Introduction
Brucella is a group of α-2
Proteobacteria that has a great impact on animal and human
health worldwide (1). Infection
with Brucella results in brucellosis, one of the most common
bacterial zoonotic diseases in humans and cattle globally (2). An estimated 500,000 cases of
brucellosis occur each year globally (3). Brucellosis can not only lead to the
reproductive failure of livestock but also decrease human
productivity. As a result, Brucella species have been
regarded as potential agricultural, animal husbandry, civilian and
even bioterrorism agents (4,5).
During chronic infection, bacteria can organize
themselves into matrix-enclosed microcolonies or aggregates, termed
biofilms (6,7). Biofilm formation is a critical
survival mechanism for bacteria in the environment (8). Altered gene and protein expression in
biofilms is responsible for cell virulence, adherence and drug
resistance (9,10). Additionally, biofilm-grown
microorganisms have an inherent lack of susceptibility to
antibiotics (11–13). Brucella melitensis (B.
melitensis) has been suggested to form biofilms during its life
cycle (14). Wild-type B.
abortus can also develop biofilms under nutritionally
deficient, microaerobic conditions (15). Previous studies have investigated
several virulence and drug resistance-associated proteins from
planktonic Brucella, such as lipopolysaccharide (16), B lymphocyte mitogen (17) and outer membrane proteins (18). However, numerous different types of
bacterial infections are presumed to be due to bacteria growing in
a biofilm state including cystic fibrosis-related lung infections,
biomaterial-related infections, chronic wounds, and endocarditis.
The USA National Institutes of Health has estimated that 80% of all
infections are biofilm-related (19). However, little is known concerning
the proteins associated with biofilm-mediated infections.
The present study investigated differences in
protein and gene expression of B. abortus cultured under
biofilm compared with planktonic conditions. The differential
proteins unique to biofilms and planktonic B. abortus were
identified by employing two-dimensional (2-D) electrophoresis and
matrix-assisted laser desorption/ionization-tandem time of
flight-mass spectrometry (MALDI-TOF/TOF-MS) analyses. The
differential genes were identified by high-throughput sequencing
and bioinformatic analysis. Findings of the current study may help
to understand the underlying molecular mechanisms that control
biofilm formation in B. abortus.
Materials and methods
Bacterial strains and culture
conditions
B. abortus strain isolate A3313 was used in
this study, which was isolated from the abortus of dairy cows in
Hohhot District, Inner Mongolia, China. The A3313 strain was grown
in Brucella broth medium (BD Biosciences) at 37°C with 5%
CO2.
All the experiments related to the cultivation of
Brucella and its biofilms, as well as the operation of
viable bacteria were conducted in a Biosafety Level 3 Laboratory in
the College of Veterinary Medicine, Huazhong Agricultural
University (Wuhan, China). For the experiments of electron
microscope observation, 2-D electrophoresis, high-throughput
sequencing and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) analysis, the Brucella and its biofilm
were effectively inactivated with glutaraldehyde or bacterial
lysate before being removed from the Biosafety Level 3
Laboratory.
Biofilm culture and microscopic
observation
Brucella broth was added to 6-well cell
culture plates. A clean coverslip sterilized by autoclaving (121°C,
20 min) was then put in each well, and the A3313 bacterial
suspension was inoculated on the coverslip at 2 ml/well. The
culture plate was placed at 37°C with 5% CO2, and the
culture medium was changed every 48 h until a complete biofilm was
formed. The coverslips were taken out, gently washed three times
with phosphate-buffered saline (PBS; 30 mM, pH 7.4), and then fixed
immediately with 2.5% glutaraldehyde for 6–8 h at 4°C. After being
washed with PBS, biofilms were stained with 200 µl 1% crystal
violet (Ding Bei Biological Technology Co., Ltd.) for 20 min at
room temperature. These procedures were conducted to protect
biofilms from falling off from the abiotic surfaces. The biofilms
were observed under a phase-contrast light microscope
(magnification, ×20) (Axiovert 135; Zeiss AG).
For scanning electron microscope observation,
biofilms were fixed with 2% osmic acid at room temperature until
black. After washing with 0.1 M PBS for three times, the samples
underwent sequential dehydration with gradient ethanol solutions
(30, 50, 70 and 90%) for 15 min each. Then, samples were dehydrated
with 100% ethanol twice (15 min each), and dried with a critical
point dryer. The dry samples were fixed on the sample stage with
conducting resin, and sprayed gold with ion sputtering equipment
(15 mA) for 2 min. The biofilms were observed under a scanning
electron microscope.
2-D electrophoresis
Biofilms and planktonic bacteria were used for 2-D
electrophoresis. For biofilm culture, the A3313 strain was grown in
Brucella broth medium in Petri dishes at 37°C and 5%
CO2. The culture medium was changed every 48 h for 8
days. After removing the supernatant, the plates were rinsed twice
with PBS. Biofilms were detached by scraping. Planktonic B.
abortus was cultured in the same condition. The culture medium
was collected and centrifugally washed twice with PBS. The
planktonic B. abortus was resuspended with PBS.
Protein was precipitated as previously described
(10,20). The biofilm and planktonic bacteria
were harvested by centrifugation (6,000 × g) at 4°C for 10 min. The
bacteria were washed four times with a solution containing 3 mM
KCl, 68 mM NaCl, 9 mM NaH2PO4 and 115 mM
KH2PO4, and then resuspended in lysis buffer
(7 M urea, 2 M thiourea, 4% CHAPS, 10 mM DTT and 2% Pharmalyte pH
3–10) with protease inhibitor. After ultrasonic decomposition on
ice at 40% maximum power for 90 cycles (5 sec on, 10 sec off; the
ultrasound conditions were the same for biofilm disruption and
planktonic cells), unbroken cells and the cell debris were
incubated at 25°C for 30 min, and removed by centrifugation (10,000
× g) for 30 min at 25°C. Proteins in the supernatant were
precipitated in 10% trichloroacetic acid (TCA) on ice for 30 min.
