Introduction
Lamin A, which is encoded by the LMNA gene,
is a major component of the nuclear lamina and nuclear skeleton
(1). Lamin A is expressed in most
adult tissues (2,3), with the expression of lamin A
increasing with age in somatic cells (4). Although studies have revealed that
lamin A serves a structural role during interphase (5), lamin A is increasingly recognized as
a mediator, and possibly a regulator, of nuclear processes through
its interactions with a variety of nuclear factors, including
double-stranded DNA, transcriptional regulators, nuclear
membrane-associated proteins and nuclear pore complexes (6,7).
Dysfunction of lamin A interrupts chromatin organization, the DNA
damage response, telomere maintenance, cellular senescence and
apoptosis (8,9). Mutations in the LMNA gene
cause a heterogeneous group of human diseases that are collectively
termed laminopathies, including progeroid syndromes and premature
aging disorders that primarily affect striated muscle, adipose,
bone and neuronal tissues, such as Hutchinson-Gilford progeria
syndrome (HGPS) (6,10,11).
Mutations leading to laminopathies are distributed throughout the
LMNA gene and show a high degree of tissue specificity
(3,12). How mutations in LMNA cause
disease and why laminopathies are highly tissue-specific remain
unclear (3). Nuclear envelope
proteomes are highly variable among tissues (13,14).
Additionally, variants of lamin A may interact differently with
proteins that are themselves expressed in a tissue-specific manner,
which could explain the tissue specificity of laminopathies
(15). However, determining the
molecular mechanisms underlying changes in lamin A protein
interactions remains a clinical challenge.
The G608G mutation in the LMNA gene causes a
truncation of lamin A, with a 50-residue region lost that includes
a second proteolytic site for zinc metallopeptidase STE24
(ZMPSTE24), resulting in an unprocessed prelamin A termed progerin
in patients with HGPS (16). The
accumulation of truncated lamin A in HGPS impedes the release of
proteins from the nuclear membrane and disrupts their regulatory
functions, thereby accelerating a subset of pathological changes
that contribute to the aging processes (17,18).
Notably, mice carrying lamin A mutations also exhibit symptoms
consistent with HGPS, including the thinning of skin, hypoplasia,
the degeneration of cardiac and skeletal muscles, and osteoporosis
(19). Increased levels of
wild-type lamin A in normal human cells result in a decreased
replicative lifespan and nuclear membrane alterations that lead to
phenotypic changes similar to those observed in HGPS fibroblasts
(13,14). These studies suggest that wild-type
lamin A, similar to mutated lamin A, is also involved in the aging
processes.
To improve understanding of the pathological
mechanisms involved in laminopathies and the aging process, the
present study sought to systematically identify lamin A-interacting
proteins in an unbiased manner. A yeast two-hybrid screen of a
human skeletal muscle cDNA library was performed using the carboxy
(C)-terminus of lamin A as bait to search for novel lamin
A-interacting factors. This screening identified copper metabolism
MURR1 domain-containing 1 (COMMD1, formerly known as MURR1) as a
novel binding partner of lamin A. Their binding affinity was
further validated using confocal colocalization and
co-immunoprecipitation experiments.
Materials and methods
Yeast two-hybrid analysis
Yeast two-hybrid analysis was conducted using a
GAL4-based system to screen a human skeletal muscle complementary
DNA (cDNA) library (Matchmaker GAL4 two-hybrid system; Clontech
Laboratories, Inc.). Briefly, a bait protein was constructed by
cloning the C-terminus of lamin A (mRNA sequence 1,413-2,241) in
frame with the GAL4 binding domain using the EcoRI and
BamHI restriction sites of the pGBKT7 vector (Clontech
Laboratories, Inc.). The yeast Saccharomyces cerevisiae
strain AH109 (Clontech Laboratories, Inc.) was sequentially
transformed with the C-terminal lamin A bait vector (pGBKT7-LA-C;
Clontech Laboratories, Inc.) and the Matchmaker human skeletal
muscle cDNA library (Clontech Laboratories, Inc.) cloned into pACT2
(Clontech Laboratories, Inc.) according to the manufacturer's
protocol (Clontech Laboratories, Inc.). Saccharomyces
cerevisiae strain AH109 transformed with pCL1 (encodes the
full-length and wild-type GAL4 protein) vector was provided as
positive control. Transformants were plated on synthetic defined
(SD)/histidine/leucine/tryptophan (TDO) medium (Clontech
Laboratories, Inc.) (low-stringency protocol); a total of
2×107 colonies were screened. Colonies were transferred
to SD/adenine/histidine/leucine/tryptophan (QDO) plates (Clontech
Laboratories, Inc.) containing
5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal)
following two rounds of selection. Positive clones were identified
under high-stringency conditions and were defined as clones that
exhibited growth on the QDO plates that were strongly positive for
galactosidase activity. The selected clones were further analyzed
by Sanger sequencing (Sangon Biotech Co., Ltd.) and compared with
known sequences in GenBank using a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Cell culture
The 293 cell line (Stem Cell Bank; Chinese Academy
of Sciences) was cultivated in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal calf serum (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin. Serial passaging was performed when the cells reached
a confluence of 80%.
