Introduction
Globally, cervical cancer has the second highest
incidence among gynecological cancers and is a major cause of
cancer-associated mortality in women (1). It is particularly widespread in
developing countries, indicating the importance of screening
programs for cervical cancer (2,3). It
is estimated that there will be 529,800 new cases and ~275,100
deaths each year worldwide (4). It
is considered that early sexual intercourse, promiscuity and human
papillomavirus (HPV) infection, particularly HPV16, are closely
associated with the development of cervical cancer (5). At present, the first-line treatments
for patients with cervical cancer are surgical resection,
chemotherapy and radiotherapy (6).
Despite advances in the diagnosis, treatment and prevention of the
disease, the prognosis of patients remains poor. The median
progression-free survival and overall survival range between 2.5
and 13.2 months, and 4.2 and 12.87 months, respectively (7,8).
Therefore, a complete understanding of the mechanisms underlying
the occurrence and development of cervical cancer are required for
the development of novel, effective therapeutic strategies.
MicroRNAs (miRNAs) are endogenous, conserved and
non-coding RNAs of 19–25 nucleotides in length that originate from
distinct hairpin precursors that are present in animals, plants and
fungi (9,10). miRNAs post-transcriptionally
downregulate the expression of various target genes via direct
interaction with the 3′-untranslated regions (UTRs) of their target
genes in a base pairing manner, which leads to mRNA degradation
and/or translational inhibition (11). Notably, increasing evidence
indicates that miRNAs are abnormally expressed in various human
cancer types and have important functions in various areas of
oncogenesis, including survival, proliferation, the cell cycle,
apoptosis, angiogenesis, migration, invasion and metastasis
(12–14). Specifically, numerous studies have
reported that miRNAs have important roles in cervical cancer
carcinogenesis and progression via the regulation of various
protein-coding genes (15–17). For example, the expression levels
of miRNA-206 (miR-206) are reduced in cervical cancer tissues and
are significantly associated with adverse clinicopathological
features, including advanced Federation of Gynecology and
Obstetrics (FIGO) stage, positive lymph node metastasis, poor
differentiation and HPV infection. Furthermore, miR-206 was
demonstrated to exhibit tumor suppressor activity in cervical
cancer by suppressing cell proliferation, migration and invasion,
and enhancing apoptosis (18).
Therefore, regarding miRNAs may be developed as novel therapeutic
targets for patients with cervical cancer.
miR-211 has been investigated in various forms of
human cancer (19–21). However, to the best of our
knowledge, there is currently no information available concerning
the role of miR-211 in cervical cancer. Therefore, the present
study aimed to investigate the expression of miR-211 in cervical
cancer tissues and cell lines, and the biological roles of miR-211
in cervical cancer cells. In addition, the potential molecular
mechanisms underlying its tumor suppressive roles were also
investigated. The results of the present study may contribute
towards identifying a novel therapeutic target for cervical
cancer.
Materials and methods
Tissue specimens
Cervical cancer tissues and corresponding adjacent
normal tissues were collected form 34 patients (age range, 39–72
years) who underwent surgical resection without radiotherapy and/or
chemotherapy in the Department of Gynaecology, Songgang People's
Hospital (Shenzhen, China) between January 2010 and July 2013. All
fresh tissues were immediately frozen in liquid nitrogen and stored
at −78°C until further experiments. The present study was approved
by the Research Ethics Committee of Songgang People's Hospital.
Written informed consent was also obtained from all patients that
participated in the study.
Cell lines and culture conditions
Four human cervical cancer cell lines (HeLa, C33A,
SiHa and CaSki), a normal human cervix epithelial cell line
(Ect1/E6E7) and the 293T cell line were obtained from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). Cells were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 mg/ml penicillin and 100 mg/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a
humidified atmosphere with 5% CO2.
Oligonucleotide transfection
The human miR-211 mimic and miRNA mimic negative
control (miR-NC) were designed and provided by Shanghai GenePharma
Co., Ltd. (Shanghai, China). The miR-211 mimics sequence was
5′-UUCCCUUUGUCAUCCUUCGCCU-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. Small interfering (si)RNA targeting
zinc finger E-box binding homeobox 1 (ZEB1) and NC siRNA were
chemically synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou,
China). The ZEB1 siRNA sequence was 5′-CACAGAUACGGCAAAAGAUdTdT-3′
and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. HeLa and
C33A cells were seeded into 6-well plates at a density of
8×105 cells per well, and transfected with miR-211
mimics (100 pmol), miR-NC (100 pmol), ZEB1 siRNA (100 pmol) or NC
siRNA (100 pmol) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at room temperature. Transfected cells
were used for subsequent experiments. Then, 48 h after
transfection, RT-qPCR was performed to detect miR-211 or ZEB1 mRNA
expression. MTT and cell migration and invasion assays were
conducted at 24 h and 48 h following transfection. Western blot
analysis was carried out 72 h following transfection.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells by
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The relative expression
of miR-211 was determined using a SYBR PrimeScript miRNA RT-PCR kit
(Takara Biotechnology Co., Ltd., Dalin, China) according to the
manufacturer's protocol, with U6 small nuclear RNA as an internal
control. The thermocycling conditions for qPCR were as follows:
42°C for 5 min, 95°C for 10 sec, followed by 40 cycles of 95°C for
5 sec, 55°C for 30 sec and 72°C for 30 sec. For ZEB1 mRNA
expression, cDNA was synthesized with an M-MLV Reverse
Transcription System (Promega Corporation, Madison, WI, USA), which
was followed by qPCR using the reagents of the SYBR Green I Mix
(Takara Biotechnology Co., Ltd., Dalian, China). The temperature
protocol for reverse transcription was as follows: 95°C for 2 min;
20 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min; and
72°C for 5 min. The thermocycling conditions for qPCR were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. The primers were designed as follows: miR-211,
5′-GATCTTCCCTTTGTCATCC-3′ (forward) and 5′-GTGTCGTGGAGTCGGCAA-3′
(reverse); U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); ZEB1,
5′-AAGTGGCGGTAGATGGTA-3′ (forward) and 5′-TTGTAGCGACTGGATTTT-3′
(reverse); and GAPDH, 5′-AACGGATTTGGTCGTATTG-3′ (forward) and
5′-GGAAGATGGTGATGGGATT-3′ (reverse). GAPDH was used as an internal
control for ZEB1 mRNA expression and mRNA levels were quantified
using the 2−ΔΔCq method (22). Each sample was performed in
triplicate.
