillin and 1 µg/ml streptomycin sulfates.
Cell transfection
For overexpression of EGOT, the polymerase chain
reaction (PCR)-amplified EGOT digested with BamHI and
EcoRI restriction enzymes was subcloned into the pcDNA3.1
mammalian expression vector (Genepharm, Inc., Sunnyvale, CA, USA).
The EGOT expression vector, pcDNA3.1-EGOT, was verified by
sequencing. Synthesized pcDNA3.1-EGOT or mock (pcDNA3.1) was
transfected (0.2 µg/well in 96-well plates; 4.0 µg/well in 6-well
plates) into 80% confluent RCC cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), which was mixed with
Opti-MEM® I Reduced Serum medium (Gibco; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
transfection assay was performed at 37°C for 24 h. Reverse
transcription-quantitative PCR (RT-qPCR) analysis was performed to
detect the expression of EGOT in cells following transfection.
RNA extraction, cDNA synthesis and the
RT-qPCR
RNAiso Plus reagent (Takara Bio, Inc., Otsu, Japan)
was used to extract total RNA from tissues and cells, according to
the manufacturer's protocol. The concentration of RNA was measured
using a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher
Scientific, Inc.). Subsequently, 1 µg RNA was used to generate a
cDNA library by performing RT with the PrimeScript™ RT reagent kit
(Takara Bio, Inc.). The expression of EGOT was detected by qPCR
with Premix Ex Taq™ II (Takara Bio, Inc.) on a Roche light cycler
480 Real-Time PCR system (Roche Applied Science, Penzberg,
Germany). The qPCR thermocycling conditions were as follows: 95°C
for 1 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30
sec, 70°C for 30 sec. GAPDH was used as an internal control and the
sequences of the primers are listed in Table II. The expression level of EGOT in
tissues and cells was analyzed via the 2−ΔΔCq method
(19). RT-qPCR analysis was
performed in triplicate.
 | Table II.Sequences of primers. |
Table II.
Sequences of primers.
| Primer name | Sequence |
|---|
| lncRNA EGOT | F:
5′-CACTGCACAGGGAAACACAAA-3′ |
|
| R:
5′-ACCCTGTTCATAAGCCCTGATG-3′ |
| GAPDH | F:
5′-GGTCTCCTCTGACTTCAACA-3′ |
|
| R:
5′-GTGAGGGTCTCTCTCTTCCT-3′ |
Cell proliferation assay
The MTT and Cell Counting Kit-8 (CCK-8) assays were
performed to assess the proliferative ability of the cells. After
~12 h seeding of ~3,000 cells/well in a 96-well plate, cells were
transfected with pcDNA3.1-EGOT or mock. For the CCK-8 assay,
following transfection, 10 µl CCK-8 (Beyotime Institute of
Biotechnology, Haimen, China) was added into the wells for 1 h,
prior to measuring the optical density (OD) value of each well
using an ELISA microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) at a wavelength of 490 nm. For the MTT assay, 20
µl MTT solution (5 mg/ml; Sigma-Aldrich, Merck KGaA, Darmstadt,
Germany) was added to the wells for 4 h. The solution was
subsequently replaced with 150 µl dimethyl sulfoxide
(Sigma-Aldrich, Merck KGaA), and the OD value was measured at a
wavelength of 490 nm following agitation of the wells for 15 min at
room temperature. The experiments were performed in replicates of
six and repeated ≥3 times.
Cell mobility assay
Cell scratch assay was performed to assess the cell
mobility of 786-O and ACHN. Cells (~3×105 cells/well)
were seeded into a 6-well plate and 24 h later were transfected
with pcDNA3.1-EGOT or mock as aforementioned. The cell monolayer
was scratched 24 h later with a sterile 200 µl pipette tip to
generate a wound. Cells were then rinsed with phosphate-buffered
saline to remove the floating cells, and then cells were cultured
in DMEM supplemented with 5% FBS. Scratches were imaged at 0 h and
after 24 h with a digital camera system. The experiments were
performed in triplicate and repeated 3 times.
Transwell invasive and migratory assays were
performed to assess the cell migratory and invasive ability.
Transwell chamber inserts with (for migration) or without (for
invasion) Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were
used according to the manufacturer's protocol. Cells were
transfected as aforementioned, resuspended in 200 µl DMEM without
serum and seeded at a density of 1×104 cells/well into
the upper channel of the inserts. The lower chamber contained DMEM
supplemented with 10% serum. Cells were allowed to migrate for 36 h
and invade for 48 h. The cells that had migrated or invaded to the
bottom of the inserts were stained with crystal violet for 15 min
at room temperature and counted using an inverted microscope. The
experiments were performed in in triplicate and repeated at least 3
times.
Flow cytometric analysis
Flow cytometry was performed to detect the
percentage of apoptotic cells following transfection with an Alexa
Fluor® 488 Annexin V/Dead Cell Apoptosis kit
(Invitrogen; Thermo Fisher Scientific, Inc.). Following seeding of
~3×105 cells/well in a six-well plate and incubation for
12 h, the cells were transfected with pcDNA3.1-EGOT or mock.
Following 48 h of transfection, all the cells were harvested and
stained according to the manufacturer's protocol, and the
percentage of apoptotic cells was determined by flow cytometry and
analyzed using FlowJo version X (FlowJo LLC, Ashland, OR, USA). The
experiments were performed in triplicate and repeated ≥3 times.