Western blotting
Whole cell extracts were prepared using RIPA buffer
[50 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, 1% NP-40, 0.5%
sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride,
1 µg/ml leupeptin and 1 µg/ml pepstatin]. Protein was quantified
using a bicinchoninic protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Subsequently, 10–20 µg protein was loaded into
each lane. Proteins were separated by 12% SDS-PAGE. Subsequently,
gels were blotted onto PVDF membranes. The PVDF membranes were
blocked for 1 h at room temperature (25°C) in Tris-buffered saline
containing 0.1% Tween 20 and 5% non-fat milk. Primary antibodies
were diluted in blocking solution and incubated with the membranes
overnight at 4°C. Anti-COMMD1 antibody (1:1,000; cat. no.
sc-166248; Santa Cruz Biotechnology, Inc.), anti-lamin A/C antibody
(1:2,000; cat. no. sc-20681; Santa Cruz Biotechnology, Inc.),
anti-hemagglutinin (HA) antibody (1:1,000; cat. no. 11867423001;
Roche Diagnostics) and anti-Flag antibody (1:1,000; cat. no. F3165;
Sigma-Aldrich; Merck KGaA) were used as primary antibodies.
Anti-GAPDH antibody (1:1,000; cat. no. 60004-1-Ig; ProteinTech
Group, Inc.) was used to detect the protein expression level of the
loading control GAPDH. Secondary antibodies horseradish peroxidase
(HRP)-conjugated goat anti-rabbit (cat. no. A120-101P; Bethyl
Laboratories, Inc.), goat anti-rat (cat. no. A110-143P; Bethyl
Laboratories, Inc.) and goat anti-mouse (cat. no. AP308P; Merck
KGaA) were diluted at 1:5,000 and incubations were performed for 1
h at room temperature (25°C). The proteins were detected using a
western chemiluminescent HRP substrate (EMD Millipore).
Fluorescence confocal microscopy
Total RNA was extracted from 293 cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The first
strand cDNA was synthesized using the PrimeScript 1st
Strand cDNA Synthesis kit (Takara Bio, Inc.) following the
manufacturer's protocol. COMMD1 coding sequence (CDS) and Lamin A
CDS were synthesized using the same first strand cDNA templates.
COMMD1 CDS was synthesized using the primers forward,
5′-CCCTCGAGATGGCGGGCGAGCTTGAG-3′ and reverse,
5′-CGGAATTCGGTTAGGCTGGCTGATC-3′. PCRs were performed using the rTaq
DNA Polymerase (Takara Bio, Inc.) in a total volume of 20 µl and
amplification protocol consisted of an initial denaturation at 94°C
for 5 min, followed by 30 cycles of denaturation for 45 sec at
94°C, annealing for 45 sec at 60°C and extension for 1 min at 72°C,
followed by a final extension at 72°C for 10 min. The green
fluorescent protein (GFP)-COMMD1 vector was generated by inserting
COMMD1 CDS into the pEGFP-N1 vector (Clontech Laboratories, Inc.),
which contains a GFP expression cassette, with XhoI and
EcoRI restriction sites. Lamin A CDS was generated with the
following thermocycling conditions: Initial denaturation at 94°C
for 5 min, followed by 30 cycles of denaturation for 45 sec at
94°C, annealing for 1 min sec at 60°C and extension for 90 sec at
72°C, followed by a final extension at 72°C for 10 min. Lamin A CDS
was amplified using the primers forward
5′-ACGCTCGAGATGGAGACCCCGTCCCAGCGGC-3′ and reverse
5′-CGGGATCCGCGGGCTCTGGGTTCGGGGGCT-3′. The red fluorescent protein
(RFP)-lamin A vector was generated by inserting lamin A CDS into
the pDsRed2-N1 vector (Clontech Laboratories, Inc.), which contains
an RFP expression cassette, using the XhoI and BamHI
restriction sites. All sequences from all plasmids were confirmed
by DNA sequencing following cloning.
The day prior to transfection, 293 cells
(2×105) were seeded into confocal dishes. When the cells
reached ~80% confluence, 2.5 µg pEGFP-N1-COMMD1 and 2.5 µg
pDsRed2-N1 or 2.5 µg pDsRed2-N1-lamin A were transfected into cells
using Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 48 h after transfection,
co-localization of COMMD1 and lamin A in transfected cells was
observed using a Leica TCS SP5 confocal microscope (Leica
Microsystems, Inc.; magnification, ×63) operated using Leica
confocal software (LAS AF lite; version 2.0; Leica Microsystems,
Inc.).
Co-immunoprecipitation
The pCMV-HA-COMMD1 vector was generated by inserting
COMMD1 cDNA into the pCMV-HA vector (Clontech Laboratories, Inc.)
with the EcoRI and BglII restriction sites. Lamin A
cDNA was inserted into the pCMV-Flag vector (Clontech Laboratories,
Inc.) with the HindIII and NaeI restriction sites.
The pCMV-Flag-lamin A, and pCMV-HA-COMMD1 or pCMV-HA plasmids were
co-transfected into 293 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For immunoprecipitation, whole cell
extracts of non-transfected or co-transfected cells were lysed with
RIPA buffer (Thermo Fisher Scientific, Inc.). Equal amounts (500
µg) of protein samples were incubated with 2 µg antibody. The
antibodies used in the co-immunoprecipitation experiment were as
follows: Anti-HA and anti-Flag antibody for the overexpression
plasmid and anti-COMMD1 or anti-lamin A/C for endogenous
co-immunoprecipitation. Normal mouse IgG (cat. no. sc-2025; Santa
Cruz Biotechnology, Inc.) and normal rabbit IgG (cat. no. sc-2027;
Santa Cruz Biotechnology, Inc.) were used as control. Antibodies
were incubated for 2 h at 4°C. The antibody-protein complex was
mixed with 100 µl of a protein A/G-agarose suspension (cat. nos.