MTT assay
Cell proliferation was assessed using an MTT assay
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Transfected HeLa
and C33A cells were collected after 24 h incubation and seeded in
96-well plates (3×103 cells/well). Subsequently, cells
were incubated at 37°C with 5 % CO2 for 24, 48, 72 and
96 h. At each time point, an MTT assay was performed by adding 20
µl MTT solution (5 mg/ml) to each well and incubating at 37°C for 4
h. Dimethyl sulfoxide (200 µl; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) was added and the absorbance at 490 nm was
determined using a microplate reader. All experiments were
performed in triplicate.
Cell migration and invasion
assays
Transwell chambers (pore size, 8 mm; Corning
Incorporated, Corning, NY, USA) were used for cell migration and
invasion assays. For the cell migration assay, 1×105
cells were suspended in 100 µl of FBS-free DMEM medium and added
into the upper Transwell chamber. For the cell invasion assay,
1×105 cells were suspended in 100 µl of FBS-free DMEM
medium and added into the upper part of a Transwell chamber coated
with 50 µl Matrigel (2 mg/ml; BD Biosciences, San Jose, CA, USA).
For the chemoattractant, 500 µl DMEM medium containing 20% FBS was
added to the lower chamber. Following incubation for 48 h, the
cells remaining on the upper surface of the membrane were carefully
removed with cotton swabs. The cells that had crossed the membrane
onto the lower surface were fixed with 100% methanol at room
temperature for 10 min, stained with 0.1% crystal violet (Beyotime
Institute of Biotechnology, Haimen, China) at room temperature for
10 min and counted under an IX51 inverted light microscope (×200
magnification; Olympus Corporation, Tokyo, Japan). The average cell
number in five fields was taken as the final result for both
migration and invasion assays. All experiments were performed in
triplicate.
Bioinformatics analysis
The putative target genes for miR-211 were predicted
by bioinformatics analysis using TargetScan (http://www.targetscan.org/) (23).
Luciferase reporter assay
Wild-type (Wt) or mutant (Mut) versions of 3′UTR of
ZEB1, containing the putative binding sites for miR-211, were
separately cloned into the pMIR-REPORT miRNA Expression firefly
Reporter vectors. pRL-CMV was obtained from Promega Corporation
(E2261). For the luciferase reporter assay, 293T cells were seeded
in 24-well plates at a density of 50–60% confluence. Following
incubation overnight at 37°C, cells were transfected with miR-211
mimics (50 pmol) or miR-NC (50 pmol), in addition to
pMIR-ZEB1-3′UTR Wt (0.2 µg) or pMIR-ZEB1-3′UTR Mut (0.2 µg), using
Lipofectamine 2000. After 48 h at 37°C, cells were harvested and
luciferase activities were determined with a Dual-Luciferase
Reporter assay system (Promega Corporation), according to the
manufacturer's protocol. Renilla luciferase activity served as an
internal reference. All experiments were performed in
triplicate.
Western blot analysis
Total proteins were isolated from cells or tissues
using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology). The concentration of total protein was
determined by a BCA protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts of protein (30 µg) were subjected to
10% SDS-PAGE and transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA). The membranes were
subsequently blocked with 5% non-fat milk in TBS at room
temperature for 2 h and incubated at 4°C overnight with primary
antibodies, followed by incubation with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) for 2 h at room temperature. The primary
antibodies used in the present study were as follows: Mouse
anti-human monoclonal ZEB1 (sc-81428; 1:1,000; Santa Cruz
Biotechnology, Inc.) and mouse anti-human GAPDH (sc-59540; 1:1,000;
Santa Cruz Biotechnology, Inc.). Protein bands were detected by
Western Chemiluminescent HRP Substrate (ECL; EMD Millipore). The
band density of each protein was quantified after normalization to
GAPDH with ImageJ version 1.49 (National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
Data are presented as the mean + standard deviation.
Data were analyzed using student's t-test or one-way analysis of
variance with the Student-Newman-Keuls multiple comparisons test.
P<0.05 was considered to indicate a statistically significant
difference. SPSS version 13.0 software (SPSS, Inc., Chicago, IL,
USA) was used for the statistical analysis.