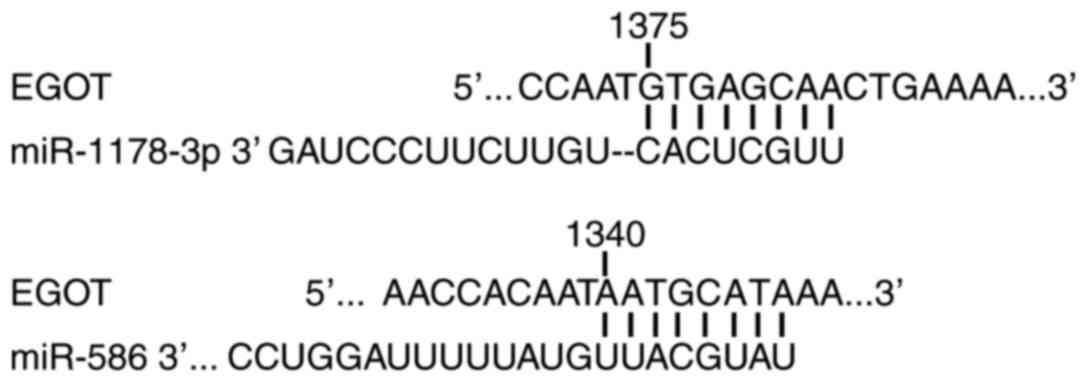
Prediction of EGOT-associated microRNA
(miRNA/miR)
Prediction of associated miRNAs was determined by
computational algorithms according to base-pairing rules between
miRNA and mRNA target sites, location of binding sequences within
the 3′-untranslated region of the target and conservation of target
binding sequences. In the present study, miRDB (mirdb.org/miRDB/index.html) and miRWalk
(http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk)
were used. Putative miRNAs were miR-1178-3p (previous name
miR-1178) and miR-586, and the binding site is displayed in
Fig. 1.
Statistical analysis
A paired t-test was used to compare the expression
levels of EGOT in matched tumor/normal tissues, and in cells
following transfection. A Student's t-test was used to characterize
the phenotypes of cells. All statistical analyses were performed
using the SPSS version 19.0 statistical software package (IBM
Corp., Armonk, NY, USA). Data are presented as the mean ± standard
error. P<0.05 was considered to indicate a statistically
significant difference.
Results
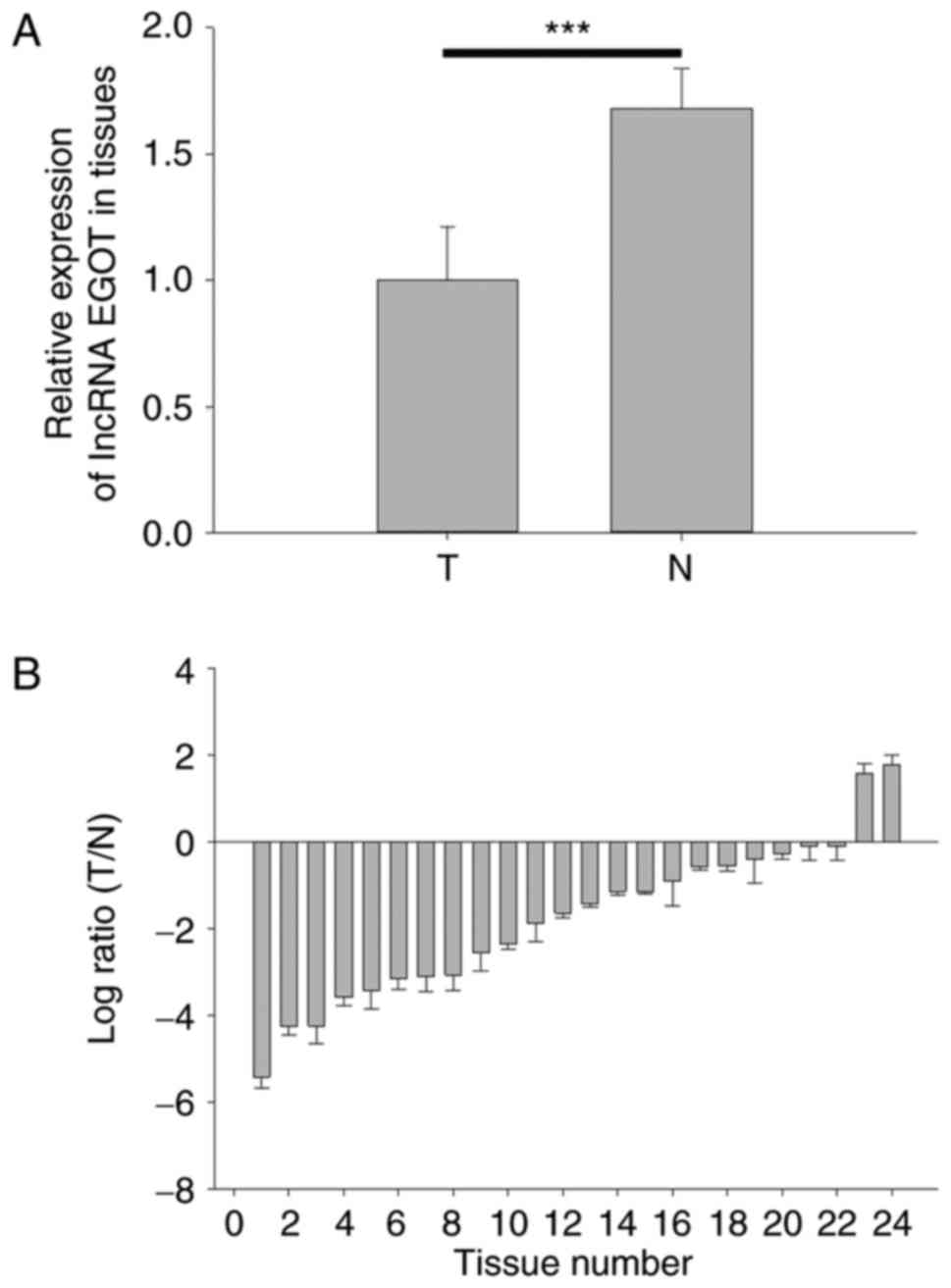
lncRNA EGOT is downregulated in RCC
tissues
To detect the expression level of lncRNA EGOT in RCC
tissues and paired normal tissues, RT-qPCR analysis was performed.