Precipitated proteins were washed with chilled acetone and then
centrifuged at 10,000 × g for 10 min at 4°C. The air-dried proteins
were dissolved in 400 µl sample preparation solution [2-D lysate:
9.5 M urea, 4% CHAPS, 2% (v/v) ampholytes, and 60 mM DTT; 10 µl 50
XProtein Inhibitor Cocktail Set I (Merck KGaA) was added into 500
µl 2-D lysate] at 25°C for 30 min and centrifuged at 10,000 × g for
20 min at 25°C. The protein concentration was determined via a
Bradford protein assay.
Proteins (200 µg) were separated by 2-D
electrophoresis. Isoelectric focusing for the first dimension was
performed in precast Immobiline DryStrips (GE Healthcare Life
Sciences) with a nonlinear gradient of pH 3 to 10 in an Ethan
IPGphor Isoelectric Focusing System (GE Healthcare Life Sciences)
according to the manufacturer's instructions. The electrophoresis
conditions were 30 V for 12 h, 500 V for 1 h, 1,000 V for 1 h,
8,000 V for 8 h and 500 V for 4 h. The second dimension (SDS-PAGE)
was conducted vertically in a Hofer SE 600 (GE Healthcare Life
Sciences) using 12.5% polyacrylamide gels. The resolved proteins
were then stained with silver for 30 min at room temperature and
scanned with UMax Powerlook 2110Xl (GE Healthcare Life Sciences).
All experiments were performed in triplicate. The gels were
analyzed with the Image Master Platinum version 5.0 software (GE
Healthcare Life Sciences). The normalized protein amount for each
protein spot was calculated as the ratio of that spot volume to the
total spot volume on the gel. Significant differences between two
groups were determined using the Student's t-test, and a fold
change ≥1.5 was considered the threshold value.
MS analysis
The differentially expressed protein spots were
excised from the 2-D gels and then subjected to MALDI-TOF/TOF-MS
analysis (Shanghai Applied Protein Technology Co. Ltd.). Before MS
analysis, the protein spots were digested in-gel by 0.1 mg/ml
trypsin for 2 h at 37°C and desalinated by Ziptip (EMD Millipore).
The digested samples were then freeze-dried. After being
re-dissolved, 1 µl samples were spotted on the target plate
(Immobiline DryStrips; 13 cm, pH 3–10 NL) with air drying, and then
0.5 µl supersaturated α-cyano-4-hydroxycinnamic acid matrixes
(Sigma-Aldrich; Merck KGaA) prepared in 50% acetonitrile with 0.1%
trifluoroacetic acid were spotted and naturally dried. All MALDI-MS
and MS/MS data were acquired in the positive reflectron ion mode on
a 4800 Plus MALDI TOF/TOF Analyzer (AB Sciex LLC). Samples were
irradiated by a 355-nm Nd:YAG laser (355 nm), and the acceleration
voltage was 2 kV. The scanned area for MS was 800–4,000 Da, and the
parent ion with signal:noise ratio >50 was selected for MS/MS
analysis. Data from MALDI-MS and MS/MS were subjected to a Database
Search from NCBI or Uniprot (NCBI preferentially) using Mascot
version 2.2 software (Matrix Science), and a Mascot score was
calculated. The MS/MS spectra were subjected to similarity searches
using the BLASTX algorithm. The parameter settings were trypsin
digestion, fixed modification of carbamidomethyl, dynamical
modification of oxidation (M), unrestricted protein mass, peptide
mass tolerance for monoisotopic data of ±100 ppm, fragment mass
tolerance of ±0.4 Da, peptide charge state of 1+, and one maximum
missed cleavage.
High-throughput sequencing
RNA sequencing of the biofilms and planktonic
bacteria were performed at Genergy Biotechnology Co., Ltd. Total
RNA was isolated and examined by NanoDrop spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and 1%
agarose gel electrophoresis (Tanon Science and Technology Co.,
Ltd.). An RNA library was then constructed and sequenced on the
Illumina HiSeq 2500 platform (Illumina, Inc.). The data are
available at National Center for Biotechnology Information under
the accession numbers SRX1604658 (biofilm conditions; http://www.ncbi.nlm.nih.gov/sra/?term=SRX1604658)
and SRX1604659 (planktonic conditions; http://www.ncbi.nlm.nih.gov/sra/SRX1604659/).
Bioinformatics analyses
The raw reads were evaluated using RSeQC 2.3.2
(http://rseqc.sourceforge.net/) (21), and sequence alignment was conducted
with TopHat 2.0.10 (http://ccb.jhu.edu/software/tophat/index.shtml)
(22). The remaining reads were
used for the following analyses.
The mRNA expression levels were detected based on
Cufflinks 2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/)
software (23). Differentially
expressed mRNAs were defined based on strict criteria [q value
≤0.05, and log2(fold change) ≥1] using the Cuffdiff program
(23). Functional annotation of
differentially expressed genes was carried out using various
bioinformatics procedures, including Gene Ontology (GO) (24) and Kyoto Encyclopedia of Genes and
Genomes (KEGG, Kolmogorov-Smirnov value <0.05) (25).
RT-qPCR
The mRNA levels of differential proteins identified
through 2-D electrophoresis and function-associated genes
identified by high-throughput sequencing and bioinformatics
analyses were detected via RT-qPCR according a previously described
method (26). In brief, total RNA
was isolated from biofilms and planktonic cells (1×106
cells) of the A3313 strain using a Takara MiniBEST Universal RNA
Extraction kit (Takara Biotechnology Co., Ltd.). The RNA quality
and concentration were determined by a NanoDrop 2000c
spectrophotometer (Thermo Fisher Scientific, Inc.). The RNA was
reverse-transcribed into cDNA using a PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd.; 42°C for 15 min). The 16S rRNA
housekeeping gene was amplified as the internal control. The
specific primers are listed in Table
SI. The SYBR Green PCR method was performed using an SYBR
Premix Ex Taq kit (Takara Biotechnology Co., Ltd.). The qPCR
reaction was carried out under the following conditions: 95°C for
30 sec, 40 cycles of 95°C for 3 sec and 60°C for 30 sec. Relative
mRNA expression ratios of selected genes were calculated with the
2−ΔΔCq method (27).
The experiment was performed with three replications.