Results
miR-211 is downregulated in cervical
cancer
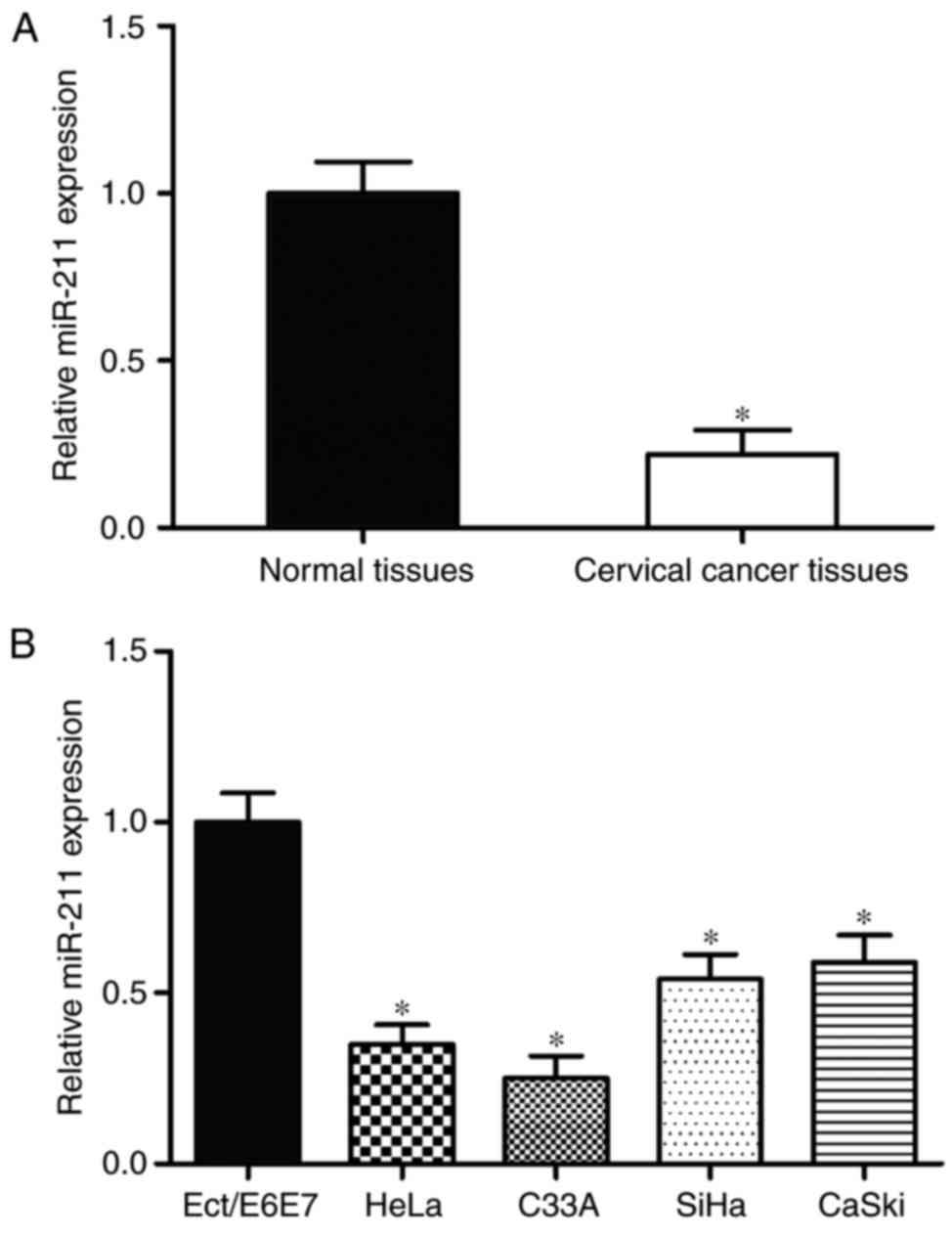
The expression of miR-211 in cervical cancer tissues
and corresponding adjacent normal tissues was measured by using
RT-qPCR. As demonstrated in Fig.
1A, miR-211 expression was significantly lower in cervical
cancer tissues compared with adjacent normal tissues (P<0.05).
Furthermore, the expression levels of miR-211 in cervical cancer
cell lines were also determined. Compared with the Ect1/E6E7 normal
human cervix epithelial cell line, HeLa, C33A, CaSki and SiHa
cervical cancer cell lines exhibited relatively low miR-211
expression (P<0.05; Fig. 1B).
Collectively, these results indicate that miR-211 is downregulated
in cervical cancer development.
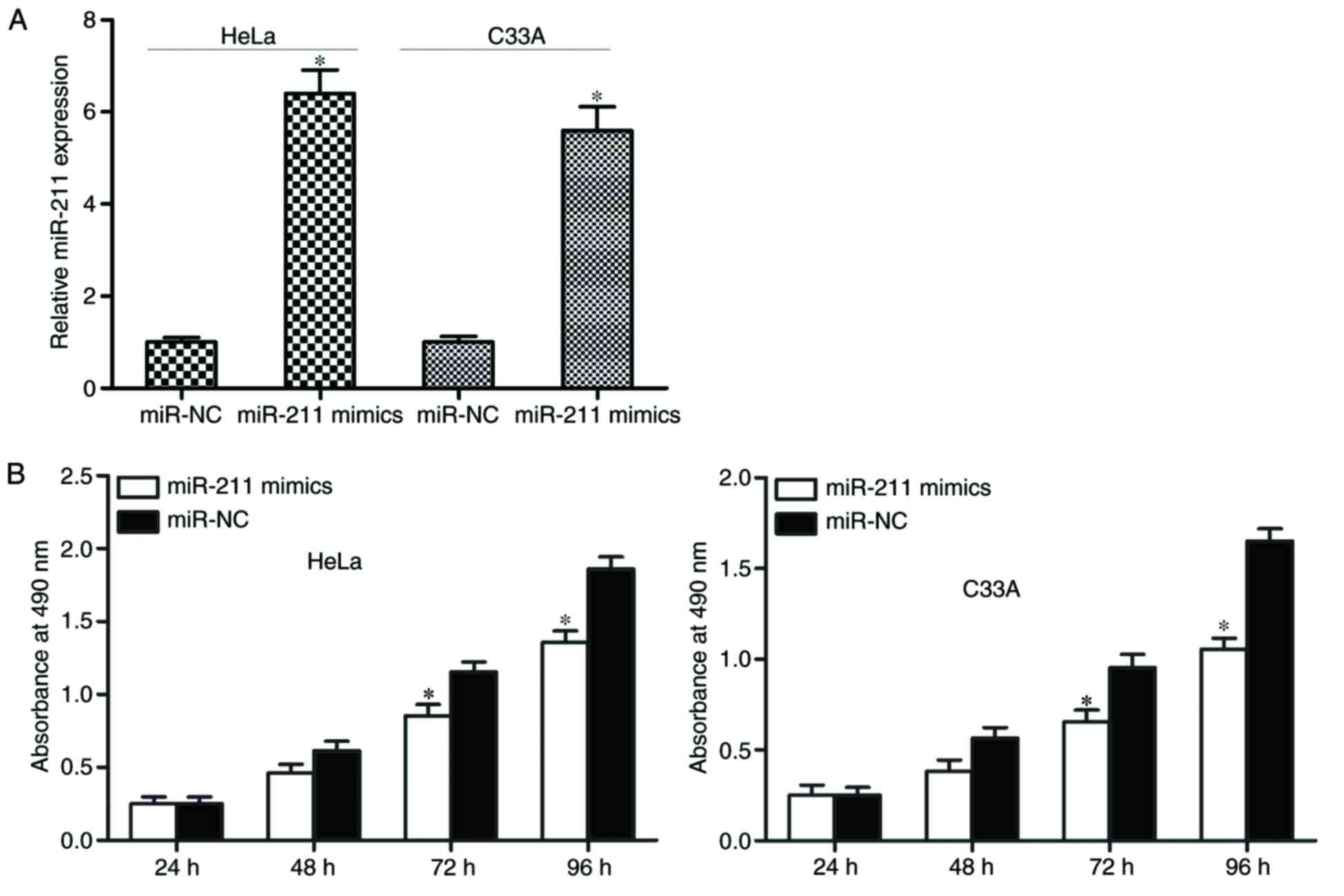
miR-211 inhibits HeLa and C33A cell
proliferation
To identify the biological roles of miR-211 in
cervical cancer, HeLa and C33A cells were transfected with miR-211
mimics or miR-NC. The transfection efficiency was evaluated using
RT-qPCR, which demonstrated that miR-211 was markedly upregulated
in miR-211 mimic-transfected HeLa and C33A cells compared with
miR-NC-transfected cells (P<0.05; Fig. 2A).