1134515 and 1243233; Roche Diagnostics, Inc.) for 24 h at 4°C.
After centrifugation at 12,000 × g for 20 sec at 4°C, the pellet
was washed twice with wash buffer 1 [50 mM Tris-HCl (pH 7.5), 150
mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, and
1 tablet complete protein inhibitor cocktail/50 ml] and once each
with wash buffer 2 (50 mM Tris-HCl, pH 7.5, 500 mM sodium chloride,
0.1% Nonidet P-40, and 0.05% sodium deoxycholate) and wash buffer 3
(50 mM Tris-HCl, pH 7.5, 0.1% Nonidet P-40 and 0.05% sodium
deoxycholate) prior to resuspension in loading buffer. The
suspension was denatured by heating for 5 min at 95–100°C and
centrifuged at 12,000 × g for 10 min at 4°C. The supernatant
fraction was used for the identification of the co-precipitated
proteins via western blotting, using the aforementioned
procedure.
Results
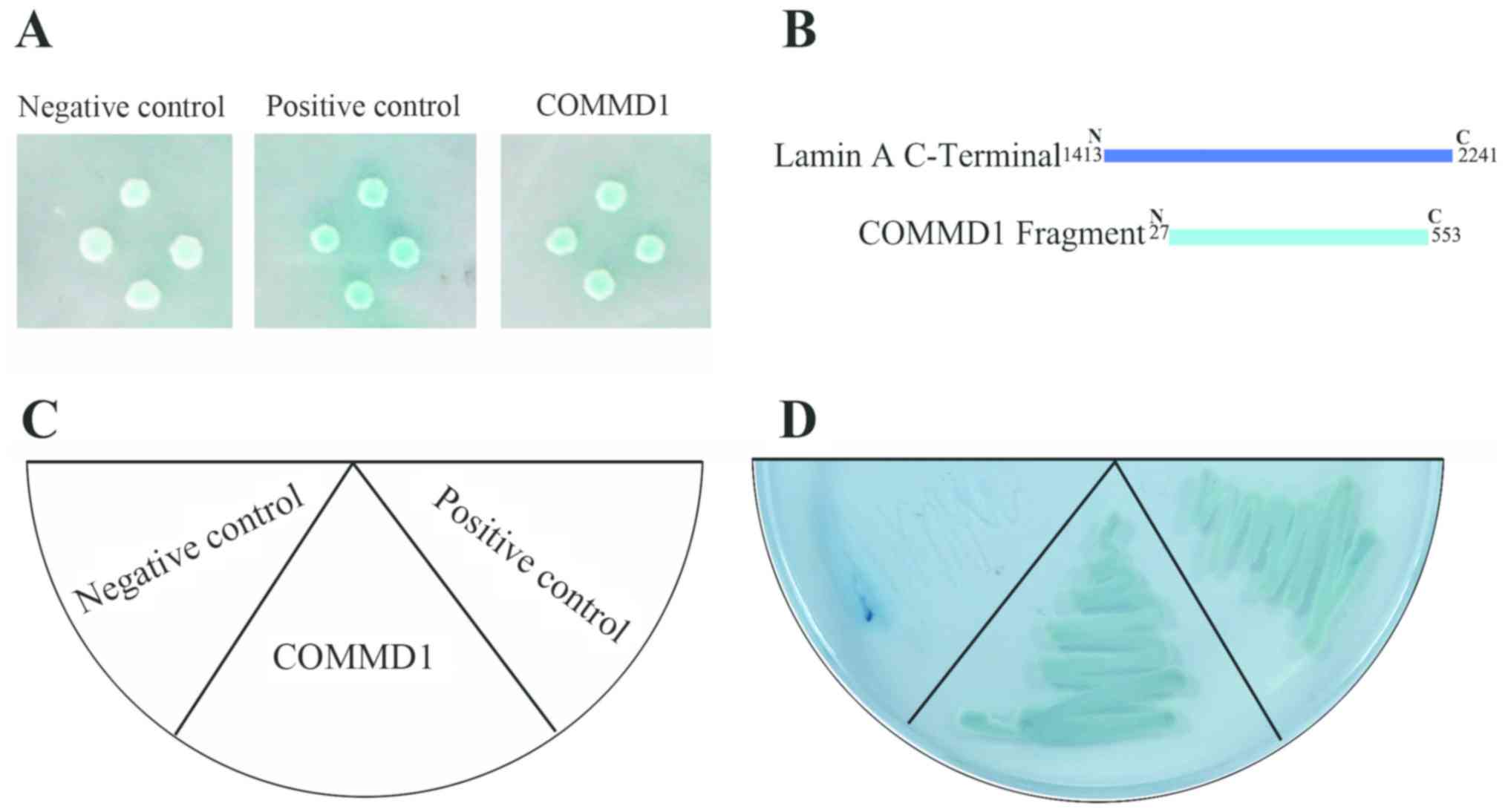
Identification of COMMD1 as a novel
lamin A-binding protein using yeast two-hybrid screening
To identify lamin A-binding proteins, a human
skeletal muscle cDNA library was screened using the yeast
two-hybrid system. Yeast cells expressing the C-terminus of lamin A
(mRNA sequence 1,413-2,241) as bait were mated with yeast cells
expressing the appropriate prey to make diploid yeast cells that
were then streaked onto low-stringency TDO medium and
high-stringency QDO medium to test for an interaction. At lower
stringency, the colonies displayed X-α-Gal activity similar to that
of the positive control (Fig. 1A).