Western blot analysis
The levels of differentially expressed proteins
identified through 2-D electrophoresis, described above, were
measured by western blotting. Briefly, cells were lysed in RIPA
buffer (Sigma-Aldrich; brand of Merck KGaA), and the protein
concentration was measured using BCA Protein Assay Kit (Thermo
Fisher Scientific, Inc.). Protein samples (20 µg) were separated
using 12% SDS-PAGE and transferred onto a polyvinylidene fluoride
membrane (GE Healthcare), and the membrane was blocked with 100 mM
Tris, 150 mM NaCl, 0.05% Tween-20 (TBST), containing 5% dry milk
powder for 2 h at room temperature. Then, the blocked membrane was
incubated with sera from primary antibodies [hypothetical protein
BAbS19_I16470 (1:1,000; cat. no. orb309412; Biorbyt Ltd.);
chaperone protein DnaJ (1:1,000; cat. no. PA3-018; Invitrogen;
Thermo Fisher Scientific, Inc.); elongation factor Tu (1:1,000;
cat. no. ab210089; Abcam); Chaperonin Cpn60/TCP-1 (1:1,000; cat.
no. 3094R-100; BioVision, Inc.); polyprenyl synthetase (1:1,000;
cat. no. ab80647; Abcam); periplasmic binding protein (1:1,000;
cat. no. M30934-1; Wuhan Boster Biological Technology, Ltd.);
enolase (1:1,000; cat. no. sc-271384; Santa Cruz Biotechnology,
Inc.); acetyl-CoA carboxylase, a subunit (1:1,000; cat. no.
MAB6898; R&D Systems, Inc.); tryptophanyl-tRNA synthetase
(1:1,000; cat. no. ab31536; Abcam); aspartate-semialdehyde
dehydrogenase (1:1,000; cat. no. EM1708-10a; Jingke Huaxue;
exosporium protein B (1:1,000; cat. no. ab92932; Abcam);
enoyl-(acyl carrier protein) reductase (1:1,000; cat. no.
abx109426; Abbexa Ltd.); Omp16 (1:1,000; cat. no. ab93127; Abcam)]
for 2 h at room temperature and then incubated with horseradish
peroxidase-labeled secondary antibodies (1:5,000; cat. no. 29139;
Invitrogen; Thermo Fisher Scientific, Inc.) in blocking buffer for
1 h at room temperature. After washing with 0.05% Tween-20 (TBST),
the membranes were incubated with DAB substrate (Tiangen Biotech