The expression levels of lncRNA EGOT in RCC tissues (1±0.213) was
significantly decreased compared with adjacent normal tissues
(1.68±0.16) (Fig. 2A; P<0.001).
The relative expression levels of lncRNA EGOT in the 24 tissues
demonstrated that lncRNA EGOT was downregulated in 22 RCC tissues
compared with normal adjacent tissues (Fig. 2B).
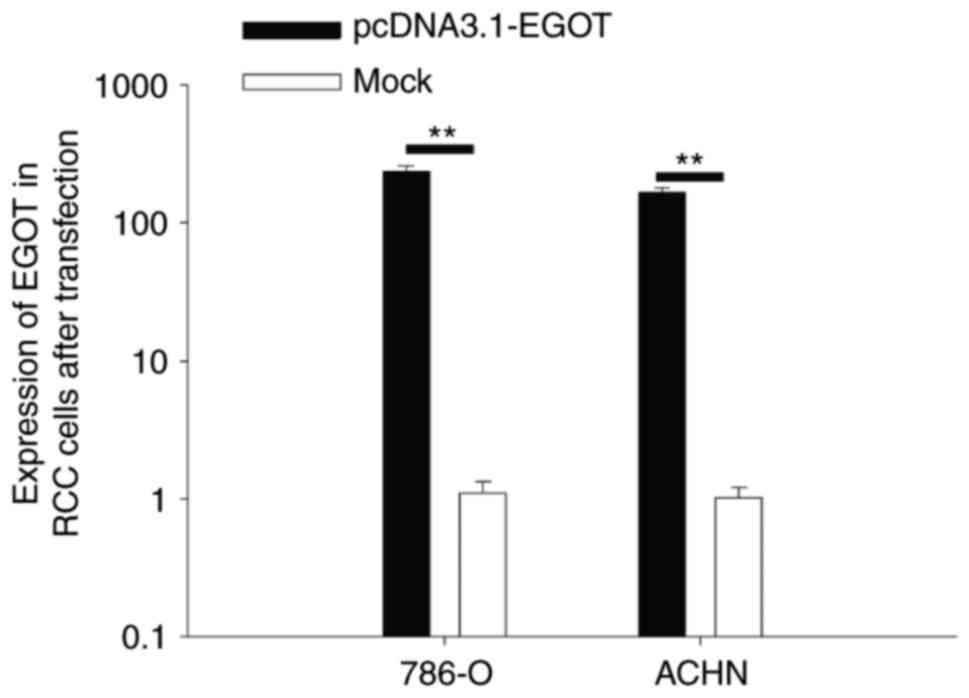
Validation of the expression of lncRNA
EGOT following transfection
RT-qPCR analysis was performed to detect the
expression of lncRNA EGOT in RCC cells following transfection. The
results demonstrated that the expression of lncRNA EGOT was
increased 236.39-fold (786-O cells) and 167.15-fold (ACHN cells) in
cells transfected with pcDNA3.1-EGOT compared with cells
transfected with mock (empty pcDNA3.1) (P<0.01; Fig. 3).