Following three rounds of selection, 9 colonies survived and were
subjected to Sanger sequencing (Table
SI). A fragment (Data S1) of the full sequence of COMMD1 (Data
S2) gene was identified as a potential binding partner of lamin A
via sequencing and a BLAST search (Fig. 1B). To validate this interaction,
the COMMD1 prey was reintroduced into yeast cells and streaked onto
QDO medium (Fig. 1C). Colonies
growing on the QDO medium displayed X-α-Gal activity similar to
that of the positive control (Fig.
1D). These results suggested that COMMD1 may interact with
lamin A.
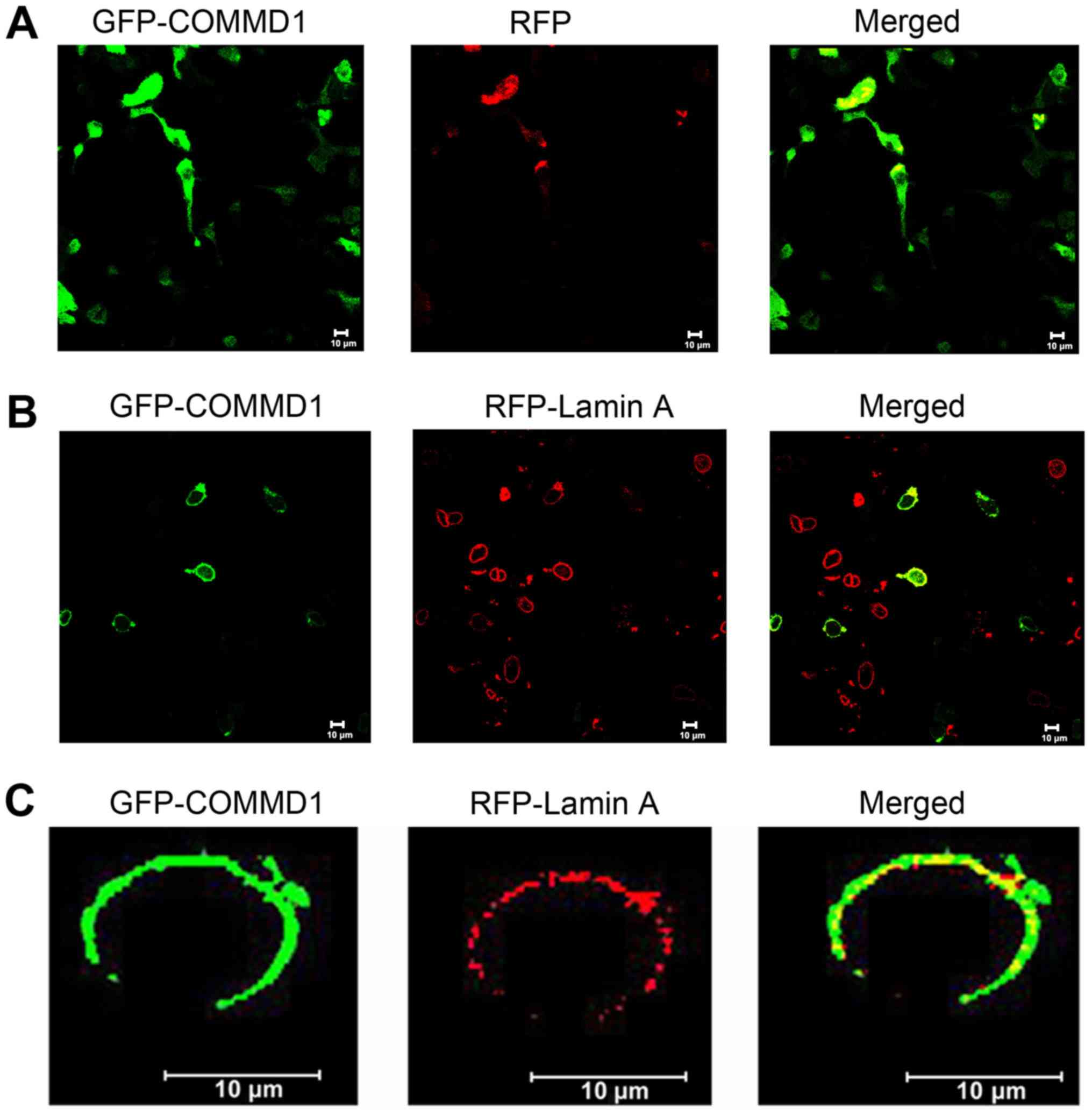
COMMD1 colocalizes with lamin A
To further verify the interaction between COMMD1 and
lamin A, the subcellular localization of COMMD1 and lamin A was
analyzed using fluorescence confocal microscopy. As shown in
Fig. 2A, COMMD1 was predominantly
cytoplasmic. The results of the co-transfection of 293 cells with
GFP-tagged COMMD1 and RFP-tagged lamin A plasmids demonstrated that
COMMD1 co-localized with lamin A in these cells (Fig. 2B and C).