Co., Ltd.) for 10 min at room temperature. Outer membrane protein
16 was used as a loading control. The western blot bands were
visualized using the Millipore ECL Western Blotting Detection
System (EMD Millipore).
Statistical analysis
All experiments were repeated three times except for
high-throughput sequencing, and the results were presented as the
mean ± standard deviation. Statistical analyses were performed
using GraphPad 6.0 (GraphPad Software, Inc.). P-values were
calculated using Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Biofilm observation
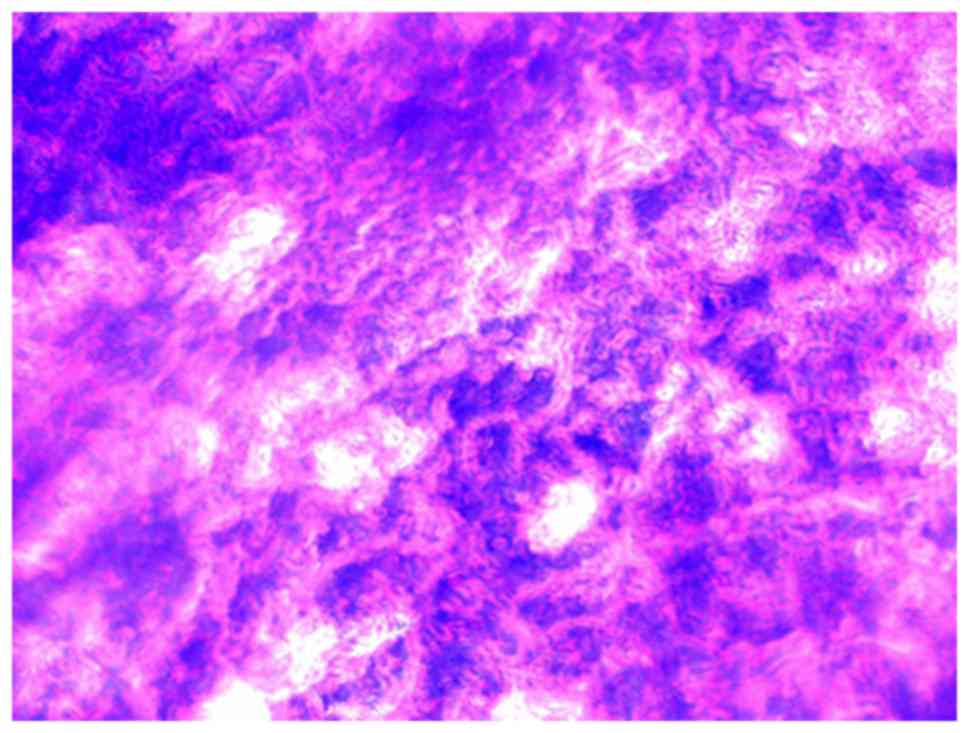
Biofilm formation was observed under a
phase-contrast light microscope. A large number of bacteria adhered
to the coverslips and formed a community (Fig. 1). The structure of biofilms on
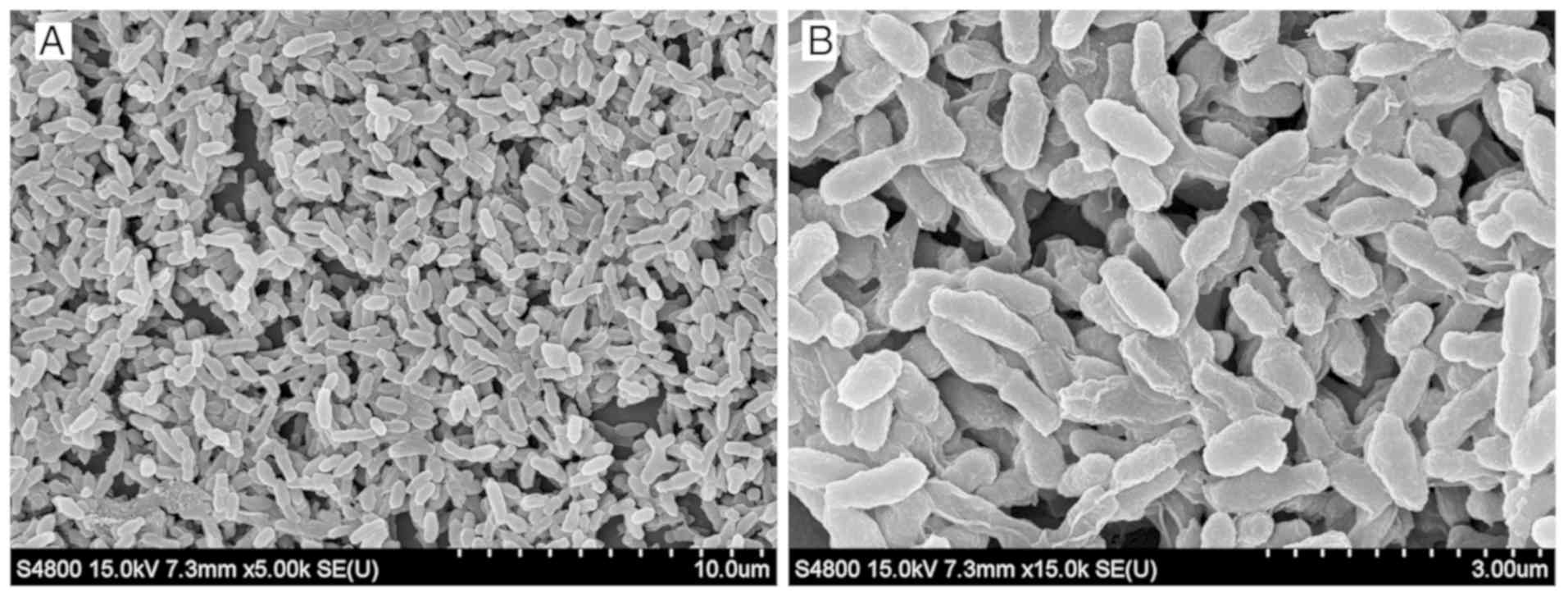
coverslips was observed under a scanning electron microscope. The
biofilms formed flake or microcolony clusters on the coverslips.
Colonies were wrapped together by the mucus they secreted, forming
an uneven, dense membrane structure (Fig. 2).
Comparative proteomics
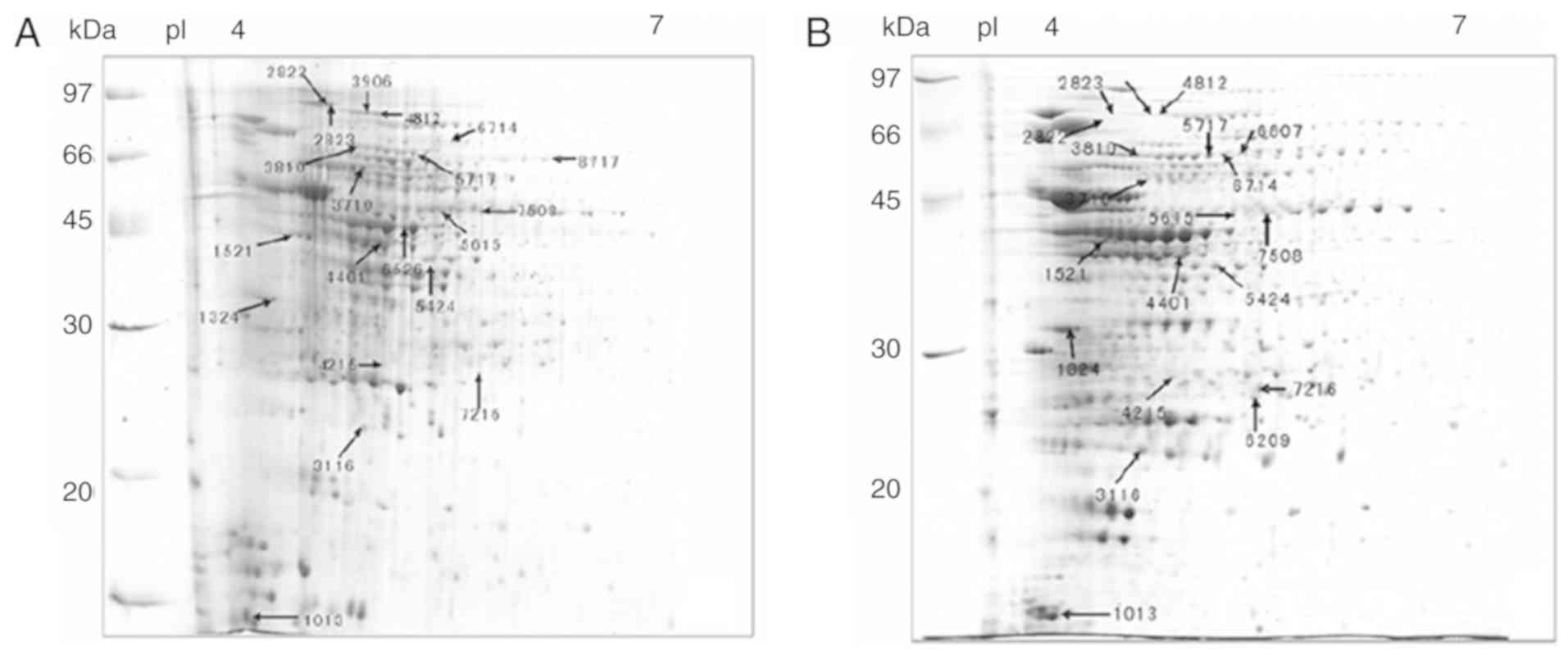
A total of 1,930 protein spots were detected in all
gels. The representative 2-D electrophoresis images of 20
differentially expressed protein spots (7 upregulated and 13
downregulated protein spots) between biofilms and planktonic cells
are presented in Fig. 3. Most
proteins were distributed in the range of isoelectric point 4–7.
MALDI MS and MS/MS analysis identified 20 protein spots
corresponding to 18 individual proteins, including 6 upregulated
(including catalase, extracellular solute-binding protein and
ubiquinol-cytochrome C reductase iron-sulfur subunit) and 12
downregulated ones (including elongation factor Tu, enolase,
isocitrate dehydrogenase and Chaperonin Cpn60/TCP-1; Table I).
 | Table I.Differentially expressed proteins in
the Brucella abortus A3313 biofilm, identified by
MALDI-TOF/TOF-MS analysis. |
Table I.