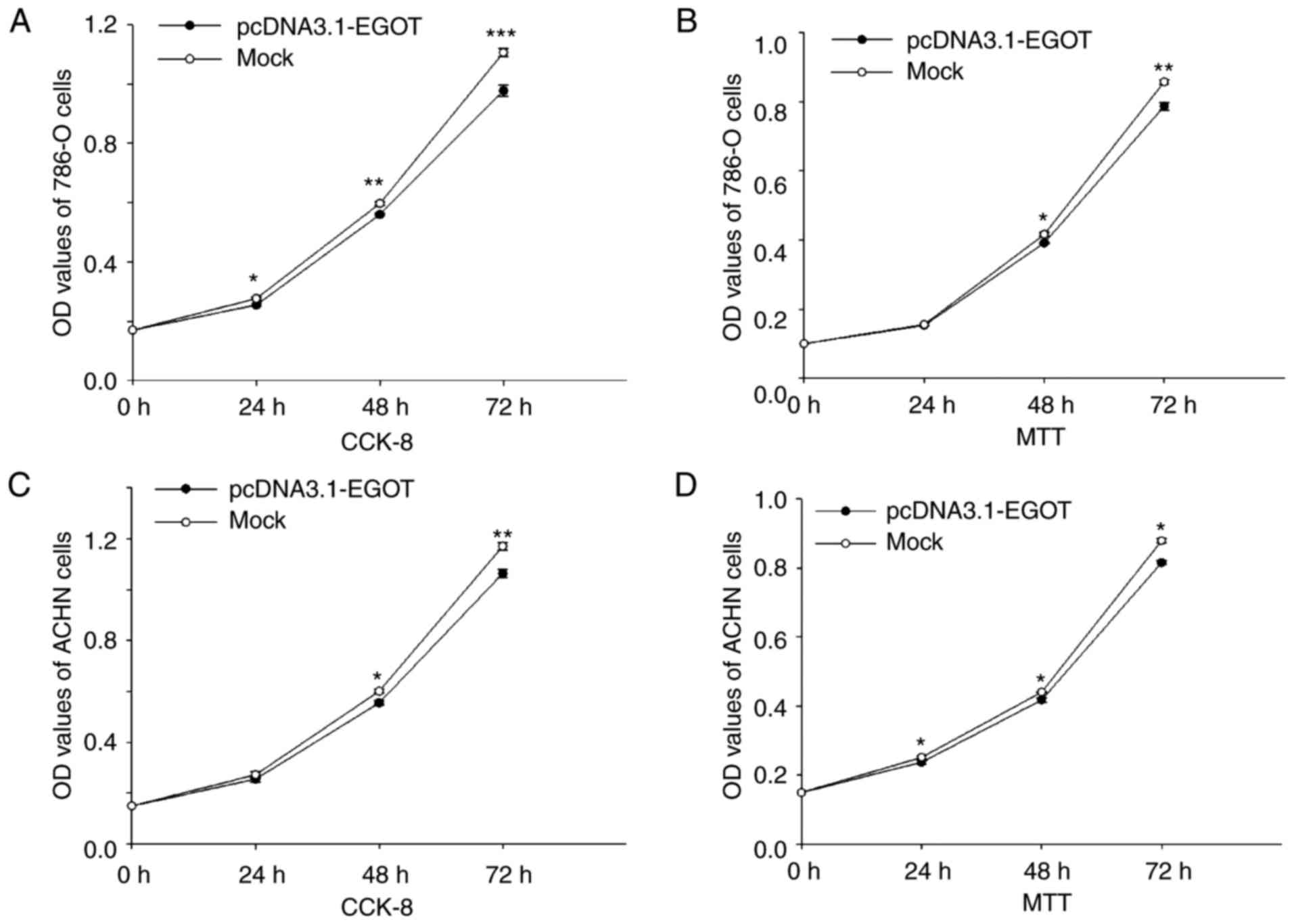
lncRNA EGOT suppresses cell
proliferation
MTT and CCK-8 assays were performed to assess cell
proliferation. In 786-O cells, compared with the mock group,
proliferation was reduced by 7.75% (24 h; P<0.05), 6.27% (48 h;
P<0.01) and 11.67% (72 h; P<0.001) when measured by the CCK-8
assay (Fig. 4A), and by 1.43% (24
h), 5.98% (48 h; P<0.05) and 8.24% (72 h; P<0.01) when
measured by the MTT assay (Fig.
4B), following upregulation of lncRNA EGOT using the
pcDNA3.1-EGOT plasmid. In addition, upregulation of lncRNA EGOT
using the pcDNA3.1-EGOT plasmid suppressed ACHN cell proliferation
by 6.26% (24 h), 7.60% (48 h; P<0.05) and 9.11% (72 h;
P<0.01) when measured by the CCK-8 assay (Fig. 4C), and by 5.58% (24 h; P<0.05),