Subsequently, the effect of miR-211 on HeLa and C33A
cell proliferation was determined by an MTT assay. As demonstrated
in Fig. 2B, upregulation of
miR-211 significantly suppressed the growth rate of HeLa and C33A
cells at 72 and 96 h.
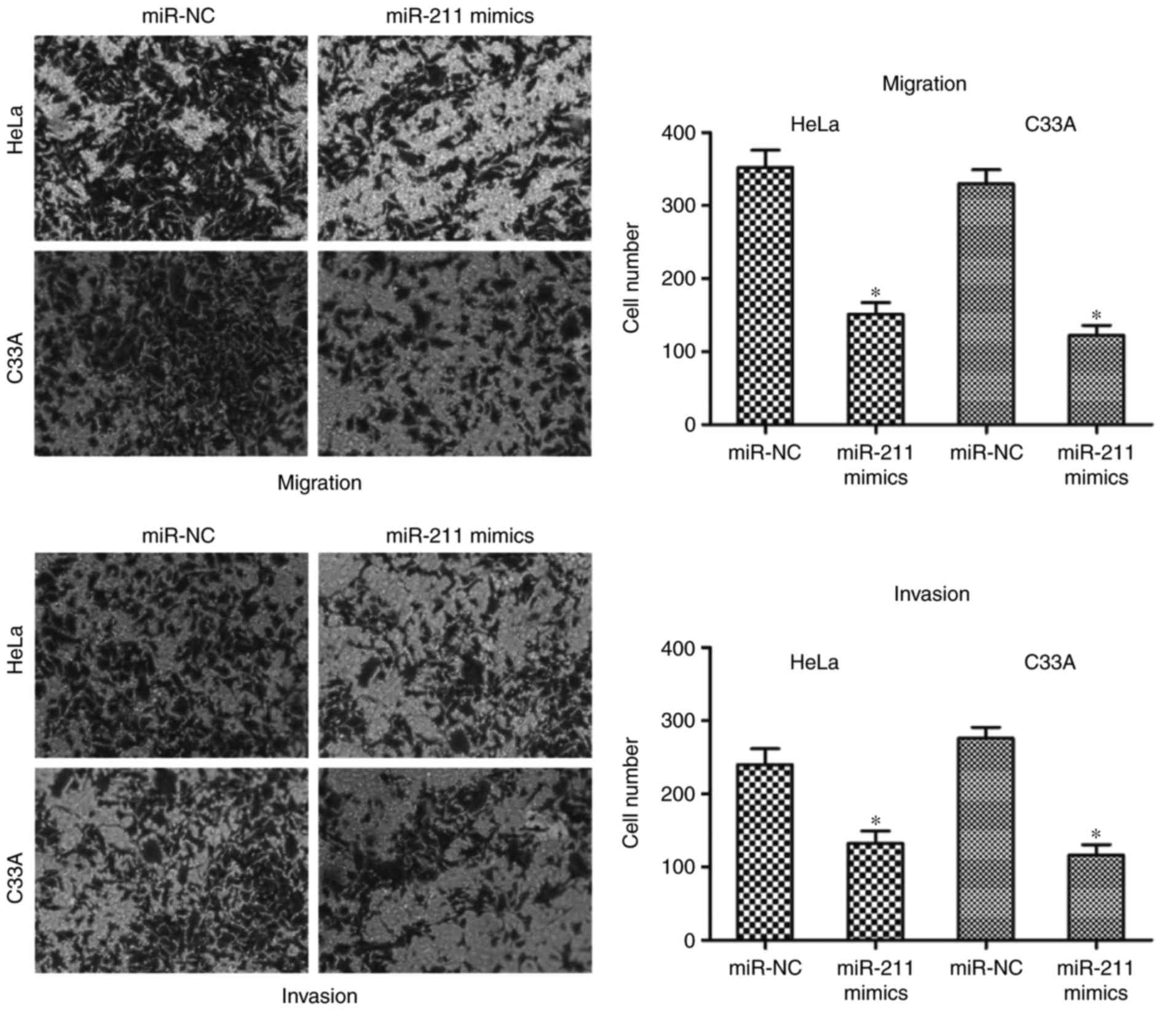
miR-211 inhibits HeLa and C33A cell
migration and invasion
The effects of miR-211 on HeLa and C33A cell
migration and invasion capacities were assessed by cell migration
and invasion assays. Results of cell migration and invasion assays
revealed that overexpression of miR-211 using mimics reduced the
migration and invasion abilities of HeLa and C33A cells compared
with miR-NC groups (P<0.05; Fig.
3). These results indicate that miR-211 may act as a tumor
suppressor in cervical cancer by inhibiting cell growth and
metastasis.