COMMD1 physically interacts with lamin
A
A co-immunoprecipitation assay was conducted to
further test the interaction between COMMD1 and lamin A. Fig. 3A shows that COMMD1 interacted with
lamin A. The interaction between endogenous COMMD1 and lamin A in
293 cells was evaluated by immunoprecipitation using anti-COMMD1
and anti-lamin A/C antibodies followed by western blotting with an
anti-lamin A/C or anti-COMMD1 antibody. Following the
co-transfection of 293 cells with the pCMV-Flag-lamin A plasmid,
and the pCMV-HA-COMMD1 or pCMV-HA plasmid, immunoprecipitation was
performed using an anti-HA antibody, followed by western blotting
using an anti-Flag and anti-HA antibody. As shown in Fig. 3B, HA-COMMD1 was present in cell
lysates from 293 cells transfected with both the pCMV-Flag-lamin A
plasmid and the pCMV-HA-COMMD1 plasmid but not in cell lysates from
293 cells cotransfected with the pCMV-Flag-lamin A and pCMV-HA
plasmids. Collectively, these results suggested that COMMD1
interacts with lamin A in vivo.
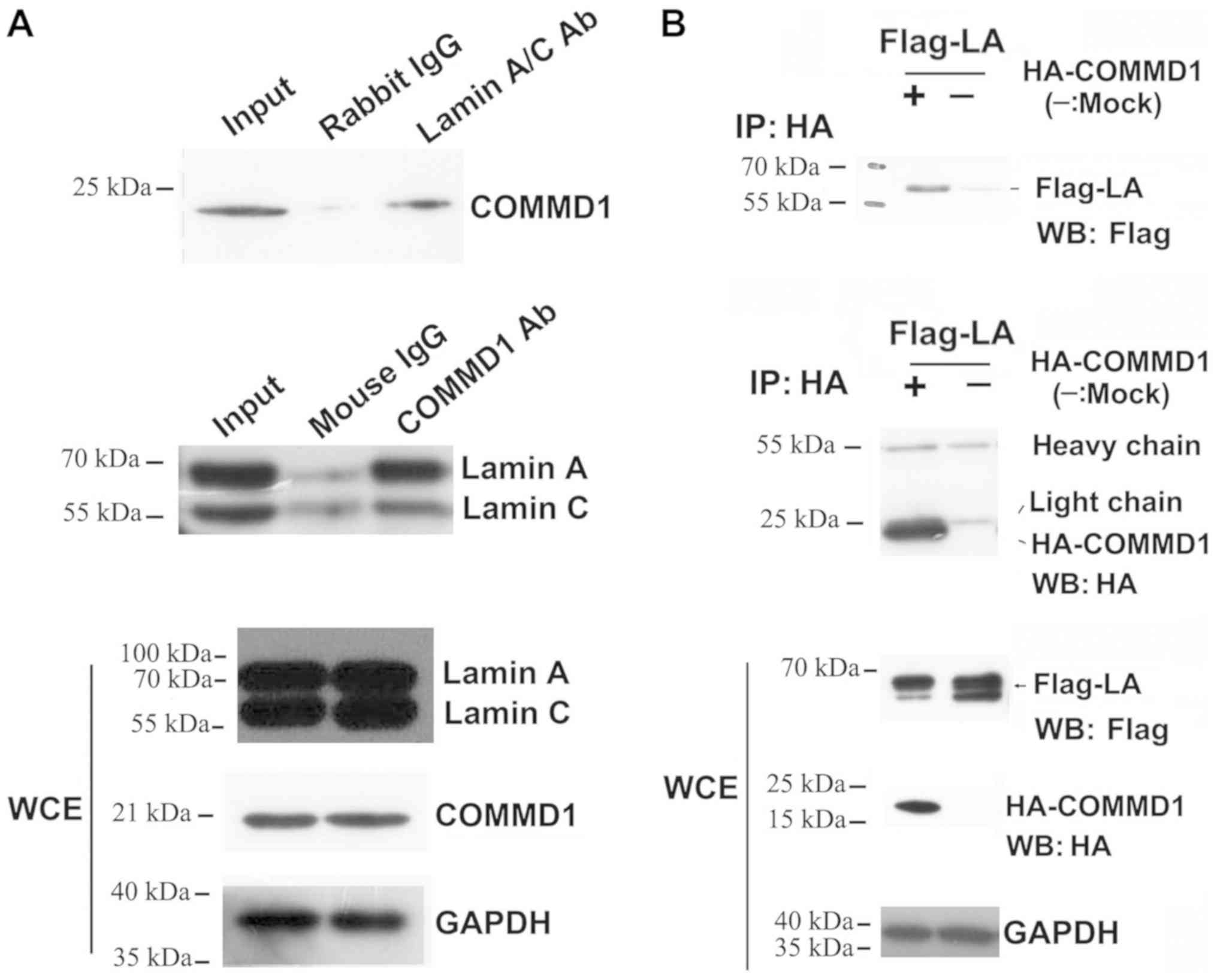 | Figure 3.COMMD1 interacts with lamin A. (A) IP
of 293 cell extracts with anti-lamin A/C or anti-COMMD1 antibodies,
followed by western blotting with anti-COMMD1 and anti-lamin A/C
antibodies. (B) Following co-transfection of 293 cells with pCMV-HA
or HA-COMMD1 and Flag-lamin A, IP was conducted with an anti-HA
antibody, followed by western blotting using an anti-Flag antibody.