Differentially expressed proteins in
the Brucella abortus A3313 biofilm, identified by
MALDI-TOF/TOF-MS analysis.
|
|
|
|
|
|
|
|
|
| Fold
changef |
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Target no. | Spot
noa | Protein
identified | BLASTX similarity
matched protein/species/identify score | Theoretical
MW/pIb | Experimental
MW/pI | Mascot
scorec | No. of peptides
matchedd | Coverage
(%)e | Mean | Standard
deviation | P-value |
|---|
| E11 | 1916 | gi|189024835 | hypothetical
protein BAbS19_I16470 | 17153.2/4.96 | 18000/5.20 | 102 | 5 | 60 | −6.84 | 0.06 | 0.005 |
| E12 | 873 | gi|189020752 | DnaJ, chaperone
protein DnaJ | 41595.3/6.51 | 43000/7.10 | 336 | 13 | 8 | −5.55 | 0.14 | 0.003 |
| E13 | 1275 | gi|189019956 | elongation factor
Tu | 42749/5.29 | 35000/5.10 | 132 | 4 | 37 | −4.32 | 0.15 | 0.005 |
| J12 | 534 | gi|189020977 | Chaperonin
Cpn60/TCP-1 | 57479.4/5.08 | 60000/5.20 | 133 | 14 | 25 | −3.16 | 0.17 | 0.008 |
| E15 | 998 | gi|189023639 | polyprenyl
synthetase | 36201.5/5.04 | 42000/5.70 | 309 | 13 | 38 | −2.39 | 0.12 | 0.015 |
| E16 | 1054 | gi|189022660 | Periplasmic binding
protein | 37595.3/5.85 | 41000/6.00 | 105 | 12 | 52 | −2.19 | 0.17 | 0.006 |
| E17 | 701 | gi|189019858 | Enolase | 45404.2/5.03 | 49000/5.20 | 430 | 15 | 57 | −1.95 | 0.05 | 0.030 |
| E18 | 1220 | gi|189020664 | Acetyl-CoA
carboxylase, a subunit | 35075.2/6.14 | 35000/7.10 | 659 | 20 | 39 | −1.95 | 0.09 | 0.014 |
| E19 | 1007 | gi|189023395 | tryptophanyl-tRNA
synthetase | 39230.9/5.49 | 42000/6.20 | 109 | 12 | 43 | −1.90 | 0.09 | 0.007 |
| E20 | 875 | gi|189022544 |
aspartate-semialdehyde dehydrogenase | 37701.4/5.37 | 43000/6.10 | 131 | 9 | 38 | −1.88 | 0.06 | 0.002 |
| E21 | 1481 | gi|189020611 | ExsB protein | 25638.7/6.1 | 30000/7.30 | 178 | 11 | 40 | −1.82 | 0.10 | 0.014 |
| C24 | 1378 | gi|189023658 | enoyl-(acyl carrier
protein) reductase | 29205/5.61 | 33000/7.00 | 191 | 7 | 23 | −1.76 | 0.04 | 0.024 |
| E4 | 467 | gi|189022983 | catalase | 56525.1/6.52 | 56000/8.00 | 423 | 18 | 35 | 2.41 | 0.26 | 0.011 |
| E6 | 984 | gi|189022336 | extracellular
solute-binding protein | 39181.9/4.88 | 42000/5.00 | 316 | 10 | 31 | 1.78 | 0.01 | 0.002 |
| E7 | 1666 | gi|189024657 |
Ubiquinol-cytochrome C reductase
iron-sulfur subunit | 20053.9/5.35 | 25000/5.60 | 162 | 6 | 36 | 1.70 | 0.04 | 0.032 |
| E8 | 2126 | gi|189020978 | Chaperonin
Cpn10 | 10386.6/5.41 | 12000/5.80 | 98 | 8 | 69 | 1.68 | 0.05 | 0.004 |
| E9 | 1308 | gi|189024343 | Tetracycline
resistance protein TetB | 34252.1/6.92 | 32000/5.80 | 390 | 10 | 28 | 1.67 | 0.03 | 0.020 |