5.14% (48 h; P<0.05) and 7.21% (72 h; P<0.05) when measured
by the MTT assay (Fig. 4D). These
results suggested that lncRNA EGOT may suppress RCC cell
proliferation.
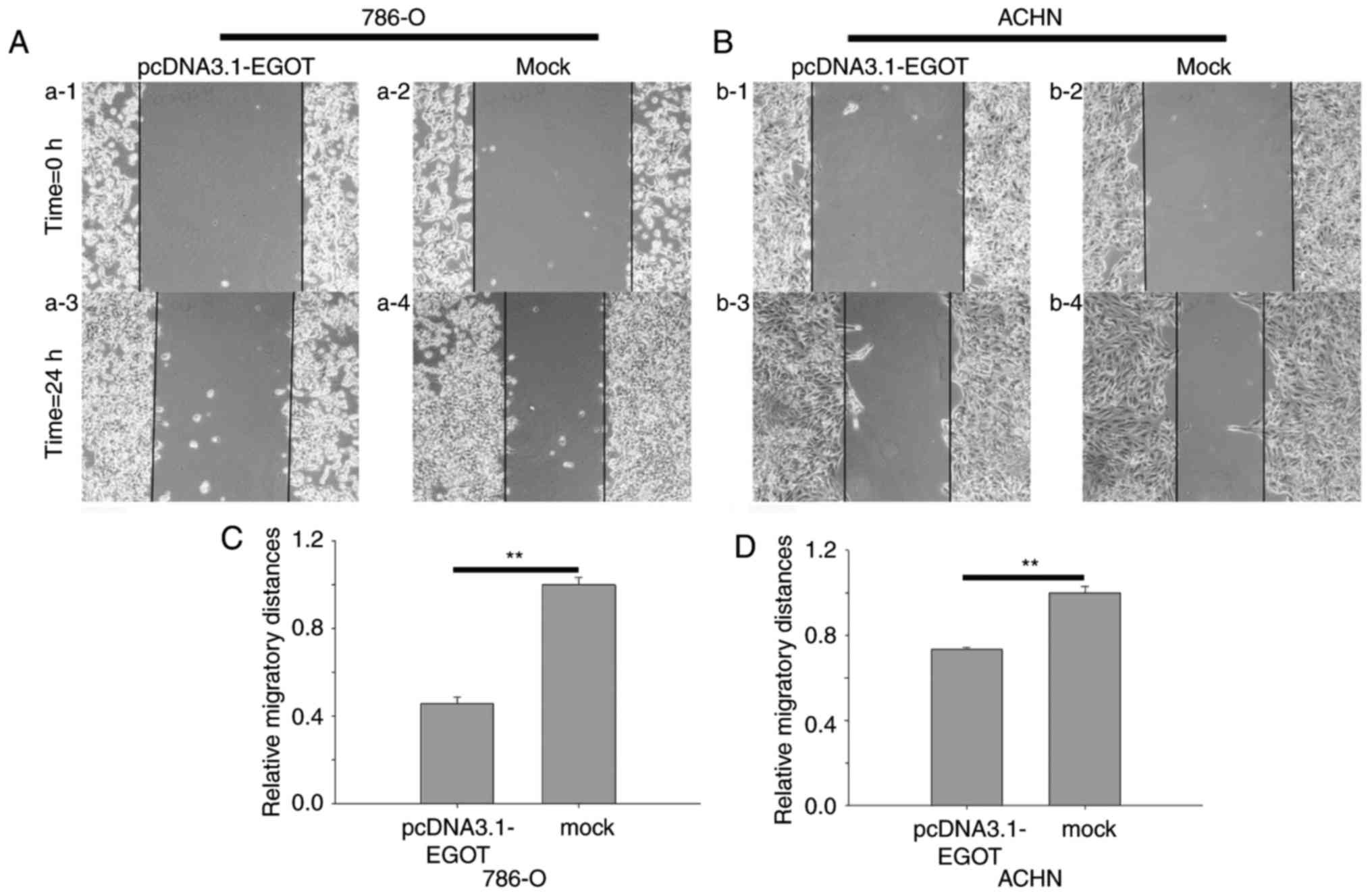
lncRNA EGOT suppresses RCC cell
mobility
Wound-healing, Transwell migratory and Transwell
invasive assays were performed in the present study. The results of
the wound-healing assay demonstrated that at 24 h, the migratory
distance of cells transfected with pcDNA3.1-EGOT was reduced by
54.23% compared with the mock group in 786-O cells (P<0.01), and
by 26.60% in ACHN cells (P<0.01) (Fig. 5).
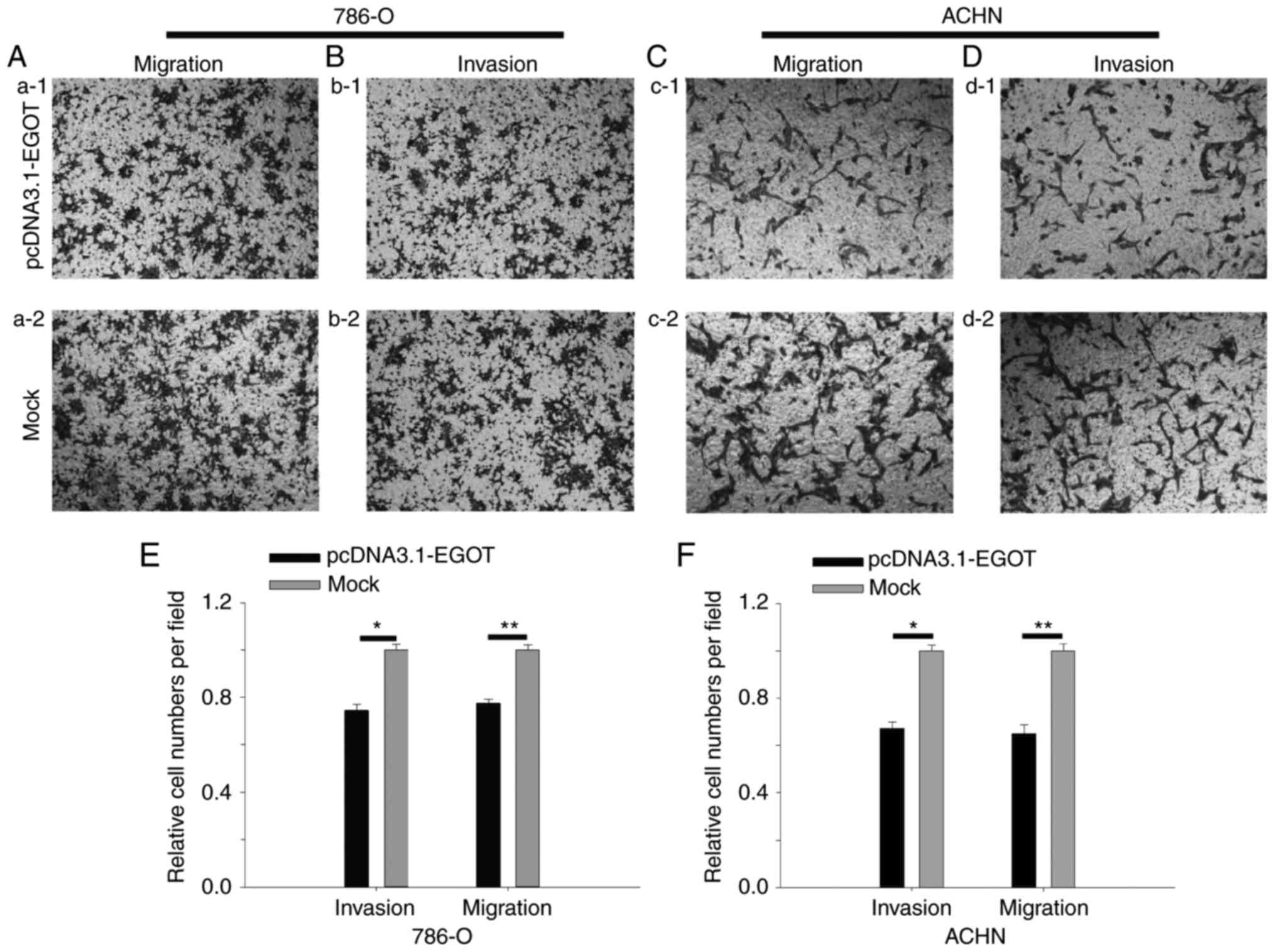
The results of the Transwell assay revealed that in
786-O cells, upregulation of lncRNA EGOT reduced invasive ability
by 25.45% (P<0.05) and migratory ability by 22.42% (P<0.01).