COMMD1, copper metabolism MURR1 domain-containing 1; HA,
hemagglutinin; Ab, antibody; IgG, immunoglobulin G; WCE, whole cell
extract; IP, immunoprecipitation; WB, western blotting. |
Discussion
In the present study, lamin A binding proteins were
screened using the yeast two-hybrid system, and a novel interaction
was identified between COMMD1 and lamin A. In addition to
interacting with lamin A, COMMD1 was also observed to interact with
prelamin A in other yeast two-hybrid screens (data not shown). The
yeast two-hybrid system is a powerful method for the identification
of proteins that interact with one another; however, it exhibits
high false positive rates and involves non-physiological conditions
(20). To overcome these
drawbacks, the interaction between COMMD1 and lamin A was further
validated using co-localization and co-immunoprecipitation
experiments. Co-localization experiments using fluorescent confocal
microscopy revealed that COMMD1 and lamin A co-localized to the
nuclear envelope, which indicated the potential functional
importance of their interaction. The interaction between COMMD1 and
lamin A was also tested in endogenous and exogenous
co-immunoprecipitation experiments. Collectively, the results of
the present study revealed that COMMD1 physically interacts with
lamin A.
COMMD1 is the best characterized member of the COMMD
protein family, which consists of 10 subgroups (COMMD1-COMMD10)
(21). A mutation in COMMD1 was
originally identified as the genetic cause of canine copper
toxicosis (22). Following that
report, COMMD1 was found to be a pleiotropic protein that
participates in several cellular processes, ranging from copper
homoeostasis and sodium transport, to the regulation of the NF-κB
pathway and the expression of hypoxia-inducible factor-1α
(HIF-1α)-mediated genes (23).
COMMD1 suppresses NF-κB activity; however, the activation of NF-κB
in Zmpste24-deficient mice resulted in premature aging
(24). COMMD1 was found to
regulate HIF-1α activity during the adaptation to hypoxia by
controlling HIF-1α protein stability (25). Furthermore, the stabilization of
HIF-1α increased the lifespan of Caenorhabditis elegans
(26,27). Another COMMD1 binding partner,
superoxide dismutase 1, which is regulated by COMMD1 (28,29),
has also been shown to regulate normal cellular lifespan (30,31).
A G608G mutation in the LMNA gene or the loss of ZMPSTE24
activity resulted in the accumulation of unprocessed prelamin A in
cells, which induced DNA damage and genomic instability, and led to
premature aging (8,9). DNA damage stimulated the
relocalization of COMMD1 into the nucleoplasm and its interaction
with alternate reading frame p14ARF (ARF) in humans and p19ARF in
mice (32). ARF stabilizes the
basal level of COMMD1 through K63-dependent polyubiquitination
(32), which also promotes
cellular senescence in mouse and human cells by regulating the p53
pathway (33,34), and mediates resistance to cell
cycle arrest in LMNA-/− cells (35).
Lamin A is first synthesized as a prelamin A
precursor with a conserved CAAX domain that is proteolytically
processed by ZMPSTE24, removing the final 15 amino acids (36). The mature lamin A protein comprises
a 28-residue positively charged globular head domain, a central
α-helical rod-shaped domain, a conserved nuclear localization
signal and one immunoglobulin (Ig)-like structure domain formed of
116 residues at the C-terminus (37). The Ig-like domain was identified as
an interaction hotspot that regulates interactions between lamin A
and its binding partners (37,38),
its destabilization was found to affect most lamin A
protein-protein interactions (3).
The yeast two-hybrid results from the present study suggested that
COMMD1 may interact with the C-terminus of lamin A. The interaction
between COMMD1 and lamin A was verified using co-localization and
co-immunoprecipitation assays. COMMD1 is highly conserved in
vertebrates (39) and ubiquitously
expressed in diverse eukaryotic tissues (40). COMMD1 predominantly localizes in
the cytoplasm surrounding the nucleus (41,42);
however, a small fraction of COMMD1 can be found in the nucleus
(43,44). COMMD1 contains a COMM domain near
its C-terminus, which serves as a scaffold for protein-protein
interactions (21), and has two
highly conserved nuclear export signals, which are necessary and
sufficient to induce nuclear export (44). In addition, in nucleocytoplasmic
shuttling, COMMD1 may interact with lamin A at the nuclear envelope
(44). The nuclear export of
COMMD1 is important in the regulation of NF-κB and HIF-1 activity
in response to cellular stress (44). The interaction between COMMD1 and
lamin A may also be involved in the regulation of the translocation
of COMMD1 to the nucleoplasm from the perinuclear regions following
DNA damage (32). In the present
study, it was found that COMMD1 interacts with the C-terminus of
lamin A; however, these results do not indicate if the interaction
between COMMD1 and lamin A is direct or indirect.
Co-immunoprecipitation and confocal fluorescence microscopy assays
could not exclude the fact that other mediators are involved in
this protein-protein interaction complex. As such, how and where
COMMD1 interacts with lamin A requires further study.
In conclusion, to the best of our knowledge, the
present study provided the first evidence that COMMD1 physically
interacts with lamin A, suggesting possible roles for COMMD1 in
cellular senescence and nucleocytoplasmic transport. Further
studies are required to elucidate the precise mechanisms underlying
the lamin A-COMMD1 interaction with regard to the aging process and
laminopathies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Brian K.
Kennedy (Buck Institute for Research on Aging), Professor Matt
Kaeberlein (University of Washington), Professor Yousin Suh (Albert
Einstein College of Medicine) and Professor Baohua Liu (Shenzheng
University) for technical assistance and discussion.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81671399 and
81170327), Ordinary University Innovation Team Construction Project
of Guangdong Province (grant no. 2015KCXTD022), Unique Innovative
Projects in Ordinary University of Guangdong Province (grant no.