| E10 | 1775 | gi|189019259 | Ribosomal protein
L9 | 20957.7/4.86 | 22000/5.70 | 353 | 14 | 57 | 1.51 | 0.09 | 0.032 |
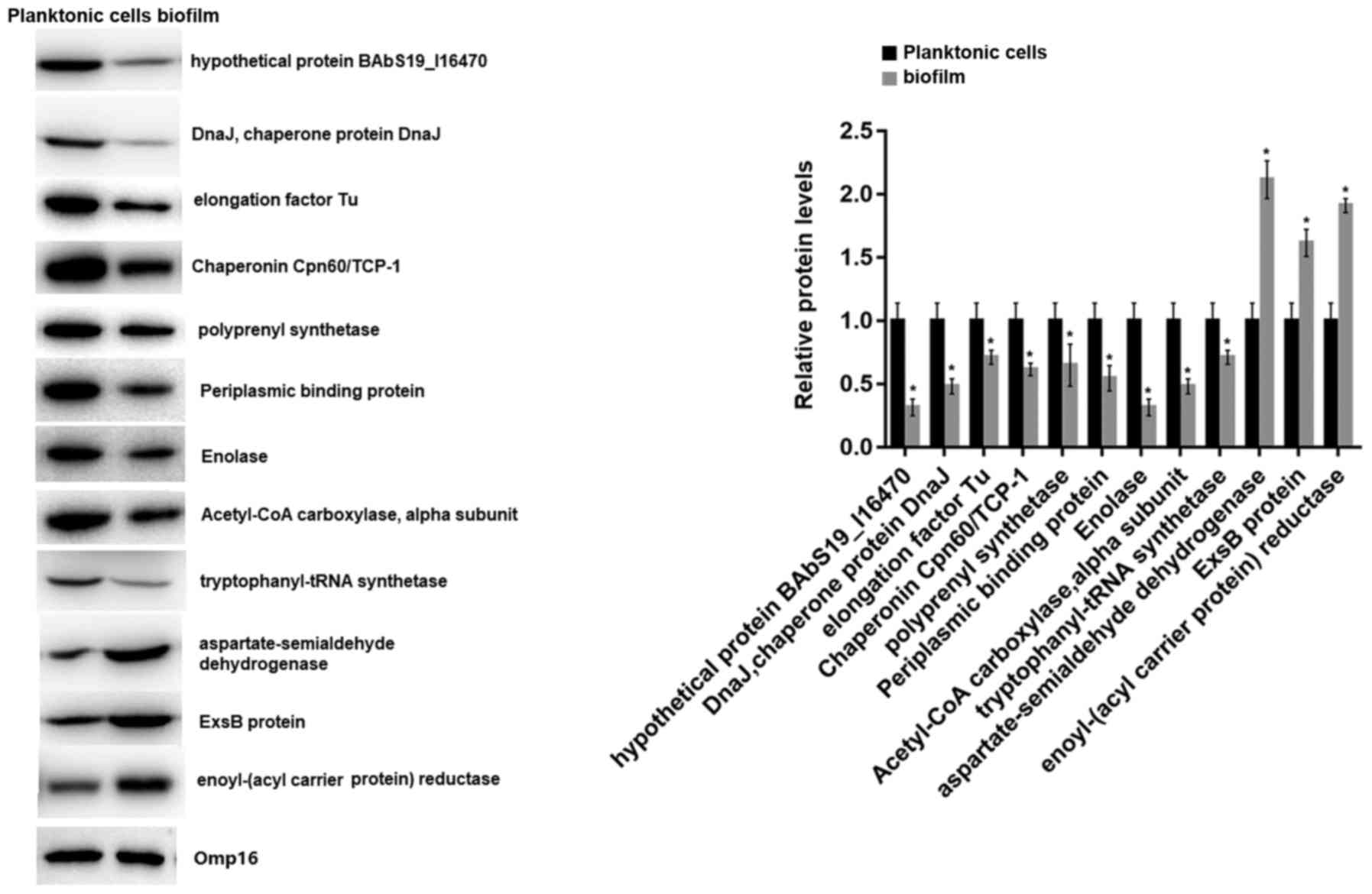
RT-qPCR and western blot analysis
The mRNA and protein expression levels of the 18
identified proteins in 2-D electrophoresis were detected by RT-qPCR
and western blot analyses, respectively. RT-qPCR analysis revealed
that all of the 18 genes were downregulated in biofilms compared
with planktonic cells, including elongation factor Tu (fold change
= −100.0), enolase (fold change = −8.6), isocitrate dehydrogenase
(fold change = −9.9), and Chaperonin Cpn60/TCP-1 (fold change =
−12.5; Fig. 4A). The consistency
rate with the 2-D electrophoresis results at the transcriptional
level was 66.67% (12/18). Thus, the proteins of the 12 genes that
had consistent expression trends in 2-D electrophoresis and RT-qPCR
were further detected by western blot analysis. The results showed
that 9 proteins were significantly downregulated and 3 were
upregulated in biofilms (Fig. 5).
Therefore, the 9 downregulated proteins were considered
differentially expressed proteins between biofilms and planktonic
cells of the A3313 strain. The fold changes of the 18 mRNAs are
shown in Table II.
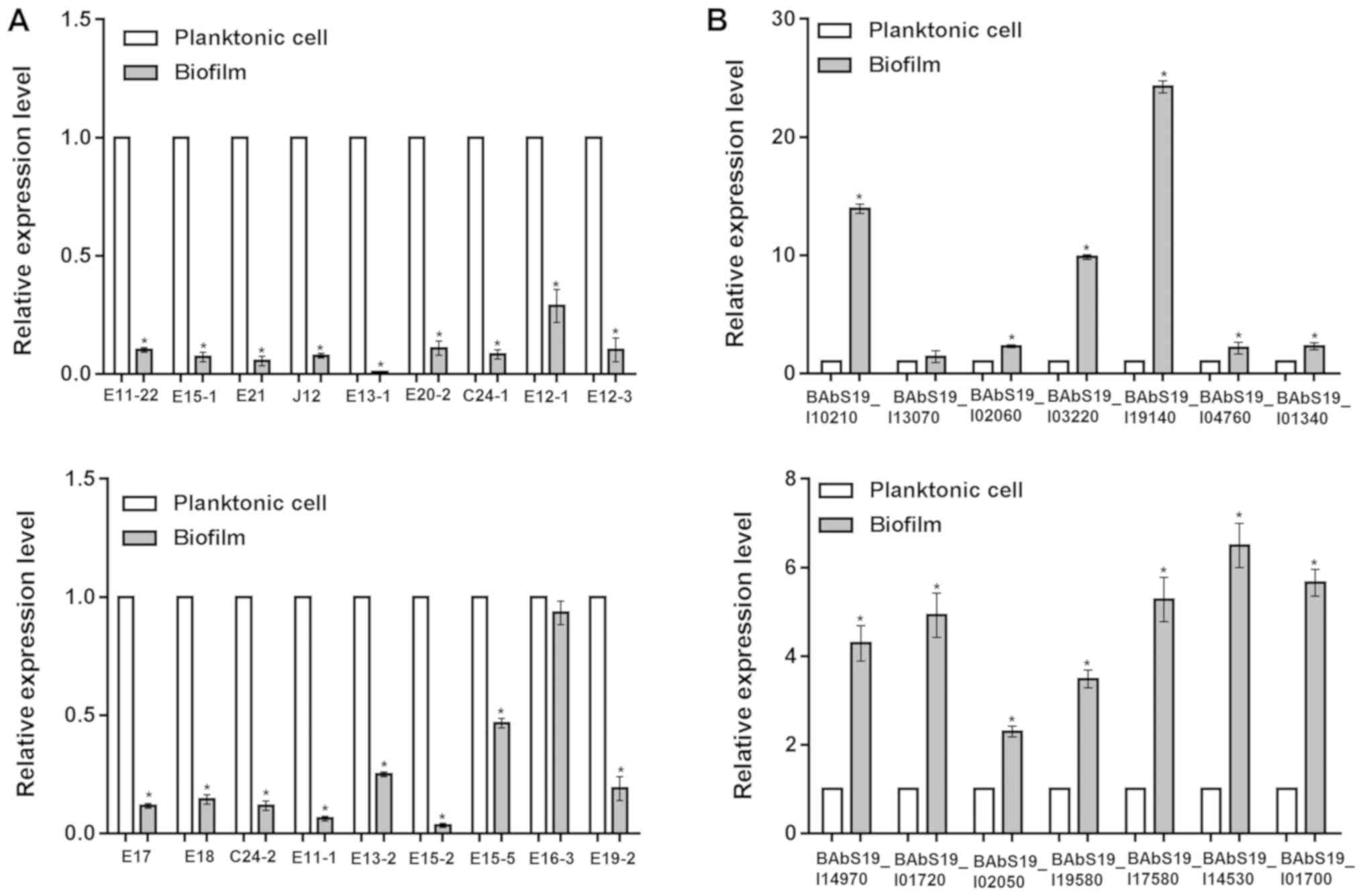 | Figure 4.Differential expression of mRNAs
between Brucella abortus cultured under planktonic or
biofilm conditions. The relative expression levels of (A) 18 genes
encoding proteins identified by 2-D electrophoresis and (B) 14
genes identified via high-throughput sequencing. 2-D,
two-dimensional. *P<0.05 vs. control (planktonic cells). E11-22,
hypothetical protein BAbS19_I16470; E15-1, polyprenyl synthetase;
E21, ExsB protein; J12, Chaperonin Cpn60TCP-1; E13-1, elongation
factor Tu; E20-2, aspartate-semialdehyde dehydrogenase; C24-1,
enoyl-(acyl carrier protein) reductase; E12-1, DnaJ, chaperone
protein DnaJ; E12-3, isocitrate dehydrogenase; E17, Enolase; E18,
Acetyl-CoA carboxylase, alpha subunit; C24-2, putative sulfite
oxidase subunit YedY; E11-1, Bacterial protein export chaperone
SecB; E13-2, Antifreeze protein, type I; E15-2, Lactatemalate
dehydrogenase; E15-5,
Phosphoribosylformylglycinamidinecyclo-ligase; E16-3, Periplasmic
binding protein; E19-2, tryptophanyl-tRNAsynthetase. |
 | Table II.RT-qPCR identification of the mRNA
expression levels of 18 proteins identified in 2-D
electrophoresis. |
Table II.
RT-qPCR identification of the mRNA
expression levels of 18 proteins identified in 2-D