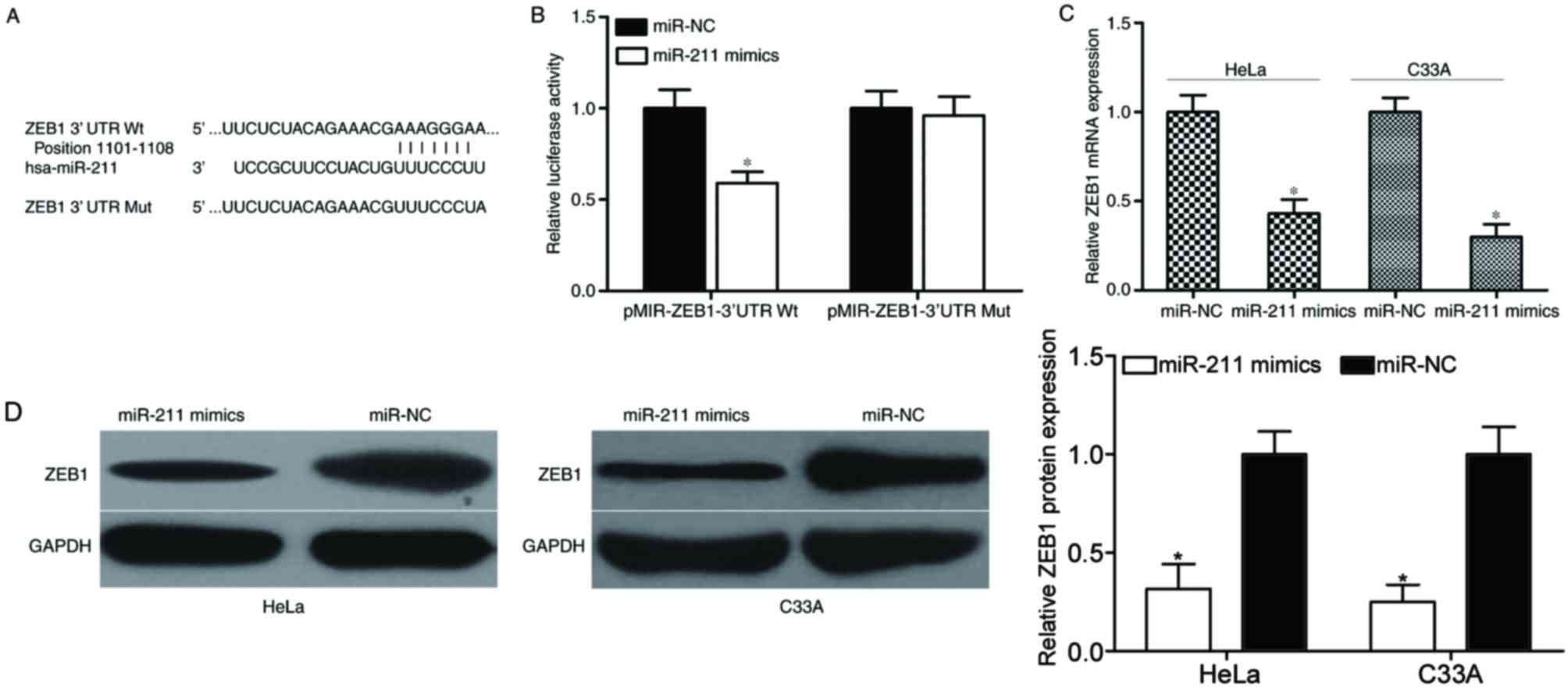
ZEB1 is a direct target of miR-211 in
cervical cancer
To investigate the potential molecular mechanism
underlying the inhibition of HeLa and C33A cell growth and
metastasis by miR-211, bioinformatics analysis was performed to
identify the potential target genes of miR-211. Based on
bioinformatics analysis, hundreds of potential targets were
identified. Among these putative targets, a number of them had
previously been reported as direct targets of miR-211 in different
cancer types, including preferentially expressed antigen in
melanoma (PRAME) as a target of miR-211 in melanoma (24), chromodomain helicase DNA binding
protein 5 (CHD5) in colorectal cancer (21), transforming growth factor β
receptor II (TGFβRII) in head and neck carcinomas (25), SATB homeobox 2 (SATB2) in
hepatocellular carcinoma (26) and
cyclin D1 in ovarian cancer (27).
The present study selected ZEB1 for further analysis as it was
previously reported to be expressed at abnormally high levels in
cervical cancer (28), and has
been implicated in the tumorigenesis and progression of cervical
cancer (29,30).
To confirm whether ZEB1 was a putative target of
miR-211, luciferase reporter assays were performed. 293T cells were
cotransfected with miR-211 mimics or miR-NC, and pMIR-ZEB1-3′UTR Wt
or pMIR-ZEB1-3′UTR Mut (Fig. 4A).
As demonstrated in Fig. 4B,
transfection with miR-211 mimics decreased the luciferase
activities of the luciferase reporter construct carrying the Wt
3′UTR, which contains the potential miR-211 binding site
(P<0.05). However, miR-211 overexpression did not affect the
luciferase activities in the luciferase reporter construct carrying
the Mt 3′UTR containing the potential miR-211 binding site.
Subsequently, RT-qPCR and western blotting analysis
was performed to measure ZEB1 mRNA and protein expression in HeLa
and C33A cells transfected with miR-211 mimics or miR-NC. The
results indicated that the mRNA (Fig.
4C) and protein (Fig. 4D)
levels of ZEB1 were significantly reduced following transfection
with miR-211 mimics, compared with the miR-NC-transfected cells
(P<0.05). The results of these experiments indicate that ZEB1
may be a direct target gene of miR-211 in cervical cancer.