2015KTSCX049), Dongguan International Science & Technology
Cooperation Project (grant no. 201650812001) and Scientific
Research Fund of Guangdong Medical University (grant nos. 2XQ11009
and 2XK14030).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, WC and ZZ designed the study. ZJ drafted the
manuscript. ZJ, JZ, QP, HZ and YY performed and analyzed the
experiments. HC, WZ and XS analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HGPS
|
Hutchinson-Gilford progeria
syndrome
|
|
COMMD1
|
copper metabolism MURR1
domain-containing 1
|
|
GFP
|
green fluorescent protein
|
|
RT-PCR
|
reverse transcription polymerase chain
reaction
|
|
NF-κB
|
nuclear factor-κB
|
|
ARF
|
alternate reading frame protein
product of the CDKN2A locus
|
References
|
1
|
Dechat T, Pfleghaar K, Sengupta K, Shimi
T, Shumaker DK, Solimando L and Goldman RD: Nuclear lamins: Major
factors in the structural organization and function of the nucleus
and chromatin. Genes Dev. 22:832–853. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Röber RA, Weber K and Osborn M:
Differential timing of nuclear lamin A/C expression in the various
organs of the mouse embryo and the young animal: A developmental
study. Development. 105:365–378. 1989.PubMed/NCBI
|
|
3
|
Dittmer TA, Sahni N, Kubben N, Hill DE,
Vidal M, Burgess RC, Roukos V and Misteli T: Systematic
identification of pathological lamin A interactors. Mol Biol Cell.
25:1493–1510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rohani L, Johnson AA, Arnold A and
Stolzing A: The aging signature: A hallmark of induced pluripotent
stem cells? Aging Cell. 13:2–7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gonzalez JM, Pla D, Perez-Sala D and
Andres V: A-type lamins and Hutchinson-Gilford progeria syndrome:
Pathogenesis and therapy. Front Biosci (Schol Ed). 3:1133–1146.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Andrés V and González JM: Role of A-type
lamins in signaling, transcription, and chromatin organization. J
Cell Biol. 187:945–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McCord RP, Nazario-Toole A, Zhang H,
Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J and Cao K:
Correlated alterations in genome organization, histone methylation,
and DNA-lamin A/C interactions in Hutchinson-Gilford progeria
syndrome. Genome Res. 23:260–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Musich PR and Zou Y: Genomic instability
and DNA damage responses in progeria arising from defective
maturation of prelamin A. Aging (Albany NY). 1:28–37. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh S, Srivastava A, Srivastava P,
Dhuriya YK, Pandey A, Kumar D and Rajpurohit CS: Advances in stem
cell research-a ray of hope in better diagnosis and prognosis in
neurodegenerative diseases. Front Mol Biosci. 3:722016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meier I and Brkljacic J: The Arabidopsis
nuclear pore and nuclear envelope. Arabidopsis Book. 8:e01392010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuo B, Yang J, Wang F, Wang L, Yin Y, Dan
J, Liu N and Liu L: Influences of lamin A levels on induction of
pluripotent stem cells. Biol Open. 1:1118–1127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maggi L, Carboni N and Bernasconi P:
Skeletal muscle laminopathies: A review of clinical and molecular
features. Cells. 5:2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Korfali N, Wilkie GS, Swanson SK, Srsen V,
de Las Heras J, Batrakou DG, Malik P, Zuleger N, Kerr AR, Florens L
and Schirmer EC: The nuclear envelope proteome differs notably
between tissues. Nucleus. 3:552–564. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wong X, Luperchio TR and Reddy KL: NET
gains and losses: The role of changing nuclear envelope proteomes
in genome regulation. Curr Opin Cell Biol. 28:105–120. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gonzalo S, Kreienkamp R and Askjaer P:
Hutchinson-Gilford Progeria Syndrome: A premature aging disease
caused by LMNA gene mutations. Ageing Res Rev. 33:18–29. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Navarro CL, De Sandre-Giovannoli A,
Bernard R, Boccaccio I, Boyer A, Geneviève D, Hadj-Rabia S,
Gaudy-Marqueste C, Smitt HS, Vabres P, et al: Lamin A and ZMPSTE24
(FACE-1) defects cause nuclear disorganization and identify
restrictive dermopathy as a lethal neonatal laminopathy. Hum Mol
Genet. 13:2493–2503. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burtner CR and Kennedy BK: Progeria
syndromes and ageing: What is the connection? Nat Rev Mol Cell
Biol. 11:567–578. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ghebre YT, Yakubov E, Wong WT,
Krishnamurthy P, Sayed N, Sikora AG and Bonnen MD: Vascular aging:
Implications for cardiovascular disease and therapy. Transl Med
(Sunnyvale). 6:2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naito M, Omoteyama K, Mikami Y, Takagi M
and Takahashi T: Suppression of lamin A/C by short hairpin RNAs
promotes adipocyte lineage commitment in mesenchymal progenitor
cell line, ROB-C26. Histochem Cell Biol. 137:235–247. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kubben N, Voncken JW, Demmers J, Calis C,
van Almen G, Pinto Y and Misteli T: Identification of differential
protein interactors of lamin A and progerin. Nucleus. 1:513–525.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burstein E, Hoberg JE, Wilkinson AS,
Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW
and Duckett CS: COMMD proteins, a novel family of structural and
functional homologs of MURR1. J Biol Chem. 280:22222–22232. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
van De Sluis B, Rothuizen J, Pearson PL,
van Oost BA and Wijmenga C: Identification of a new copper
metabolism gene by positional cloning in a purebred dog population.