electrophoresis.
| Protein spot | Protein | FCa | FCb |
|---|
| E11 | hypothetical
protein BAbS19_I16470 | −10.0 | −6.84 |
| E21 | ExsB protein | −20.0 | −1.82 |
| C24 | enoyl-(acyl carrier
protein) reductase | −12.5 | −1.76 |
| J12 | Chaperonin
Cpn60TCP-1 | −12.5 | −3.16 |
| E20 |
aspartate-semialdehyde dehydrogenase | −9.1 | −1.88 |
| E13 | elongation factor
Tu | −100 | −4.32 |
| E15 | polyprenyl
synthetase | −14.3 | −2.39 |
| E12 | DnaJ, chaperone
protein DnaJ | −3.5 | −5.55 |
| E17 | Enolase | −8.6 | −1.95 |
| E18 | Acetyl-CoA
carboxylase, a subunit | −7.0 | −1.95 |
| E16 | Periplasmic binding
protein | −1.1 | −2.19 |
| E19 |
tryptophanyl-tRNAsynthetase | −5.3 | −1.90 |
| E4 | catalase | −33.3 | 2.41 |
| E6 | extracellular
solute-binding protein | −22.6 | 1.78 |
| E7 |
Ubiquinol-cytochrome C reductase
iron-sulfur subunit | −25.0 | 1.70 |
| E8 | Chaperonin
Cpn10 | −14.3 | 1.68 |
| E9 | Tetracycline
resistance protein TetB | −12.5 | 1.67 |
| E10 | Ribosomal protein
L9 | −33.3 | 1.51 |
High-throughput sequencing and
bioinformatics analyses
A total of 22,039,653 and 37,506,048 reads were
generated from biofilms and planktonic cells, respectively. After
pre-processing, 16,624,091 and 34,222,222 aligned reads were
obtained from biofilms and planktonic cells respectively.
Based on the thresholds of q value ≤0.05 and
log2(fold change) ≥1, 157 differentially expressed mRNAs were
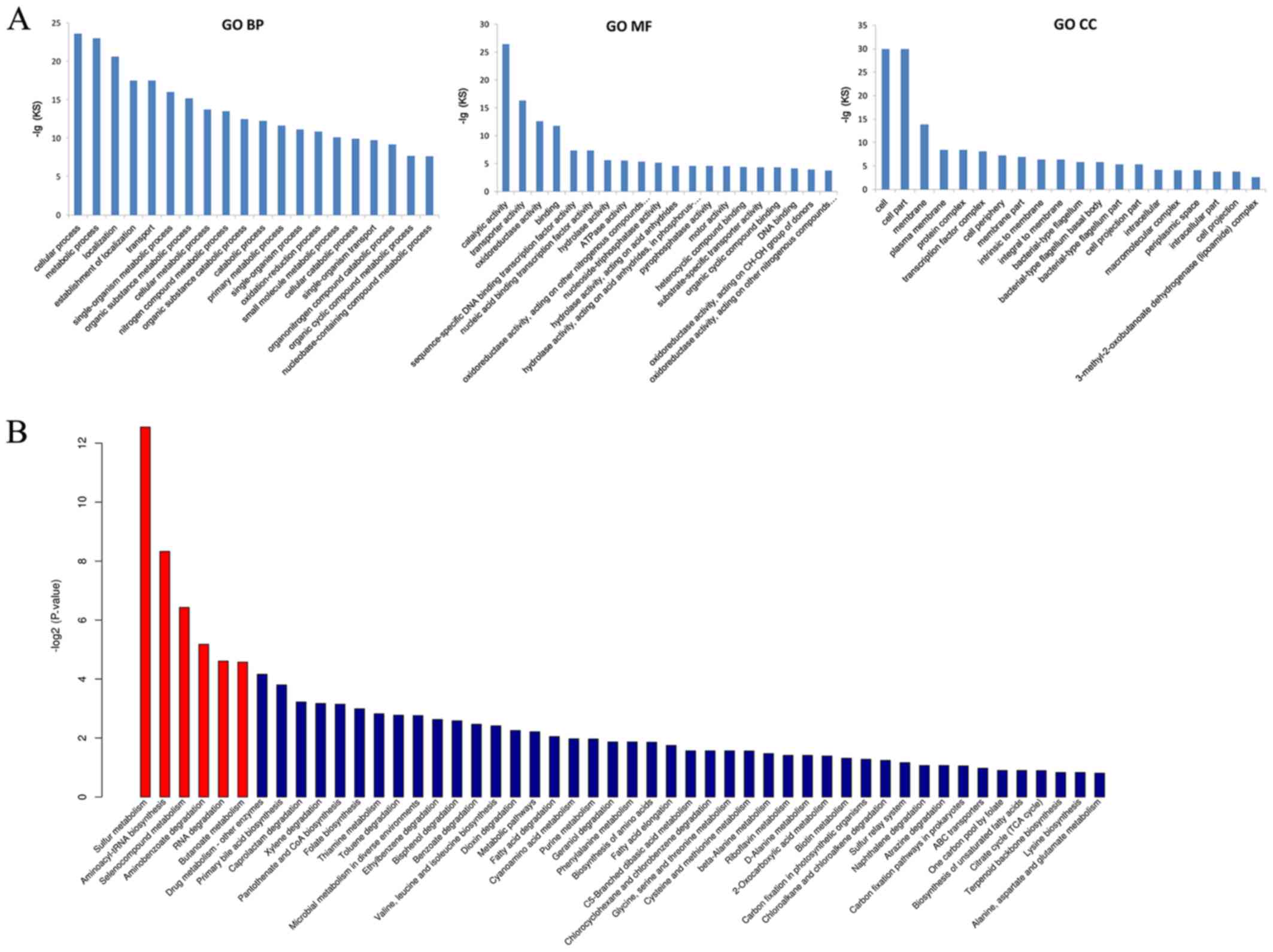
identified. These mRNA species were grouped in three categories
defined by GO, including biological processes (including protein
glycosylation, nitrogen compound metabolic process, and cell wall
organization), cellular compartment (such as integral component of
membrane, plasma membrane, and cytoplasm), and molecular function
(including DNA binding, ATP binding, and oxidoreductase activity;
Fig. 6A). In addition, these
differentially expressed mRNAs were significantly involved in six
pathways (annotated by KEGG), including RNA degradation, sulfur
metabolism, butanoate metabolism, aminoacyl-tRNA biosynthesis,
aminobenzoate degradation, and selenocompound metabolism (Fig. 6B).
Confirmation of function and
pathway-associated genes
RT-qPCR was performed to confirm 14 function and
pathway-associated genes identified by bioinformatics analyses,
including BAbS19_I10210 [log2(fold change) = 2.67], BAbS19_I13070
[log2(fold change) = 2.51], BAbS19_I02060 [log2(fold change) =
1.92], BAbS19_I03220 [log2(fold change) = 2.79], and the results
demonstrated that all of the 14 genes were upregulated in biofilms,
