Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2)
is a transcription factor that serves an important role in
protecting cells from oxidative stress (1). Nrf2 protein contains the Kelch-like
ECH-associated protein 1 (Keap1) binding domain. Under normal
conditions, Nrf2 forms a complex with Keap1 and is of a low
concentration in the cytoplasm (2). Researchers have reported that
reactive oxygen species (ROS), key factors that activates Nrf2,
induces the dissociation of Keap1 from Nrf2, permitting the
translocation of Nrf2 into the nucleus (3). Once in the nucleus, Nrf2
heterodimerizes with small muscle aponeurotic fibrosarcoma (Maf)
protein and then binds to the antioxidant response element (ARE) of
downstream antioxidant genes, activating their transcription
(4). An increasing body evidence
indicates that exercise upregulates the expression of Nrf2 in
various tissues, which protects cells from ROS-induced damage
(5–9). Our recent pilot study using mouse
skeletal muscle revealed no significant changes in the expression
levels of Nrf2 protein following eight weeks of aerobic exercise
training; however, this may be due to relatively low exercise
intensities. Notably, the expression levels of Nrf2 mRNA increased
significantly in the experiment. This discrepancy may suggest that
post-transcriptional control and/or post-translational modification
are involved in the regulation of Nrf2 expression in mouse skeletal
muscle post-exercise. Among the multiple factors that may affect
Nrf2 protein expression, microRNAs (miRNAs/miRs) have been proposed
as key post-transcriptional regulators of gene expression (10).
miRNAs are short non-coding RNAs of ~21 nucleotides
in length and serve a central role in many aspects of cell biology
(11,12). Typically, by binding to the 3′
untranslated region (3′UTR) of a complementary mRNA sequence,
miRNAs can directly degrade the transcript or inhibit translation
(11–13). Previous studies of normal and
cancer cells have reported that few miRNAs could directly or
indirectly regulate Nrf2 (14–19).
It has been suggested that miR-144 (15), miR-28 (14,20),
miR-153, miR-27a and miR-142-5p could directly regulate Nrf2
(18); miR-200a (16), miR-23a (17) and miR-141 (18,19)
targeted Keap1 (a negative regulator of Nrf2) to indirectly promote
the degradation of Nrf2 protein. In addition, miR-155 disrupted the
Nrf2-dependent signaling pathway in hepatic cells; however, whether
this occurs in a direct or indirect manner remains unknown
(21). Nevertheless, miRNAs
modulating the expression of Nrf2 induced by aerobic exercise
training have not been well studied. However, accumulating evidence
suggests that the expression levels of some miRNAs, including
miR-1, miR-107, miR-161, miR-23, miR-133 and miR-486, known as the
skeletal muscle-specific microRNAs (myomiRs), tend to be regulated
in response to exercise training (22,23).
On this basis, the aim of the present study was to
screen and identify the potential miRNAs that directly regulate
Nrf2 expression within the skeletal muscle of mice following
exercise. The present study first evaluated the effects of an
8-week aerobic exercise training program on the expression of Nrf2
mRNA and protein, and miRNAs in mouse skeletal muscle. Secondly,
the profile of differentially expressed miRNAs in the exercise and
non-exercise control groups was analyzed and filtered to determine
putative miRNAs that target the 3′UTR of Nrf2. Finally, in
vitro analysis was conducted to verify the selected candidate
miRNAs. The present study aimed to improve understanding of the
regulatory mechanism of miRNAs in the effects of aerobic exercise
training on the expression Nrf2 protein in skeletal muscle.
Materials and methods
Animal and exercise program
The present study was approved by the Animal Care
and Use Committee of Beijing Sport University (Beijing, China). A
total of 20 male C57BL/6J mice (20±2 g, 8-weeks-old) were purchased
from Charles River Development, Inc. (Beijing, China) with a body
weight of 18±2 g. The animals were housed indoors under a
temperature of 20–25°C, humidity of 50–70%, 12-h light/dark cycles,
and had ad libitum access to deionized water and standard
chow. They were randomly divided into the control (C; n=10) and
exercise (E; n=10) groups. Mice in the C group were housed without
exercise training. Mice in the E group were trained according to an
aerobic exercise protocol, which was adopted from a previous study
with some modifications (24).
Briefly, two days prior to the formal exercise protocol, the
animals were familiarized to running on a treadmill at a low
intensity for 10 min/day, following which the animals were made to
run on the treadmill for 1 h at 12 m/min. Mice of the E group were
trained 6 days per week for a total of 8 weeks. To avoid acute
effects from the last exercise session, the trained mice were able
to recover for at least 48 h with free access to food and water
prior to tissue collection. The mice were euthanized by cervical
dislocation and whole hind limb muscle samples (a mixture of
muscles) were excised, cleaned of blood and connective tissue,
rapidly frozen in liquid nitrogen, and stored at −80°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of Nrf2 mRNA
Total RNA was extracted from C2C12 cells (China
Agricultural University, Beijing, China) or 50 mg of whole
hind-limb muscles (a mixture of muscles) with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) following the manufacturer's instructions. RT-PCR was carried
out using the ReverTra Ace Qpcr RT kit (Toyobo Life Science, Osaka,
Japan), and was performed in a reaction volume of 20 µl containing
2 µl total RNA, 1 µl primer mix, 1 µl RT enzyme mix, 4 µl 5X RT
buffer and 6 µl nuclease-free water. For the synthesis of cDNA, the
reaction mixtures were incubated at 65°C for 5 min, 37°C for 15 min
and 98°C for 5 min. qPCR was performed on an ABI 7500 Real-time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
the SYBR Green Real-time PCR Master Mix kit (Toyobo Life Science)
with the previously synthesized cDNA template (FSQ-101 Toyobo Life
Science) in a reaction volume of 20 µl. The primers obtained from
Qiagen GmbH (Hilden, Germany) were as follows: Nrf2 (cat. no.
QT00095270) and 18S gene (cat. no. QT010036875) as a reference
gene, which were confirmed with software (ABI 7500RT PCR). qPCR was
performed with a final volume of 20 µl containing 10 µl SYBR Green
Real-time PCR Master Mix, 2 µl primers, 2 µl cDNA and 6 µl
nuclease-free water. The thermocycling conditions were as follows:
95°C for 10 min, then 40 cycles of 95°C for 15 sec, 60°C for 60 sec
and 72°C for 30 sec. The difference in expression between the E and
C groups was calculated using the 2−∆∆Cq method, as
described previously (25). To
assess the specificity of the amplified PCR products, melting curve
analysis was conducted following the last cycle.