Hum Mol Genet. 11:165–173. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bartuzi P, Hofker MH and van de Sluis B:
Tuning NF-κB activity: A touch of COMMD proteins. Biochim Biophys
Acta. 1832:2315–2321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Osorio FG, Bárcena C, Soria-Valles C,
Ramsay AJ, de Carlos F, Cobo J, Fueyo A, Freije JM and López-Otín
C: Nuclear lamina defects cause ATM-dependent NF-κB activation and
link accelerated aging to a systemic inflammatory response. Genes
Dev. 26:2311–2324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van de Sluis B, Muller P, Duran K, Chen A,
Groot AJ, Klomp LW, Liu PP and Wijmenga C: Increased activity of
hypoxia-inducible factor 1 is associated with early embryonic
lethality in Commd1 null mice. Mol Cell Biol. 27:4142–4156. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mehta R, Steinkraus KA, Sutphin GL, Ramos
FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D and Kaeberlein M:
Proteasomal regulation of the hypoxic response modulates aging in
C. elegans. Science. 324:1196–1198. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leiser SF, Begun A and Kaeberlein M: HIF-1
modulates longevity and healthspan in a temperature-dependent
manner. Aging Cell. 10:318–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vonk WI, Kakkar V, Bartuzi P, Jaarsma D,
Berger R, Hofker MH, Klomp LW, Wijmenga C, Kampinga HH and van de
Sluis B: The Copper metabolism MURR1 domain protein 1 (COMMD1)
modulates the aggregation of misfolded protein species in a
client-specific manner. PLoS One. 9:e924082014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vonk WI, Wijmenga C, Berger R, van de
Sluis B and Klomp LW: Cu,Zn superoxide dismutase maturation and
activity are regulated by COMMD1. J Biol Chem. 285:28991–29000.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blander G, de Oliveira RM, Conboy CM,
Haigis M and Guarente L: Superoxide dismutase 1 knock-down induces
senescence in human fibroblasts. J Biol Chem. 278:38966–38969.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun X, Komatsu T, Lim J, Laslo M, Yolitz
J, Wang C, Poirier L, Alberico T and Zou S: Nutrient-dependent
requirement for SOD1 in lifespan extension by protein restriction
in Drosophila melanogaster. Aging Cell. 11:783–793. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang Y, Wu M and Li HY: Tumor suppressor
ARF promotes non-classic proteasome-independent polyubiquitination
of COMMD1. J Biol Chem. 283:11453–11460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Larsson LG: Oncogene- and tumor suppressor
gene-mediated suppression of cellular senescence. Semin Cancer
Biol. 21:367–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Evan GI and d'Adda di Fagagna F: Cellular
senescence: Hot or what? Curr Opin Genet Dev. 19:25–31. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nitta RT, Smith CL and Kennedy BK:
Evidence that proteasome-dependent degradation of the
retinoblastoma protein in cells lacking A-type lamins occurs
independently of gankyrin and MDM2. PLoS One. 2:e9632007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barrowman J, Wiley PA, Hudon-Miller SE,
Hrycyna CA and Michaelis S: Human ZMPSTE24 disease mutations:
Residual proteolytic activity correlates with disease severity. Hum
Mol Genet. 21:4084–4093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu B and Zhou Z: Lamin A/C, laminopathies
and premature ageing. Histol Histopathol. 23:747–763.
2008.PubMed/NCBI
|
|
38
|
Goldman RD, Goldman AE and Shumaker DK:
Nuclear lamins: Building blocks of nuclear structure and function.
Novartis Found Symp. 264:3–16; discussion 16–21, 227–230.
2005.PubMed/NCBI
|
|
39
|
Maine GN and Burstein E: COMMD proteins
and the control of the NF kappa B pathway. Cell Cycle. 6:672–676.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Klomp AE, van de Sluis B, Klomp LW and
Wijmenga C: The ubiquitously expressed MURR1 protein is absent in
canine copper toxicosis. J Hepatol. 39:703–709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lian M and Zheng X: HSCARG regulates
NF-kappaB activation by promoting the ubiquitination of RelA or
COMMD1. J Biol Chem. 284:17998–18006. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Drevillon L, Tanguy G, Hinzpeter A, Arous
N, de Becdelièvre A, Aissat A, Tarze A, Goossens M and Fanen P:
COMMD1-mediated ubiquitination regulates CFTR trafficking. PLoS
One. 6:e183342011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
de Becdelièvre A, Rocca J, Aissat A,
Drévillon L, Moutereau S, Le Gouvello S, Hinzpeter A, Tarze A and
Fanen P: COMMD1 modulates noxious inflammation in cystic fibrosis.
Int J Biochem Cell Biol. 45:2402–2409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Muller PA, van de Sluis B, Groot AJ,
Verbeek D, Vonk WI, Maine GN, Burstein E, Wijmenga C, Vooijs M,
Reits E and Klomp LW: Nuclear-cytosolic transport of COMMD1
regulates NF-kappaB and HIF-1 activity. Traffic. 10:514–527. 2009.
View Article : Google Scholar : PubMed/NCBI
|