Downregulation of ZEB1 inhibits HeLa
and C33A cell proliferation, migration and invasion
Finally, to investigate whether ZEB1 knockdown
exhibits similar tumor suppressive roles to miR-211 overexpression
in HeLa and C33A cells, siRNA targeting ZEB1 was employed to
downregulate ZEB1 expression. Following transfection, ZEB1
expression in HeLa and C33A cells was detected by western blot
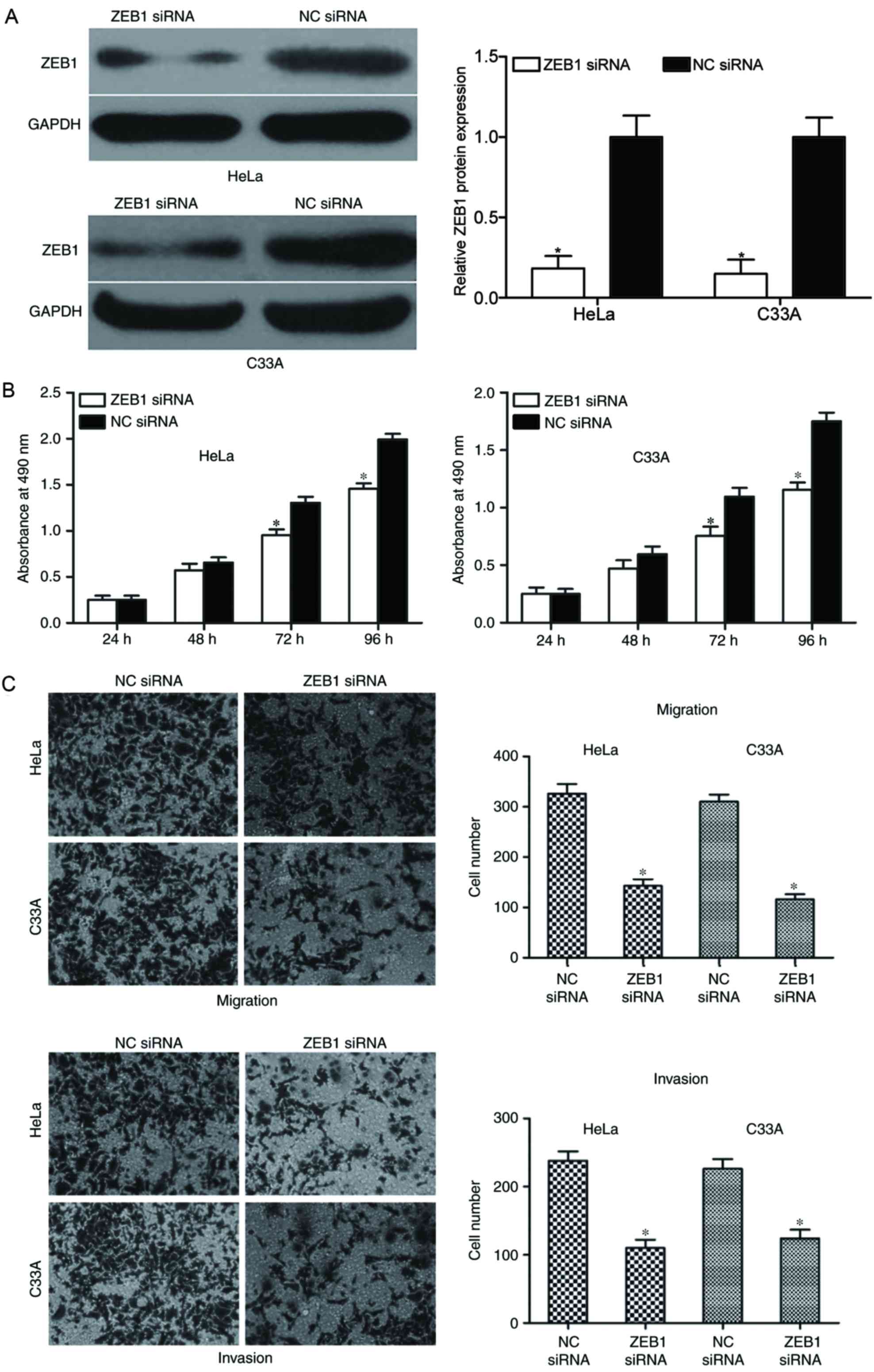
analysis. As demonstrated in Fig.
5A, the expression levels of ZEB1 were significantly reduced in
HeLa and C33A cells following transfection with ZEB1 siRNA,
compared with NC siRNA-transfected cells (P<0.05).
Subsequently, an MTT assay was performed to
determine the effect of ZEB1 downregulation on the proliferation of
HeLa and C33A cells. The results demonstrated that downregulation
of ZEB1 suppressed the proliferation of HeLa and C33A cells
compared with NC siRNA-transfected cells at 72 and 96 h (P<0.05;
Fig. 5B). Furthermore, the present
study also investigated the effect of ZEB1 knockdown on HeLa and
C33A cell motility by cell migration and invasion assays. The
results demonstrated that in ZEB1 downregulated HeLa and C33A
cells, the migration and invasion abilities were significantly
reduced compared with NC siRNA-transfected groups (P<0.05;
Fig. 5C). Collectively, these
results indicate that ZEB1 knockdown exhibited similar
tumor-suppressive effects to miR-211 overexpression in cervical
cancer, which further confirms that ZEB1 may be a direct functional
target gene of miR-211.
Discussion
miR-211 is located at chromosome 15q13, which is a
locus that is frequently deleted in cancer (31–33).
Numerous studies have reported that miR-211 is abnormally expressed
in various cancer types. For example, miR-211 was demonstrated to
be upregulated in oral carcinoma samples and high miR-211
expression was associated with advanced nodal metastasis, vascular
invasion and a poor prognosis (19). Expression levels of miR-211 were
also reported to be increased in colon cancer tissues and
statistically associated with age. Survival analysis revealed that
patients with colon cancer that exhibited high miR-211 expression
had a shorter survival time compared with patients with lower
miR-211 expression. Furthermore, miR-211 was validated as a risk
factor for colon cancer prognosis (20). An upregulation of miR-211 has also
been reported in colorectal cancer (21) and head and neck carcinomas
(25). However, in melanoma, Mazar
et al (34) reported that
miR-211 was downregulated in tumor cell lines compared with normal
melanocytes. In addition, expression levels of miR-211 were reduced
in melanoma tissues. In addition, Maftouh et al (35) demonstrated that miR-211 was
downregulated in pancreatic cancer and significantly associated
with prognosis, and miR-211 was also reported to be downregulated
in breast cancer (36),
hepatocellular carcinoma (26) and
ovarian cancer (27). These
studies indicate that the expression levels of miR-211 in human
cancers exhibits tissue specificity and may have important roles in
these types of human cancer.
Dysregulation of miR-211 expression is reported to
be implicated in the initiation and progression of human cancers.
In oral carcinoma, a high expression of miR-211 was associated with
increases in cell proliferation, migration and the formation of
anchorage-independent colonies (19). Cai et al (21) reported that miR-211 overexpression
enhanced the cell growth and invasion of colorectal cancer in
vitro and in vivo. Additionally, in non-small cell lung
cancer, miR-211 promoted cell proliferation, colony formation and
invasion (37). These results
indicate that miR-211 may exhibit oncogenic roles in human cancer.