Western blotting analysis
Once total protein (20 µg) from 50 mg mouse skeletal
muscle samples or total protein (10 µg) from C2C12 cells
(2.5×105 per well) was extracted using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China), the protein concentration was
determined with a bicinchoninic acid kit (Thermo Fisher Scientific,
Inc.). Proteins were separated on a Bolt Bis-Tris plus 4–12% gel
(Thermo Fisher Scientific, Inc.) by electrophoresis at 200 V for 35
min; the fractionated proteins were then transferred to a
nitrocellulose transfer membrane using the iBlot Gel Transfer
System (Invitrogen; Thermo Fisher Scientific, Inc.) at 20 V for 7
min and 30 sec. The membrane was blocked for 60 min at the room
temperature in Tris-buffered saline with 0.01% Tween-20 (TBST)
containing 5% nonfat milk. The samples were then incubated
overnight at 4°C using the following primary antibodies: Anti-Nrf2
antibodies (1:200, cat. no. sc-722; and 1:2,000, cat. no.
sc-365949; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), and
β-actin antibody (1:1,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.). Of the primary antibodies against NRF2, the
sc-722 antibody was used in animal experiments and the sc-365949
antibody was used in cell experiments. The blots were washed 3
times with 1X TBST, then incubated with an anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat. no.
zb-2301; OriGene Technologies, Inc., Beijing, China) at 37°C for 1
h. Following washing with 1X TBST, the blots were visualized with
an ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.).
The density of the protein bands was analyzed using the Molecular
Imager® ChemiDoc™ XRS + with Image Lab™ Software version
6.0.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The protein
expression levels were normalized to that of β-actin and then
expressed as the fold change of the control group.
miScript miRNA PCR array
Total RNA of skeletal muscle from 50 mg muscle
samples was extracted using an miRNeasy Mini kit (Qiagen GmbH). The
total RNA was assessed with a NanoDrop 2000 spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington,
DE, USA), and its quality fulfilled the requirements of total RNA
absorbance ratios: >2.0 (260/280 nm) and >1.8 (260/230 nm).
Then, 1.5 ng RNA was employed to conduct RT-PCR to produce cDNA
with the miScript II RT kit (Qiagen GmbH). RT-PCR was performed in
a reaction volume of 20 µl containing 2 µl total RNA, 4 µl 5X
miScript HiSpec Buffer, 2 µl 10X miScript Nucleics Mix, 2 µl
miScript Reverse Transcriptase Mix and 10 µl nuclease-free water.
The reaction mixtures were incubated at 37°C for 60 min and 95°C
for 5 min. miRNA expression was analyzed using a miScript miRNA PCR
Array (Qiagen GmbH) according to the manufacturer's instructions.
Prior to analysis, the miScript miRNA quality control PCR Array was
employed to determine the quality of the extracted cDNA template,
which met the criterion for conducting miScript miRNA PCR array
analysis. In addition, 96-well plates were used to determine the
expression profile of 940 miRNAs from one mouse muscle sample via
array analysis. A reaction mix with a final volume of 25 µl
containing 12.5 µl 2X QuantiTect SYBR Green Master Mix, 2.5 µl 10X
miScript Universal Primer, 1 µl cDNA and 9 µl nuclease-free water
was employed. The thermocycling conditions were as follows: 95°C
for 15 min, followed by 40 cycles of 94°C for 15 sec, 55°C for 30
sec and 70°C for 30 sec. The Cq values were obtained using an ABI
7500 Real-time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and were analyzed using online data analysis
tools based on the ΔΔCq method (relative gene
expression=2−(ΔCq sample−ΔCq control) (25). The P-value and log2 fold change
(log2 FC) between the C and E groups were calculated; P<0.05 and
|log2 FC| ≥1 were regarded to indicate differentially expressed
miRNAs.
Bioinformatics prediction
TargetScan v7.1 (www.targetscan.org/) (26–28),
miRanda (released August 2010, www.microrna.org/) (29,30)
and DIANA v4.0
(diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index)
(31,32) were used to predict the putative
miRNAs that target the 3′UTR of Nrf2. To reduce the number of false
positives and increase fidelity, the intersection of the prediction
results from these three tools was considered to indicate the
putative miRNAs. In addition, by analyzing the selected
differentially expressed miRNAs with the miScript miRNA PCR array,
miR-340-5p and miR-101a-3p were proposed to be the miRNAs that may
target Nrf2 mRNA following 8 weeks of aerobic exercise.
miRNA mimics and plasmid
construction
miRNA mimics are innovative molecules designed for
gene silencing approaches that contain non-natural or artificial
double stranded miRNA-like RNA fragments (33). These RNA fragments are constructed
to contain a sequence motif on its 5′-end that is partially
complementary to the target sequence in the 3′UTR of the transcript
(33). On this basis, miRNA mimics
and a control were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The sequences were as follows: miR-101a-3p
mimic, 5′-UACAGUACUGUGAUAACUGAA-3′; miR-340-5p mimic,
5′-UUAUAAAGCAAUGAGACUGAUU-3′; and mimic control,
5′-UUUGUACUACACAAAAGUACUG-3′.
The pMIR-Report luciferase and the control reporter
pRL-TK plasmids were purchased from Ambion (Thermo Fisher
Scientific, Inc.). The full length 3′UTR sequence of Nrf2
containing the putative target site for miR-101a-3p or miR-340-5p
was synthesized, and then inserted into the XhoI and HindIII sites
of the pMIR-Report luciferase vector by GENEWIZ Beijing (Beijing,
China); the construct was named pMIR-Nrf2. A mutation of Nrf2 3′UTR
in the putative binding site of miR-101a-3p or miR-340-5p was
introduced into the putative seed-matching sequence of pMIR-Nrf2
(Fig. 1A and B); these constructs
were named pMIR-Nrf2-Mut1 for miR-101a-3p and pMIR-Nrf2-Mut2 for
miR-340-5p. As determined from the miRanda database (www.microrna.org/microrna/home.do),
miR-340-5p was predicted to target two binding sites within the
3′UTR of Nrf2 at 84–90 (mirSVR score=−0.9843) and 366–380 (mirSVR
score=−0.1789), while miR-101a-3p had two binding sites located at
258–263 (mirSVR score=−0.3732) and 350–355 (mirSVR score=−0.3981).
MirSVR scoring is a machine learning method for ranking miRNA
target sites, in which a lower mirSVR score indicates a stronger
association between a miRNA and a target gene (34). Thus, the miRNA with the lowest
mirSVR score was chosen as the target miRNA in the present study. A
base in the target site was changed to generate a mutation in the
3′UTR of Nrf2 (Fig. 1).
Cell culture, transfection and dual
luciferase reporter assay
293T cells were obtained from China Agricultural
University (Beijing, China), and were cultured in complete growth
medium [Dulbecco's modified Eagle's medium (DMEM) supplemented with
10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences,
Logan, UT, USA)], 100 µg/ml penicillin and 100 µg/ml streptomycin
(HyClone; GE Healthcare Life Sciences) at 37°C in a humidified
incubator containing 5% CO2.
A total of ~2×105 293T cells were seeded
in a 24-well plate in DMEM containing 10% FBS 24 h prior to
transfection. The experiment was set for four groups as follows:
pMIR-Nrf2+miRNA mimics control, pMIR-Nrf2+miRNA mimics (miR-101a-3p
or miR-340-5p), pMIR-Nrf2-Mutation+miRNA mimics control, and
pMIR-Nrf2-Mutation+miRNA mimics (Mutation1/Mutation2 corresponds to
mutations in the putative binding sequence of miR-101a-3a and
miR-340-5p, respectively). Cell transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions.