For ACHN cells, upregulation of lncRNA EGOT reduced invasive the
number of cells by 32.80% (P<0.05) and migratory ability by
34.84% (P<0.05) (Fig. 6). The
results suggested that lncRNA EGOT suppressed RCC cell
mobility.
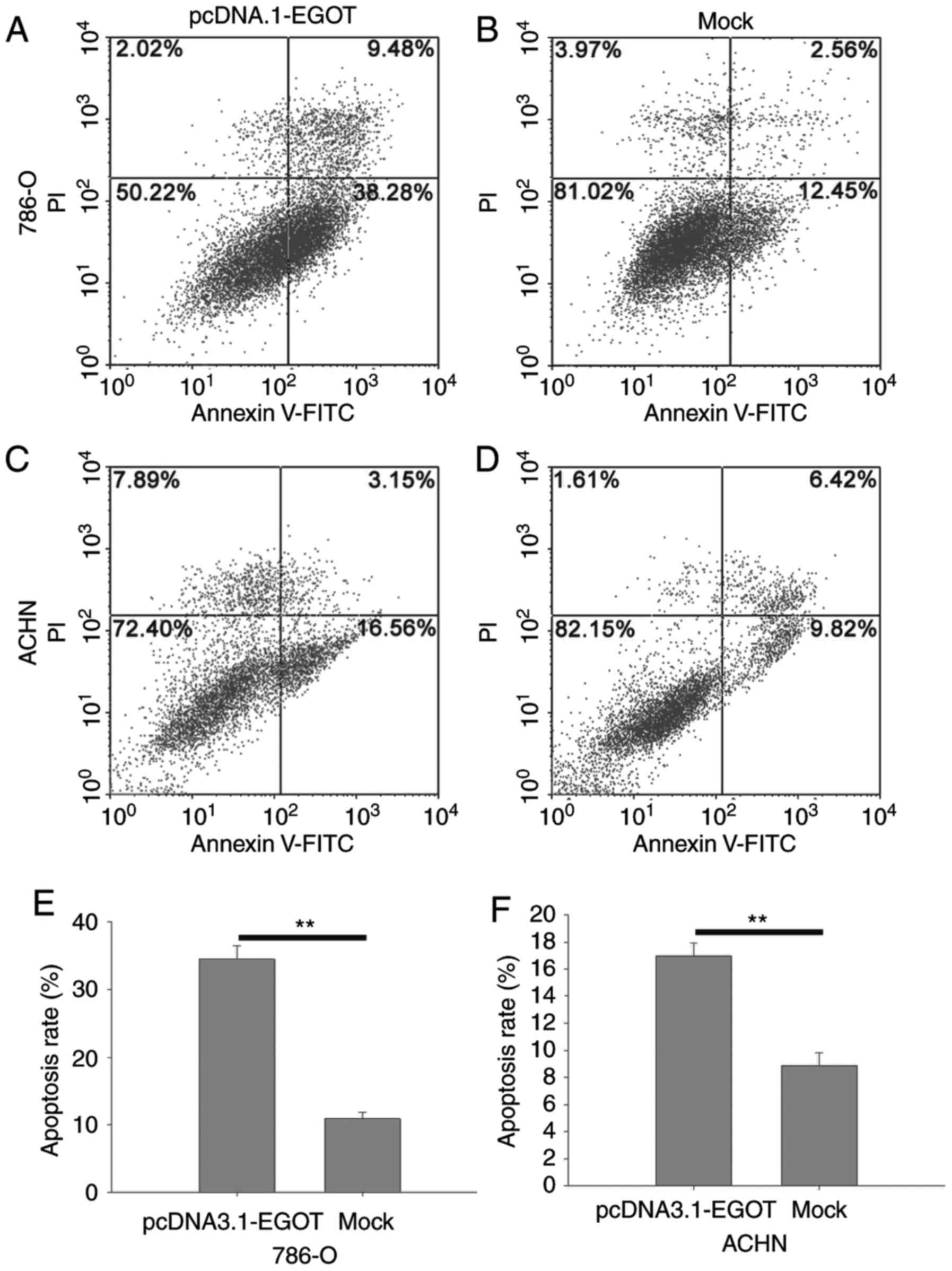
lncRNA EGOT induces cell
apoptosis
The percentage of apoptotic cells was quantified by
flow cytometry (Fig. 7). The
results demonstrated that the apoptotic rate in 786-O cells
transfected with pcDNA3.1-EGOT was 34.50%, compared with an
apoptotic rate of 10.93% in mock cells (P<0.01). In ACHN cells,
the apoptosis rate of cells transfected with pcDNA3.1-EGOT was
16.96% compared with an apoptotic rate of 8.89% in mock cells
(P<0.01). These results suggested that EGOT may induce cellular
apoptosis in RCC cell lines.