However, in melanoma, miR-211 functioned as a tumor suppressor with
suppressive roles in cell growth and invasion (34). In pancreatic cancer, induction of
miR-211 expression reduced the migration and invasion of pancreatic
cancer cells. Furthermore, enhanced miR-211 expression enhanced the
chemosensitivity of pancreatic cancer cells to gemcitabine
(35). In breast cancer,
overexpression of miR-211 suppressed the growth, cell cycle,
migration and invasion of triple-negative breast cancer cells
(36). Additionally, in ovarian
cancer, enforced miR-211 expression inhibited the cell
proliferation, enhanced apoptosis and arrested cells in the
G0/G1-phase (27). Furthermore,
miR-211 has been identified as a tumor suppressor in numerous types
of human cancer, including hepatocellular carcinoma (26) and gastric cancer (38). These studies reported contradictory
results in that miR-211 was reported to be an oncogene in certain
cancer types and a tumor suppressor in others. These conflicting
results may be explained by the ‘imperfect complementarity’ of the
interactions between miRNAs and target genes (39).
To the best of our knowledge, the present study was
the first to provide sufficient evidence that the expression levels
of miR-211 were reduced in cervical cancer tissues and cell lines
compared with those in corresponding adjacent normal tissues and
the normal human cervix epithelial cell line, respectively.
Therefore, it may be hypothesized that miR-211 acts as a tumor
suppressor in cervical cancer carcinogenesis and progression.
Notably, overexpression of miR-211, using miR-211 mimics, inhibited
the cell proliferation, migration and invasion of cervical cancer
cells. These results indicate that miR-211 may provide potential
therapeutic targets for cervical cancer treatment.
miRNAs exert functional roles through incomplete
pairing with the 3′UTR of their target genes and regulating the
expression level of target genes. Therefore, it is important to
validate the direct target genes of miR-211. Previously, various
target genes of miR-211 were identified, including lacZ in oral
carcinoma (19), SRC kinase
signaling inhibitor 1 in non-small cell lung cancer (37), PRAME in melanoma (24), CHD5 in colorectal cancer (21), TGFβRII in head and neck carcinomas
(25), cell division cycle 25B in
breast cancer (36), SATB2 in
hepatocellular carcinoma (26),
and cyclin D1 and cyclin-dependent kinase 6 in ovarian cancer
(27). In the present study, a
molecular association between miR-211 and ZEB1 was demonstrated.
ZEB1, a member of the zinc finger family, is located on the short
arm of human chromosome 10 (40).
Previous studies have reported that ZEB1 was upregulated in breast,
ovarian, endometrial and prostate cancer. Upregulation of ZEB1 was
also reported to be associated with poor differentiation,
aggressive disease, the development of metastases and a poor
clinical prognosis in abovementioned cancer types (41–44).
Additionally, Ma et al (28) demonstrated that ZEB1 expression was
increased in cervical cancer tissues, and was significantly
associated with differentiation status, the occurrence of vascular
invasion and metastasis to lymph nodes in cervical cancer.
Furthermore, Chen et al (29) demonstrated that the expression of
ZEB1 was associated with FIGO stage and lymph node metastasis.
Functionally, downregulation of ZEB1 decreased the proliferation
and metastasis capacities of cervical cancer cells (30). These results indicate that ZEB1 may
be considered a valuable therapeutic target for cervical cancer
treatment.
In conclusion, the present study investigated
miR-211 expression and its functional roles in regulating the
growth and metastasis of cervical cancer cells at the cellular
level. The present study reported that miR-211 may act as a tumor
suppressor in cervical cancer, providing a theoretical basis for
its application in the treatment of cervical cancer.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nagamitsu Y, Nishi H, Sasaki T, Takaesu Y,
Terauchi F and Isaka K: Profiling analysis of circulating microRNA
expression in cervical cancer. Mol Clin Oncol. 5:189–194. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schiffman MH and Castle P: Epidemiologic
studies of a necessary causal risk factor: Human papillomavirus
infection and cervical neoplasia. J Natl Cancer Inst. 95:E22003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dueñas-Gonzalez A, Cetina L, Mariscal I
and de la Garza J: Modern management of locally advanced cervical
carcinoma. Cancer Treat Rev. 29:389–399. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roque DR, Wysham WZ and Soper JT: The
surgical management of cervical cancer: An overview and literature
review. Obstet Gynecol Surv. 69:426–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang N, Wei H, Yin D, Lu Y, Zhang Y, Zhang
Q, Ma X and Zhang S: MicroRNA-195 inhibits proliferation of
cervical cancer cells by targeting cyclin D1a. Tumour Biol.
37:4711–4720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rhoades MW, Reinhart BJ, Lim LP, Burge CB,
Bartel B and Bartel DP: Prediction of plant microRNA targets. Cell.
110:513–520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu Y, Zhou Y, Qu W, Deng M and Zhang C: A
Lasso regression model for the construction of microRNA-target
regulatory networks. Bioinformatics. 27:2406–2413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siomi H and Siomi MC: Posttranscriptional
regulation of microRNA biogenesis in animals. Mol Cell. 38:323–332.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gardiner AS, Twiss JL and
Perrone-Bizzozero NI: Competing interactions of RNA-binding
proteins, MicroRNAs, and their targets control neuronal development
and function. Biomolecules. 5:2903–2918. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Guan DH, Bi RX, Xie J, Yang CH
and Jiang YH: Prognostic value of microRNAs in gastric cancer: A
meta-analysis. Oncotarget. 8:55489–55510. 2017.PubMed/NCBI
|
|
14
|
Drusco A and Croce CM: MicroRNAs and
cancer: A long story for short RNAs. Adv Cancer Res. 135:1–24.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou D, Zhou Q, Wang D, Guan L, Yuan L and
Li S: The downregulation of MicroRNA-10b and its role in cervical
cancer. Oncol Res. 24:99–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang T, Zou P, Wang T, Xiang J, Cheng J,
Chen D and Zhou J: Down-regulation of miR-320 associated with
cancer progression and cell apoptosis via targeting Mcl-1 in
cervical cancer. Tumour Biol. 37:8931–8940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling S, Ruiqin M, Guohong Z, Bing S and
Yanshan C: Decreased microRNA-206 and its function in cervical
cancer. Eur J Gynaecol Oncol. 36:716–721. 2015.PubMed/NCBI
|
|
19
|
Chang KW, Liu CJ, Chu TH, Cheng HW, Hung
PS, Hu WY and Lin SC: Association between high miR-211 microRNA
expression and the poor prognosis of oral carcinoma. J Dent Res.