Briefly, the culture medium was replaced with Opti-minimum
essential medium (MEM) from the kit. The pMIR-Report or the mutant
luciferase constructs were mixed with pRL-TK at a ratio of 20:1,
and miRNA mimics or the control were diluted with 50 µl Opti-MEM to
a final concentration of 50 nM. These diluted components were
combined and incubated for 20 min at room temperature. The mixture
(100 µl) was then added to each well and the medium was replaced
with DMEM containing 10% FBS following culture for 6 h;
transfection was carried out in triplicate. Following a 48 h
incubation, the cells were harvested and lysed in the passive lysis
buffer included in the Dual-Luciferase Reporter Assay System
(Promega Corporation, Madison, WI, USA). The luciferase activity of
firefly and Renilla were measured using the
Dual-Luciferase® Reporter Assay System in a
GloMax® 20/20 luminometer (Promega Corporation); the
firefly luciferase activity was normalized to that of
Renilla. The relative luciferase activity was used to
validate whether miR-340-5p and miR-101a-3p regulate the expression
of Nrf2 by binding to the 3′UTR.
Overexpression and knockdown of
miR-340-5p in C2C12 cells
miRNA inhibitors are chemically modified,
single-stranded nucleic acids designed to specifically bind to and
inhibit endogenous miRNA molecules (35). In the present study, C2C12 cells
(1.5×105 per well) were cultured in DMEM under the same
conditions as those used aforementioned for 293T cells. The cells
were seeded in 6-well plates upon reaching 70% confluence the day
prior to transfection. miR-340-5p mimics
(5′-UUAUAAAGCAAUGAGACUGAUU-3′), inhibitors
(5′-AAUCAGUCUCAUUGCUUUAUAA-3′), mimics control
(5′-UUUGUACUACACAAAAGUACUG-3′) and inhibitor control
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′) (Guangzhou RiboBio Co., Ltd.) were
transfected into C2C12 cells at a concentration of 150 nM using
Lipofectamine 3000™ (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions. The medium was changed
24 h post-transfection; C2C12 cells were harvested following 48 h
to determine the mRNA and protein expression levels of Nrf2.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Cell experiments were performed in triplicate, and
independently repeated at least three times. Statistical
calculations were performed using SPSS 13.0 (SPSS, Inc., Chicago,
IL, USA). Data were analyzed by an Independent Samples t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
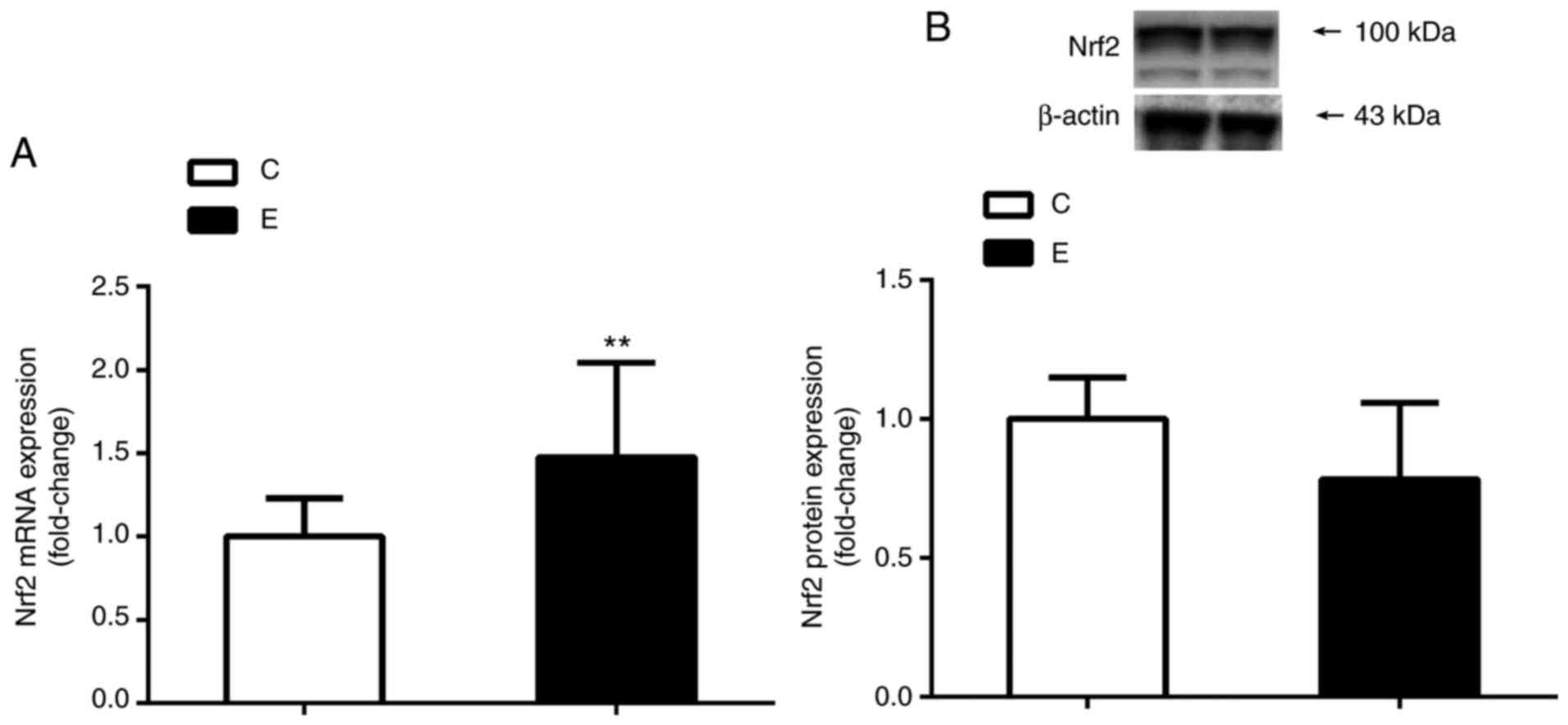
Effects of exercise training on Nrf2
mRNA and protein expression
The results of RT-qPCR analysis revealed that the
mRNA expression levels of Nrf2 in the E group were significantly
increased when compared with the C group (P<0.01). Additionally,
no significant difference was observed in the levels of Nrf2
protein expression between the two groups following 8 weeks of
aerobic exercise training as determined by western blotting
(Fig. 2).