Discussion
lncRNAs have been implicated in multiple molecular
mechanisms, including transcriptional regulation,
post-transcriptional regulation and processing of other short
lncRNAs (20). Accumulating
evidence has demonstrated that lncRNA serves an important role in
tumorigenesis and the development of RCC. Malouf et al
(18) identified 1,934 expressed
lncRNAs by next-generation deep sequencing in 475 cases of primary
clear cell (cc)RCC, and the expression of 1,070 lncRNAs
demonstrated a strong association with the mRNA expression of their
neighboring genes. In another study, Blondeau et al
(21) reported 1,308 dysregulated
lncRNAs in ccRCC. Another microarray assay revealed 27,279 lncRNAs
in RCC samples, of which 897 were markedly dysregulated (22). To date, it appears that only a
small number of studies have investigated the function or
underlying mechanism of lncRNAs in RCC (23–25),
and the way in which lncRNAs regulate RCC cell behavior remains to
be identified.
lncRNA EGOT, located at 3p26.1, is nested within an
intron of the gene encoding inositol triphosphate receptor type 1,
and may be involved in eosinophil development and in the maturation
of eosinophils, in addition to regulating protein and
eosinophil-derived neurotoxin expression (26). A subsequent study (17) revealed that lncRNA EGOT was
downregulated in breast cancer, and that decreased expression
levels of EGOT were significantly associated with larger tumor
size, lymph node metastasis and poor prognosis. However, it appears
that no studies have investigated the role of EGOT in RCC. Based on
previously published studies (17,18,26),
the expression and function of EGOT was examined in RCC in the
present study.
In the present study, the expression levels of EGOT
in RCC and adjacent normal tissues was detected by RT-qPCR
analysis, and the results revealed that EGOT was downregulated in
RCC tissues compared with adjacent normal tissues. The function of
EGOT in RCC cells (786-O and ACHN) was additionally examined. The
results revealed that EGOT upregulation partially suppressed RCC
cell proliferation, migration and invasion, and induced RCC cell
apoptosis. EGOT upregulation resulted in slight alterations in cell
proliferation, although this was not statistically significant,
which suggested that EGOT primarily affects RCC cell migration,
invasion and apoptosis. Therefore, EGOT may serve as a tumor
suppressor in RCC.
There is only a small number of studies that have
investigated the expression or function of lncRNAs in RCC. lncRNA
urothelial carcinoma-associated 1 (UCA1), metastasis-associated
lung adenocarcinoma transcript-1 (MALAT1), homeobox transcript
antisense RNA (HOTAIR) and renal cell carcinoma related
transcript-1 (RCCRT1) have been demonstrated to be upregulated in
RCC (27–31). In particular, the expression levels
of RCCRT1 and MALAT1 were associated with poor prognosis, and UCA1,
HOTAIR and RCCRT1 were associated with RCC cell migration, invasion
or proliferation (27–31). lncRNA growth arrest-specific
transcript 5, maternally expressed 3, cancer susceptibility
candidate 2 and TRIM52 antisense RNA 1 were downregulated in RCC,
affecting cell function (23,25,32,33).
Another study revealed that lncRNA-suppressing androgen receptor in
renal cell carcinoma may suppress hypoxic cell cycle progression in
von Hippel-Lindau (VHL)-mutant RCC cells; however, SARCC may induce
hypoxic cell cycle progression in VHL-restored RCC cells (34), which suggested that lncRNA SARCC
may be a potential novel therapeutic biomarker. Therefore, the
roles of lncRNAs in RCC remain to be identified and a greater
number of studies that focus on this are required.
In conclusion, lncRNA EGOT was downregulated in RCC
tissues compared with normal tissues. EGOT may affect RCC cell
proliferation, migration, invasion and apoptosis, which indicated
that EGOT may serve an important role in the tumorigenesis of RCC.
In a future study, a dual-luciferase assay will be performed to
explore the association between EGOT and miR-1178-3p or miR-586,
which were identified in the present study to be putative
regulators of EGOT. The possible downstream effector would be
screened by microarray analysis and western blotting.
Acknowledgements
The present study was supported by the Guangdong
Natural Science Foundation (grant nos. 2014A030313709 and
2015A030313889), the Fund of Shenzhen Key Laboratory (grant no.
ZDSYS201504301045406) and the ‘San-ming’ Project of Medicine in
Shenzhen.
References
|
1
|
Chow WH and Devesa SS: Contemporary
epidemiology of renal cell cancer. Cancer J. 14:288–301. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iliopoulos O: Molecular biology of renal
cell cancer and the identification of therapeutic targets. J Clin
Oncol. 24:5593–5600. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu WS, Liu YD, Fu Q, Zhang WJ, Xu L,
Chang Y and Xu JJ: Prognostic significance of ubiquinol-cytochrome
c reductase hinge protein expression in patients with clear cell
renal cell carcinoma. Am J Cancer Res. 6:797–805. 2016.PubMed/NCBI
|
|
4
|
Xiang C, Cui SP and Ke Y: MiR-144 inhibits
cell proliferation of renal cell carcinoma by targeting MTOR. J
Huazhong Univ Sci Technolog Med Sci. 36:186–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu Z, Ni L, Chen D, Su Z, Yu W, Zhang Q,
Wang Y, Li C, Gui Y and Lai Y: Expression and clinical significance
of RCDG1 in renal cell carcinoma: A novel renal cancer-associated
gene. Mol Med Rep. 10:1583–1589. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang Z, Chu PG, Woda BA, Rock KL, Liu Q,
Hsieh CC, Li C, Chen W, Duan HO, McDougal S and Wu CL: Analysis of
RNA-binding protein IMP3 to predict metastasis and prognosis of
renal-cell carcinoma: A retrospective study. Lancet Oncol.