87:1063–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai K, Shen F, Cui JH, Yu Y and Pan HQ:
Expression of miR-221 in colon cancer correlates with prognosis.
Int J Clin Exp Med. 8:2794–2798. 2015.PubMed/NCBI
|
|
21
|
Cai C, Ashktorab H, Pang X, Zhao Y, Sha W,
Liu Y and Gu X: MicroRNA-211 expression promotes colorectal cancer
cell growth in vitro and in vivo by targeting tumor suppressor
CHD5. PLoS One. 7:e297502012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sakurai E, Maesawa C, Shibazaki M,
Yasuhira S, Oikawa H, Sato M, Tsunoda K, Ishikawa Y, Watanabe A,
Takahashi K, et al: Downregulation of microRNA-211 is involved in
expression of preferentially expressed antigen of melanoma in
melanoma cells. Int J Oncol. 39:665–672. 2011.PubMed/NCBI
|
|
25
|
Chu TH, Yang CC, Liu CJ, Lui MT, Lin SC
and Chang KW: miR-211 promotes the progression of head and neck
carcinomas by targeting TGFbetaRII. Cancer Lett. 337:115–124. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang G, Cui Y, Yu X, Wu Z, Ding G and Cao
L: miR-211 suppresses hepatocellular carcinoma by downregulating
SATB2. Oncotarget. 6:9457–9466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia B, Yang S, Liu T and Lou G: miR-211
suppresses epithelial ovarian cancer proliferation and cell-cycle
progression by targeting Cyclin D1 and CDK6. Mol Cancer. 14:572015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Zheng X, Zhou J, Zhang Y and Chen K:
ZEB1 promotes the progression and metastasis of cervical squamous
cell carcinoma via the promotion of epithelial-mesenchymal
transition. Int J Clin Exp Pathol. 8:11258–11267. 2015.PubMed/NCBI
|
|
29
|
Chen Z, Li S, Huang K, Zhang Q, Wang J, Li
X, Hu T, Wang S, Yang R, Jia Y, et al: The nuclear protein
expression levels of SNAI1 and ZEB1 are involved in the progression
and lymph node metastasis of cervical cancer via the
epithelial-mesenchymal transition pathway. Hum Pathol.
44:2097–2105. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ran J, Lin DL, Wu RF, Chen QH, Huang HP,
Qiu NX and Quan S: ZEB1 promotes epithelial-mesenchymal transition
in cervical cancer metastasis. Fertil Steril. 103(1606–14): e1–2.
2015.PubMed/NCBI
|
|
31
|
Feenstra M, Veltkamp M, van Kuik J,
Wiertsema S, Slootweg P, van den Tweel J, de Weger R and Tilanus M:
HLA class I expression and chromosomal deletions at 6p and 15q in
head and neck squamous cell carcinomas. Tissue Antigens.
54:235–245. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Natrajan R, Louhelainen J, Williams S,
Laye J and Knowles MA: High-resolution deletion mapping of
15q13.2-q21.1 in transitional cell carcinoma of the bladder. Cancer
Res. 63:7657–7662. 2003.PubMed/NCBI
|
|
33
|
Lipton L and Tomlinson I: The genetics of
FAP and FAP-like syndromes. Fam Cancer. 5:221–226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mazar J, DeYoung K, Khaitan D, Meister E,
Almodovar A, Goydos J, Ray A and Perera RJ: The regulation of
miRNA-211 expression and its role in melanoma cell invasiveness.
PLoS One. 5:e137792010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maftouh M, Avan A, Funel N, Frampton AE,
Fiuji H, Pelliccioni S, Castellano L, Galla V, Peters GJ and
Giovannetti E: miR-211 modulates gemcitabine activity through
downregulation of ribonucleotide reductase and inhibits the
invasive behavior of pancreatic cancer cells. Nucleosides
Nucleotides Nucleic Acids. 33:384–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song GQ and Zhao Y: MicroRNA-211, a direct
negative regulator of CDC25B expression, inhibits triple-negative
breast cancer cells' growth and migration. Tumour Biol.
36:5001–5009. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ye L, Wang H and Liu B: miR-211 promotes
non-small cell lung cancer proliferation by targeting SRCIN1.
Tumour Biol. 37:1151–1157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang CY, Hua L, Sun J, Yao KH, Chen JT,
Zhang JJ and Hu JH: MiR-211 inhibits cell proliferation and
invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp
Pathol. 8:14013–14020. 2015.PubMed/NCBI
|
|
39
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shen A, Zhang Y, Yang H, Xu R and Huang G:
Overexpression of ZEB1 relates to metastasis and invasion in
osteosarcoma. J Surg Oncol. 105:830–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singh M, Spoelstra NS, Jean A, Howe E,
Torkko KC, Clark HR, Darling DS, Shroyer KR, Horwitz KB, Broaddus
RR and Richer JK: ZEB1 expression in type I vs type II endometrial
cancers: A marker of aggressive disease. Mod Pathol. 21:912–923.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Graham TR, Zhau HE, Odero-Marah VA,
Osunkoya AO, Kimbro KS, Tighiouart M, Liu T, Simons JW and O'Regan
RM: Insulin-like growth factor-I-dependent up-regulation of ZEB1
drives epithelial-to-mesenchymal transition in human prostate
cancer cells. Cancer Res. 68:2479–2488. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Drake JM, Strohbehn G, Bair TB, Moreland
JG and Henry MD: ZEB1 enhances transendothelial migration and
represses the epithelial phenotype of prostate cancer cells. Mol
Biol Cell. 20:2207–2217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hurt EM, Saykally JN, Anose BM, Kalli KR
and Sanders MM: Expression of the ZEB1 (deltaEF1) transcription
factor in human: Additional insights. Mol Cell Biochem. 318:89–99.
2008. View Article : Google Scholar : PubMed/NCBI
|