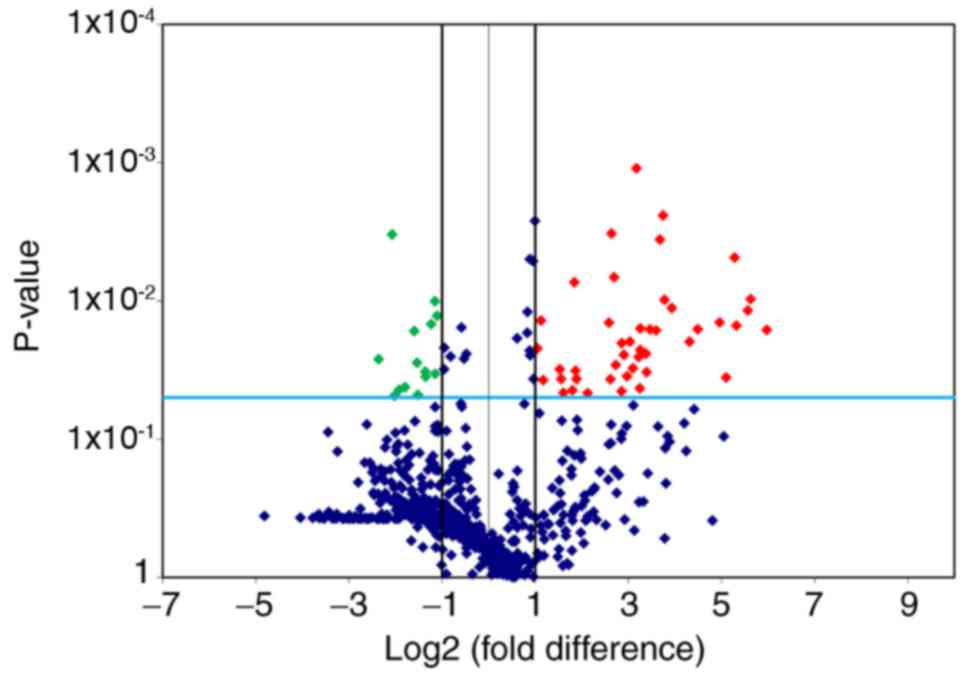
Effects of exercise training on miRNAs
expression
According to the standards of P<0.05 and |log2
FC| ≥1, 58 significantly differentially expressed miRNAs between
the C and E groups were identified in the present study. Among
them, the expression levels of 14 miRNAs were downregulated (log2
FC≤-1), whereas 44 miRNAs were upregulated (log2 FC≥1) following 8
weeks of aerobic exercise training (Table I; Fig.
3).
 | Table I.Differential expression of microRNAs
between two groups by miScript microRNA polymerase chain reaction
array analysis. |
Table I.
Differential expression of microRNAs
between two groups by miScript microRNA polymerase chain reaction
array analysis.
| miRNA ID | t-test | Fold change |
|---|
| miR-142-3p | 0.005 | 38.83 |
| miR-126a-3p | 0.020 | 19.90 |
| miR-15a-5p | 0.004 | 12.84 |
| miR-29b-3p | 0.016 | 22.48 |
| miR-27a-3p | 0.020 | 8.19 |
| miR-30e-5p | 0.033 | 10.50 |
| miR-22-3p | 0.024 | 10.43 |
| miR-30a-5p | 0.045 | 7.22 |
| miR-140-5p | 0.001 | 9.02 |
| miR-17-5p | 0.020 | 7.27 |
| miR-29a-3p | 0.025 | 7.48 |
| miR-19b-3p | 0.010 | 49.43 |
| miR-20a-5p | 0.016 | 11.05 |
| miR-106b-5p | 0.016 | 12.10 |
| miR-99a-5p | 0.011 | 15.27 |
| miR-19a-3p | 0.012 | 47.25 |
| miR-199a-5p | 0.016 | 9.57 |
| miR-411-5p | 0.031 | 2.88 |
| miR-425-5p | 0.032 | 3.65 |
| miR-335-5p | 0.049 | 4.26 |
|
miR-101a-3pa | 0.016 | 62.88 |
| miR-744-5p | 0.034 | −2.21 |
| miR-29c-3p | 0.007 | 6.46 |
| miR-30b-5p | 0.014 | 31.18 |
| miR-148b-3p | 0.046 | 3.04 |
| miR-106a-5p | 0.014 | 6.00 |
| miR-714 | 0.015 | −2.35 |
| miR-376b-5p | 0.044 | 3.49 |
| miR-20b-5p | 0.026 | 9.32 |
| miR-337-3p | 0.003 | 6.22 |
| miR-338-3p | 0.010 | 13.73 |
| miR-148a-3p | 0.043 | 9.49 |
| miR-497-5p | 0.029 | 6.64 |
| let-7i-3p | 0.007 | 3.58 |
| miR-1187 | 0.044 | −3.78 |
| miR-145a-5p | 0.037 | 3.70 |
| miR-190a-5p | 0.031 | 8.56 |
| miR-193a-5p | 0.013 | −2.15 |
| miR-199b-5p | 0.023 | 9.57 |