7:556–564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou X, Chen J and Tang W: The molecular
mechanism of HOTAIR in tumorigenesis, metastasis, and drug
resistance. Acta Biochim Biophys Sin (Shanghai). 46:1011–1015.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iranpour M, Soudyab M, Geranpayeh L,
Mirfakhraie R, Azargashb E, Movafagh A and Ghafouri-Fard S:
Expression analysis of four long noncoding RNAs in breast cancer.
Tumour Biol. 37:2933–2940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan WG, Xu DH, Xu C, Ding CL, Ning FL,
Zhou YL, Ma LB, Liu CM and Han X: Silencing of long non-coding RNA
ANRIL inhibits the development of multidrug resistance in gastric
cancer cells. Oncol Rep. 36:263–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Melissari MT and Grote P: Roles for long
non-coding RNAs in physiology and disease. Pflugers Arch.
468:945–958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui M, You L, Ren X, Zhao W, Liao Q and
Zhao Y: Long non-coding RNA PVT1 and cancer. Biochem Biophys Res
Commun. 471:10–14. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nowacki M, Vijayan V, Zhou Y, Schotanus K,
Doak TG and Landweber LF: RNA-mediated epigenetic programming of a
genome-rearrangement pathway. Nature. 451:153–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pandey RR, Mondal T, Mohammad F, Enroth S,
Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D and Kanduri C:
Kcnq1ot1 antisense noncoding RNA mediates lineage-specific
transcriptional silencing through chromatin-level regulation. Mol
Cell. 32:232–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Autuoro JM, Pirnie SP and Carmichael GG:
Long noncoding RNAs in imprinting and X chromosome inactivation.
Biomolecules. 4:76–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu SP, Zhang JF, Sui SY, Bai NX, Gao S,
Zhang GW, Shi QY, You ZL, Zhan C and Pang D: Downregulation of the
long noncoding RNA EGOT correlates with malignant status and poor
prognosis in breast cancer. Tumour Biol. 36:9807–9812. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malouf GG, Zhang J, Yuan Y, Compérat E,
Rouprêt M, Cussenot O, Chen Y, Thompson EJ, Tannir NM, Weinstein
JN, et al: Characterization of long non-coding RNA transcriptome in
clear-cell renal cell carcinoma by next-generation deep sequencing.
Mol Oncol. 9:32–43. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kunej T, Obsteter J, Pogacar Z, Horvat S
and Calin GA: The decalog of long non-coding RNA involvement in
cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 51:344–357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blondeau JJ, Deng M, Syring I, Schrödter
S, Schmidt D, Perner S, Müller SC and Ellinger J: Identification of
novel long non-coding RNAs in clear cell renal cell carcinoma. Clin
Epigenetics. 7:102015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin C, Han Z, Qian J, Bao M, Li P, Ju X,
Zhang S, Zhang L, Li S, Cao Q, et al: Expression pattern of long
non-coding RNAs in renal cell carcinoma revealed by microarray.
PLoS One. 9:e993722014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Z, Yan HY, Xia SY, Zhang C and Xiu YC:
Downregulation of long non-coding RNA TRIM52-AS1 functions as a
tumor suppressor in renal cell carcinoma. Mol Med Rep.
13:3206–3212. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang M, Huang T, Luo G, Huang C, Xiao XY,
Wang L, Jiang GS and Zeng FQ: Long non-coding RNA MEG3 induces
renal cell carcinoma cells apoptosis by activating the
mitochondrial pathway. J Huazhong Univ Sci Technolog Med Sci.
35:541–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wagner LA, Christensen CJ, Dunn DM,
Spangrude GJ, Georgelas A, Kelley L, Esplin MS, Weiss RB and Gleich
GJ: EGO, a novel, noncoding RNA gene, regulates eosinophil granule
protein transcript expression. Blood. 109:5191–5198. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Wang T, Li Y, Chen D, Yu Z, Jin L,
Ni L, Yang S, Mao X, Gui Y and Lai Y: Identification of long-non
coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol Med
Rep. 13:3326–3334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pei CS, Wu HY, Fan FT, Wu Y, Shen CS and
Pan LQ: Influence of curcumin on HOTAIR-mediated migration of human
renal cell carcinoma cells. Asian Pac J Cancer Prev. 15:4239–4243.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Y, Liu J, Zheng Y, You L, Kuang D and
Liu T: Suppressed expression of long non-coding RNA HOTAIR inhibits
proliferation and tumourigenicity of renal carcinoma cells. Tumour
Biol. 35:11887–11894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song S, Wu Z, Wang C, Liu B, Ye X, Chen J,
Yang Q, Ye H, Xu B and Wang L: RCCRT1 is correlated with prognosis
and promotes cell migration and invasion in renal cell carcinoma.
Urology. 84:730.e1–e7. 2014. View Article : Google Scholar
|
|
32
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao Y, Xu R, Xu X, Zhou Y, Cui L and He X:
Downregulation of lncRNA CASC2 by microRNA-21 increases the
proliferation and migration of renal cell carcinoma cells. Mol Med
Rep. 14:1019–1025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz
TA, Zheng J and Chang C: Differential regulation of lncRNA-SARCC
suppresses VHL-mutant RCC cell proliferation yet promotes
VHL-normal RCC cell proliferation via modulating androgen
receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 35:4866–4880.
2016. View Article : Google Scholar : PubMed/NCBI
|