| miR-24-2-5p | 0.015 | 5.75 |
| miR-29a-5p | 0.037 | 6.13 |
|
miR-340-5pa | 0.037 | 2.95 |
| miR-425-3p | 0.042 | −3.46 |
| miR-709 | 0.026 | −5.14 |
| miR-450a-5p | 0.035 | 7.86 |
| miR-1249-3p | 0.033 | −2.57 |
| miR-3069-3p | 0.028 | −2.89 |
| miR-486-3p | 0.010 | −2.22 |
| miR-136-5p | 0.002 | 13.43 |
| miR-190a-3p | 0.047 | 4.37 |
| miR-3106-5p | 0.048 | −2.86 |
| miR-1a-2-5p | 0.007 | 100.92 |
| miR-1947-3p | 0.003 | −4.20 |
| miR-667-5p | 0.049 | −4.05 |
| miR-3076-5p | 0.035 | −2.55 |
| miR-1a-1-5p | 0.036 | 34.30 |
| miR-143-3p | 0.015 | 39.94 |
| miR-486-3p | 0.017 | −3.02 |
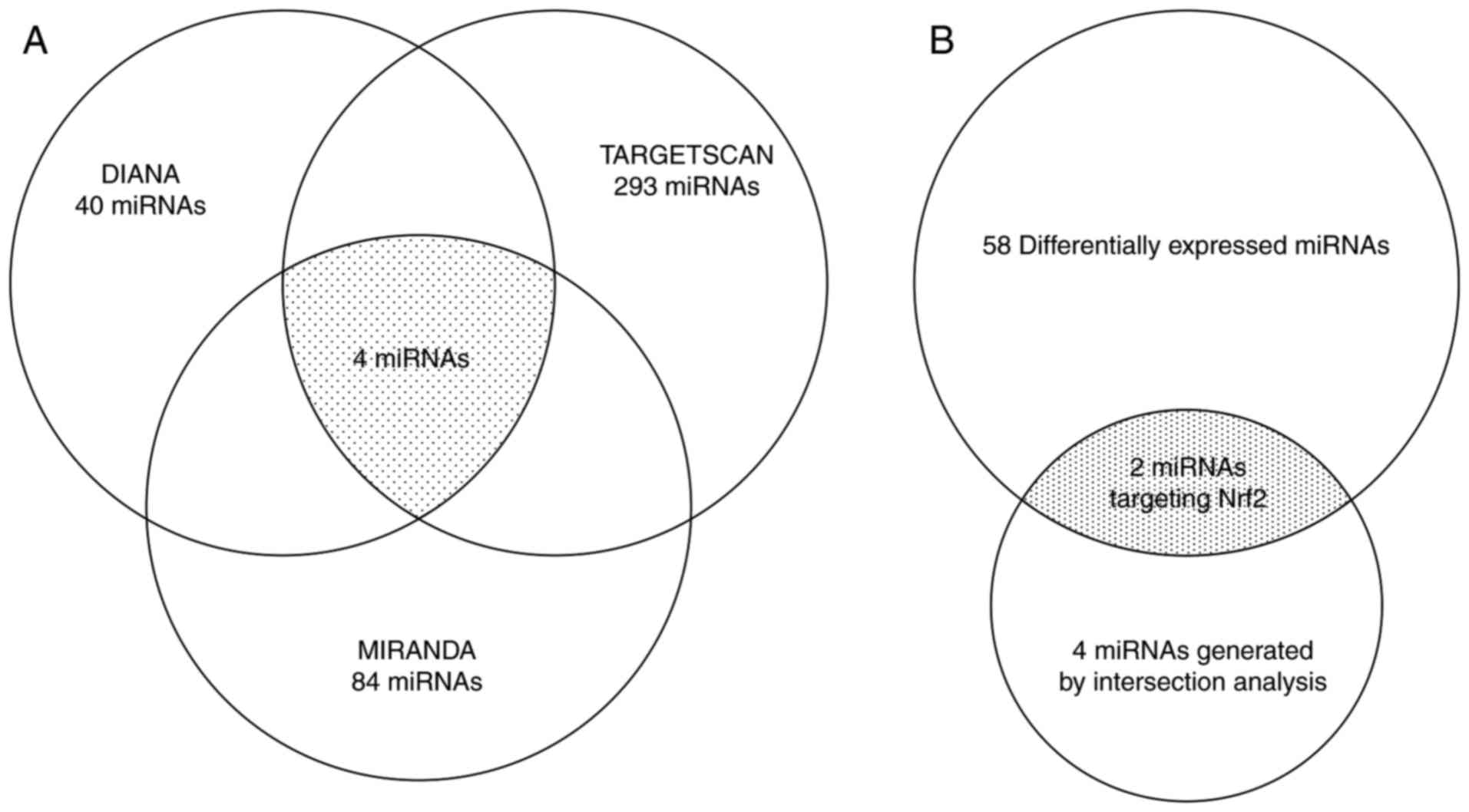
Bioinformatics prediction of the
putative miRNAs that target the 3′UTR of Nrf2
DIANA, miRanda and TargetScan prediction tools
identified 40, 84 and 293 miRNAs that target the 3′UTR of Nrf2,
respectively; 4 miRNAs (miR-101a-3p, miR-142-5p, miR-1950 and
miR-340-5p) were identified when combining the results of
prediction analysis with these three tools (Fig. 4A). In addition, by combining the 58
differentially expressed miRNAs between the E and C groups
(Table I), miR-101a-3p and
miR-340-5p (Fig. 4B; Table I) were selected as candidate miRNAs
that targeted the 3′UTR of Nrf2 and chosen for further study.
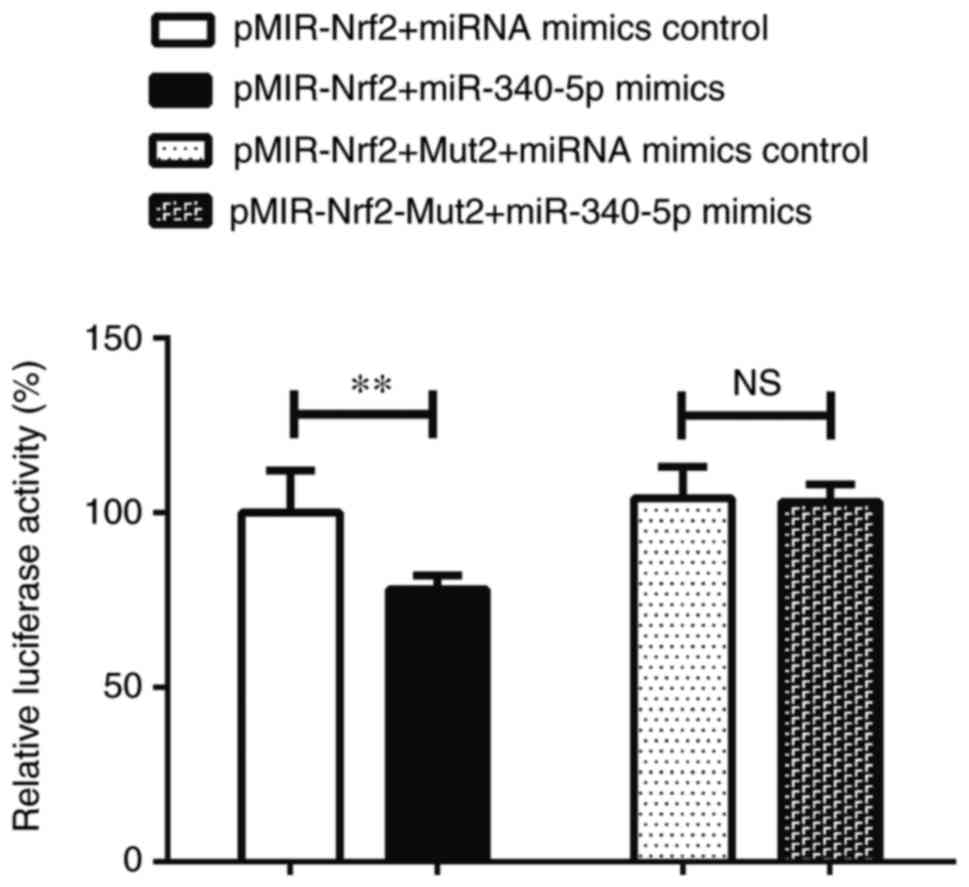
miR-340-5p may directly target 3′UTR
of Nrf2 gene
As shown in Fig. 5,
the relative luciferase activity in the pMIR-Nrf2+miR-340-5p mimics
group was significantly lower than that of the pMIR-Nrf2+miRNA
mimics control group (P<0.01). This indicated that miR-340-5p
was capable of binding to the pMIR-Nrf2 plasmid and suppressed
luciferase activity. Additionally, no significant difference was
observed in relative luciferase activity between the
pMIR-Nrf2-Mut2+miRNA mimics control and pMIR-Nrf2-Mut2+miR-340-5p
mimics groups (P>0.05; Fig. 5).
These results further indicated that the binding site of miR-340-5p
was located within the 3′UTR of Nrf2.
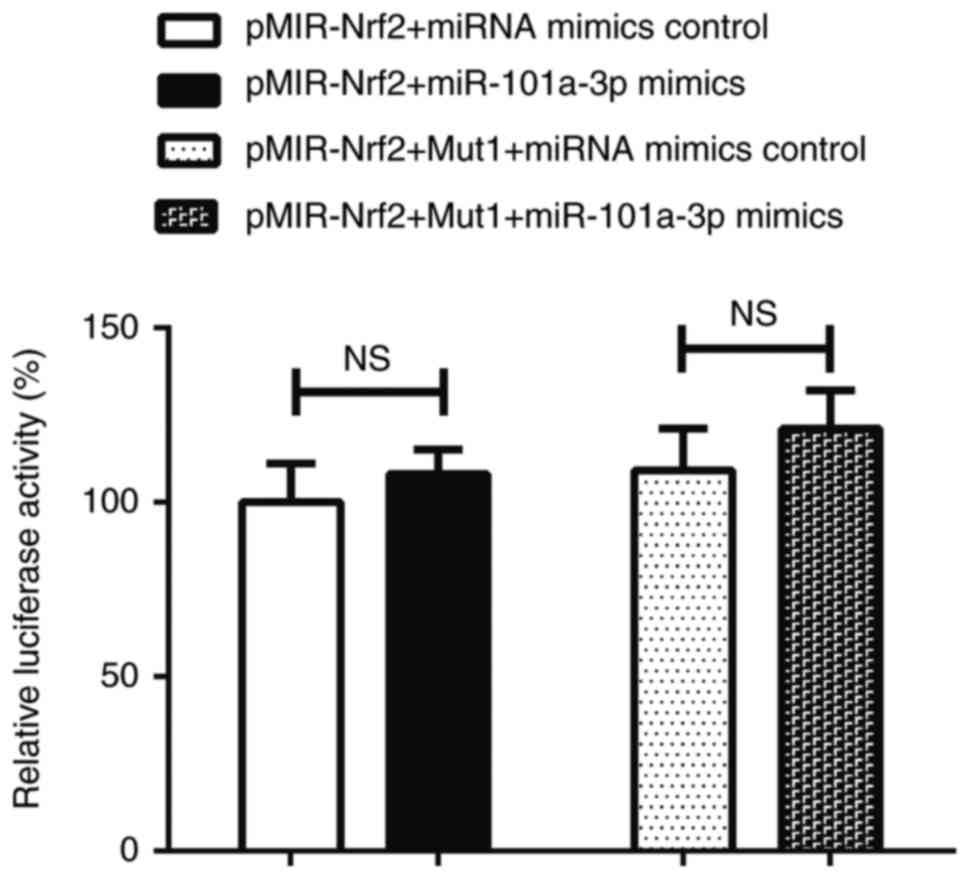
miR-101a-3p does not directly target
3′UTR of Nrf2 gene
There was no significant difference in the relative
luciferase activity between the pMIR-Nrf2+miR-101a-3p mimics and
the pMIR-Nrf2+miRNA mimics control groups (P>0.05). In addition,
there was no significant difference in relative luciferase activity
between the pMIR-Nrf2-Mut1+ miRNA mimics control and
pMIR-Nrf2-Mut1+miR-101a-3p mimics groups (P>0.05; Fig. 6). Therefore, these data indicated
that miR-101a-3p did not suppress luciferase activity and could not
bind the pMIR-Nrf2 plasmid.
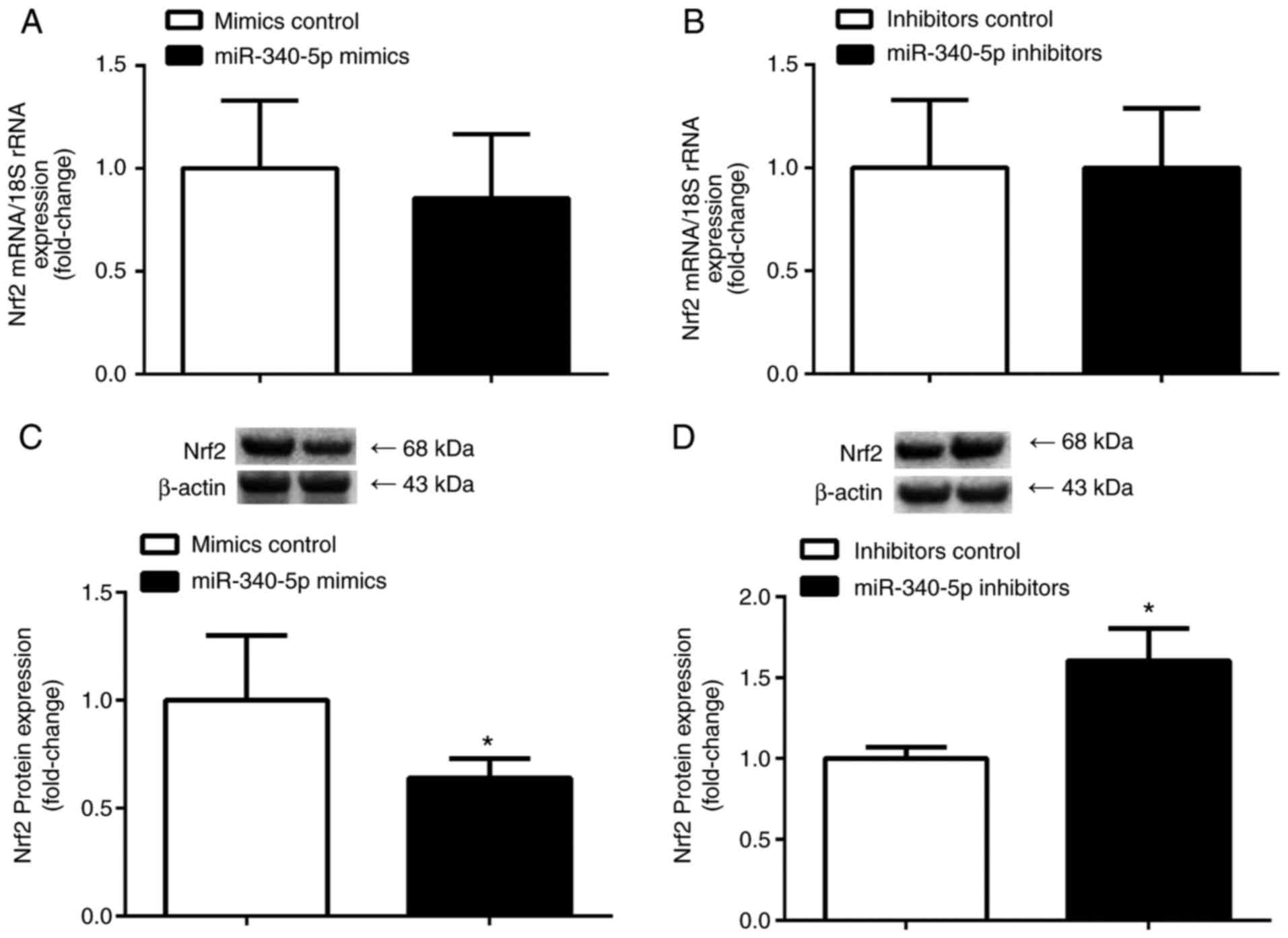
Effect of miR-340-5p overexpression
and knockdown on endogenous Nrf2 mRNA and protein expression
levels
To demonstrate the effect of miR-340-5p on
endogenous Nrf2 mRNA and protein expression levels, the present
study transiently transfected C2C12 cells with miR-340-5p mimics,
inhibitors or control, and the expression of Nrf2 mRNA and protein
was determined. Transfection of miR-340-5p mimics or inhibitors
significantly decreased and increased Nrf2 protein expression,
respectively (P<0.05); however, the mRNA expression levels
remained unchanged (P>0.05; Fig.
7).
Discussion
The main finding of the present study was that the
mRNA expression levels of Nrf2 in skeletal muscle significantly
increased following 8 weeks of aerobic exercise training, but the
protein expression levels did not increase. One miRNA, miR-340-5p,
was revealed to directly bind to the 3′UTR of Nrf2 and may be a
direct post-transcriptional regulator of Nrf2 in skeletal muscle
following aerobic exercise training.
It has been demonstrated that aerobic exercise
training increased the expression levels of Nrf2 mRNA and protein
in the cardiomyocytes of rats following myocardial infarction,
which protected against myocardial infarction-induced oxidative
injury (36); physical exercise of
moderate intensity also protected against experimental
6-hydroxydopamine-induced hemi-parkinsonism through the Nrf2-ARE
signaling pathway (20). Some
studies have confirmed that Nrf2 protein expression in whole cell
or nuclear fractions was associated with exercise intensity. For
example, the effects of varying intensities of treadmill exercise
on hippocampal Nrf2 protein expression in adult C57B1 male mice
were studied. The results revealed that the greater the intensity
of exercise, the higher the protein expression levels of Nrf2
(37). Another experiment examined
the association between exercise intensity and the protein
expression levels of Nrf2 in blood cells, and revealed that the
greater the intensity of exercise, the higher the Nrf2 protein
levels in the nucleus (38). In
addition, the expression of miRNAs that are affected by exercise
have been investigated. miRNA expression profiling revealed that
exercise significantly increased the expression of miR-181, miR-1
and miR-107, but reduced that of miR-23; no changes in the
expression of miR-133 were noted (22). In the present study, the expression
levels of Nrf2 mRNA were increased, while no significant changes in
Nrf2 protein expression within mouse skeletal muscle following
aerobic exercise for 8 weeks were reported. It was speculated that
no changes in the expression of Nrf2 protein in trained mice may be
associated with lower exercise intensities. In addition, the
regulation of miRNAs may be involved in targeting Nrf2 mRNA. It has
been reported that miRNAs may serve important roles in
post-transcriptional gene regulation by targeting mRNAs for
cleavage or translational repression (39). It is well known that if a miRNA is
completely complementary with an mRNA transcript, the miRNA will
cleave and degrade its target; otherwise, the miRNA will bind the
mRNA to execute translational inhibition (40). These results suggest that miRNA
translational repression may be one of factors that lead to this
discrepancy in the expression levels of Nrf2 mRNA and protein.
A total of 58 miRNAs exhibiting significant
differential expression following 8 weeks of aerobic exercise
intervention were studied in the present study. By combining the
results of prediction analysis, and the profiles of differentially
expressed miRNAs between the C and E groups, miR-340-5p and
miR-101a-3p were determined to be the miRNAs that target the 3′UTR
of Nrf2. To verify this, the present study measured the luciferase
signals of these two miRNAs, which revealed that the luciferase
activity of the pMIR-Nrf2+miR-340-5p mimics group was suppressed,
while that of the pMIR-Nrf2+ miR-101a-3p mimics group was
unchanged, compared with their corresponding control groups.
Additionally, the luciferase activity of the plasmid with mutant
type Nrf2 3′UTR was not affected by miR-340-5p or miR-101a-3p
mimics. These results indicated that only miR-340-5p could target
the 3′UTR of Nrf2 directly, and may be involved in downregulating
the protein expression of Nrf2 following 8 weeks of aerobic
exercise training; miR-101a-3p was revealed to not directly
regulate Nrf2. However, it has been shown that Cullin 3 is a
ubiquitous ligase that degrades Nrf2 (41). miR-101a-3p was reported to bind to
the 3′UTR of Cullin 3 to suppress expression and stabilize Nrf2 via
the inhibition of the proteasomal degradation pathway (4,42).
Thus, miR-101a-3p may also serve an important role in targeting
Nrf2 expression as an indirect regulator; however, further research
is required in the future to verify this.
To investigate if miR-340-5p had an effect on the
endogenous mRNA and protein expression levels of Nrf2, the present
study detected the expression of Nrf2 in C2C12 cells transfected
with miR-340-5p mimics or inhibitors. The results revealed that
miR-340-5p mimics decreased the expression levels of Nrf2 protein,
whereas the expression levels increased in response to miR-340-5p
inhibitors. However, miR-340-5p mimics and inhibitors had no effect
on Nrf2 mRNA expression. Taken together, it could by concluded that
miR-340-5p directly regulated the expression of Nrf2 by inhibiting
the translation of Nrf2 mRNA. These data further supported that in
the present in vivo study, aerobic exercise may have induced
the expression of miR-340-5p, inhibiting the expression of Nrf2
protein, which may explain the discrepancy in the mRNA and protein
expression levels of Nrf2.
In addition, Keap1 (43), Cullin-associated and
neddylation-dissociated 1 (CAND1) (44), and small Maf proteins (45) have also been reported to be
important regulators of Nrf2 expression. miRNAs may indirectly
affect the expression of Nrf2 by targeting Keap1, CAND1 and small
Maf proteins.
In conclusion, the results of the present study
provide evidence that miR-340-5p may be a miRNA that directly
targets the 3′UTR of Nrf2. Therefore, miR-340-5p may be regarded as
a potential direct regulator of Nrf2 expression in the
post-exercise skeletal muscle of mice. These observations may
provide insight into the miRNA-associated mechanisms underlying the
effects of aerobic exercise training on Nrf2 protein expression in
skeletal muscle.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 31471134), and the
Fundamental Research Funds for the Central Universities (grant no.
2015SYS005).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TM and YZ conceived and designed the experiments. TM
and YL performed the experiments and analyzed the data. YZ and JW
contributed to the reagents/materials/analysis. TM, YZ and JW gave
final approval of the version to be published.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Use Committee of Beijing Sport University (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kobayashi A, Ohta T and Yamamoto M: Unique
function of the Nrf2-Keap1 pathway in the inducible expression of
antioxidant and detoxifying enzymes. Methods Enzymol. 378:273–286.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wakabayashi N, Itoh K, Wakabayashi J,
Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F,
Roop DR, et al: Keap1-null mutation leads to postnatal lethality
due to constitutive Nrf2 activation. Nat Genet. 35:238–245. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heiss EH, Schachner D, Werner ER and
Dirsch VM: Active NF-E2-related factor (Nrf2) contributes to keep
endothelial NO synthase (eNOS) in the coupled state: Role of
reactive oxygen species (ROS), eNOS, and heme oxygenase (HO-1)
levels. J Biol Chem. 284:31579–31586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi A, Kang MI, Okawa H, Ohtsuji M,
Zenke Y, Chiba T, Igarashi K and Yamamoto M: Oxidative stress
sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to
regulate proteasomal degradation of Nrf2. Mol Cell Biol.
24:7130–7139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang P, Li CG, Qi Z, Cui D and Ding S:
Acute exercise stress promotes Ref1/Nrf2 signalling and increases
mitochondrial antioxidant activity in skeletal muscle. Exp Physiol.
101:410–420. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsou YH, Shih CT, Ching CH, Huang JY, Jen
CJ, Yu L, Kuo YM, Wu FS and Chuang JI: Treadmill exercise activates
Nrf2 antioxidant system to protect the nigrostriatal dopaminergic
neurons from MPP+ toxicity. Exp Neurol. 263:50–62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Merry TL and Ristow M: Nuclear factor
erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced
mitochondrial biogenesis and the anti-oxidant response in mice. J
Physiol. 594:5195–5207. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kumar RR, Narasimhan M, Shanmugam G, Hong
J, Devarajan A, Palaniappan S, Zhang J, Halade GV, Darley-Usmar VM,
Hoidal JR and Rajasekaran NS: Abrogation of Nrf2 impairs
antioxidant signaling and promotes atrial hypertrophy in response
to high-intensity exercise stress. J Transl Med. 14:862016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Done AJ and Traustadottir T: Nrf2 mediates
redox adaptations to exercise. Redox Biol. 10:191–199. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ying SY, Chang DC and Lin SL: The microRNA
(miRNA): Overview of the RNA genes that modulate gene function. Mol
Biotechnol. 38:257–268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang M, Yao Y, Eades G, Zhang Y and Zhou
Q: miR-28 regulates Nrf2 expression through a Keap1-independent
mechanism. Breast Cancer Res Treat. 129:983–991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou S, Ye W, Zhang Y, Yu D, Shao Q, Liang
J and Zhang M: miR-144 reverses chemoresistance of hepatocellular
carcinoma cell lines by targeting Nrf2-dependent antioxidant
pathway. Am J Transl Res. 8:2992–3002. 2016.PubMed/NCBI
|
|
16
|
Narasimhan M, Patel D, Vedpathak D,
Rathinam M, Henderson G and Mahimainathan L: Identification of
novel microRNAs in post-transcriptional control of Nrf2 expression
and redox homeostasis in neuronal, SH-SY5Y cells. PLoS One.
7:e511112012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khan AUH, Rathore MG, Allende-Vega N, Vo
DN, Belkhala S, Orecchioni S, Talarico G, Bertolini F, Cartron G,
Lecellier CH and Villalba M: Human leukemic cells performing
oxidative phosphorylation (OXPHOS) generate an antioxidant response
independently of reactive oxygen species (ROS) production.
EBioMedicine. 3:43–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van Jaarsveld MT, Helleman J, Boersma AW,
van Kuijk PF, van Ijcken WF, Despierre E, Vergote I, Mathijssen RH,
Berns EM, Verweij J, et al: miR-141 regulates KEAP1 and modulates
cisplatin sensitivity in ovarian cancer cells. Oncogene.
32:4284–4293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F,
Zheng F and Lin X: miR-141 activates Nrf2-dependent antioxidant
pathway via down-regulating the expression of keap1 conferring the
resistance of hepatocellular carcinoma cells to 5-fluorouracil.
Cell Physiol Biochem. 35:2333–2348. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aguiar AS Jr, Duzzioni M, Remor AP,
Tristão FS, Matheus FC, Raisman-Vozari R, Latini A and Prediger RD:
Moderate-intensity physical exercise protects against experimental
6-hydroxydopamine-induced hemiparkinsonism through Nrf2-antioxidant
response element pathway. Neurochem Res. 41:64–72. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wan C, Han R, Liu L, Zhang F, Li F, Xiang
M and Ding W: Role of miR-155 in fluorooctane sulfonate-induced
oxidative hepatic damage via the Nrf2-dependent pathway. Toxicol
Appl Pharmacol. 295:85–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Safdar A, Abadi A, Akhtar M, Hettinga BP
and Tarnopolsky MA: miRNA in the regulation of skeletal muscle
adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS
One. 4:e56102009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aoi W, Ichikawa H, Mune K, Tanimura Y,
Mizushima K, Naito Y and Yoshikawa T: Muscle-enriched microRNA
miR-486 decreases in circulation in response to exercise in young
men. Front Physiol. 4:802013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gong H, Xie J, Zhang N, Yao L and Zhang Y:
MEF2A binding to the Glut4 promoter occurs via an AMPKα2-dependent
mechanism. Med Sci Sports Exerc. 43:1441–1450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
ELife. 4:2015. View Article : Google Scholar
|
|
27
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:(Database Issue). D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41:(Web Server Issue). W169–W173. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reczko M, Maragkakis M, Alexiou P, Grosse
I and Hatzigeorgiou AG: Functional microRNA targets in protein
coding sequences. Bioinformatics. 28:771–776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Z: The guideline of the design and
validation of miRNA mimics. Methods Mol Biol. 676:211–223. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang L, Chen HY, Hao NB, Tang B, Guo H,
Yong X, Dong H and Yang SM: microRNA inhibitors: Natural and
artificial sequestration of microRNA. Cancer Lett. 407:139–147.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang HK, Miao Y, Wang YH, Zhao M, Feng
ZH, Yu XJ, Liu JK and Zang WJ: Aerobic interval training protects
against myocardial infarction-induced oxidative injury by enhancing
antioxidase system and mitochondrial biosynthesis. Clin Exp
Pharmacol Physiol. 41:192–201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tutakhail A, Nazary QA, Lebsir D,
Kerdine-Romer S and Coudore F: Induction of brain Nrf2-HO-1 pathway
and antinociception after different physical training paradigms in
mice. Life Sci. 209:149–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Done AJ, Newell MJ and Traustadottir T:
Effect of exercise intensity on Nrf2 signalling in young men. Free
Radic Res. 51:646–655. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Furukawa M and Xiong Y: BTB protein Keap1
targets antioxidant transcription factor Nrf2 for ubiquitination by
the Cullin 3-Roc1 ligase. Mol Cell Biol. 25:162–171. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim JH, Lee KS, Lee DK, Kim J, Kwak SN, Ha
KS, Choe J, Won MH, Cho BR, Jeoung D, et al: Hypoxia-responsive
microRNA-101 promotes angiogenesis via heme oxygenase-1/vascular
endothelial growth factor axis by targeting cullin 3. Antioxid
Redox Signal. 21:2469–2482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sihvola V and Levonen AL: Keap1 as the
redox sensor of the antioxidant response. Arch Biochem Biophys.
617:94–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Villeneuve NF, Lau A and Zhang DD:
Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin
proteasome system: An insight into cullin-ring ubiquitin ligases.
Antioxid Redox Signal. 13:1699–1712. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li W, Yu S, Liu T, Kim JH, Blank V, Li H
and Kong AN: Heterodimerization with small Maf proteins enhances
nuclear retention of Nrf2 via masking the NESzip motif. Biochim
Biophys Acta. 1783:1847–1856. 2008. View Article : Google Scholar : PubMed/NCBI
|