in accordance with the sequencing results (Fig. 4B). For instance, the fold changes
for the genes above (BAbS19_I10210, BAbS19_I13070, BAbS19_I02060,
and BAbS19_I03220) were 13.9, 1.4, 2.3 and 9.9, respectively
(Table III). These genes were
considered differentially expressed genes between biofilms and
planktonic cells of A3313 strain.
 | Table III.Confirmation of the expression levels
of function-associated genes identified by high-throughput
sequencing. |
Table III.
Confirmation of the expression levels
of function-associated genes identified by high-throughput
sequencing.
| Target gene | Gene name | Enriched function
(cellular component) | Enriched function
(molecular function) | Enriched function
(biological process) | Pathway | FCa | log2
(FC)b |
|---|
| 16S rRNA |
|
|
|
|
|
|
|
| BAbS19_I10210 | Phage
integrase |
| DNA binding
[GO:0003677] | DNA integration
[GO:0015074]; DNA recombination [GO:0006310] |
| 13.9 | 2.67 |
| BAbS19_I13070 | Ketol-acid
reductoisomerase (EC 1.1.1.86; acetohydroxy-acid isomeroreductase;
α-keto-β-hydroxylacyl reductoisomerase) |
| ketol-acid
reductoisomerase activity [GO:0004455] | isoleucine
biosynthetic process [GO:0009097]; valine biosynthetic process
[GO:0009099] |
| 1.4 | 2.51 |
| BAbS19_I02060 |
Phosphomethylpyrimidine kinase |
| ATP binding
[GO:0005524]; phosphomethylpyrimidine kinase activity
[GO:0008972] | thiamine
biosynthetic process [GO:0009228] |
| 2.3 | 1.92 |
| BAbS19_I03220 | Uncharacterized
protein |
| sialyltransferase
activity [GO:0008373] | protein
glycosylation [GO:0006486] |
| 9.9 | 2.79 |
| BAbS19_I19140 | Short-chain
dehydrogenase/reductase SDR |
| oxidoreductase
activity [GO:0016491] |
|
| 24.3 | 3.71 |
| BAbS19_I04760 | EAL domain
protein |
|
|
| Aminobenzoate
degradation [path:ko00627] | 2.1 | 2.38 |
| BAbS19_I01340 | Putative lipid II
flippase MurJ | integral component
of membrane [GO:0016021]; plasma membrane [GO:0005886] |
| cell wall
organization [GO:0071555]; peptido glycan biosynthetic process
[GO:0009252]; regulation of cell shape [GO:0008360]; transport
[GO:0006810] |
| 2.3 | 2.87 |
| BAbS19_I14970 |
Binding-protein-dependent transport
systems inner membrane component | integral component
of membrane [GO:0016021]; plasma membrane [GO:0005886] |
| transport
[GO:0006810] |
| 4.3 | 3.12 |
| BAbS19_I01720 | Sulfite reductase
(NADPH) hemoprotein β-component |
| 4 iron, 4 sulfur
cluster binding [GO:0051539]; heme binding [GO:0020037]; metal ion
binding [GO:0046872]; oxidoreductase activity [GO:0016491] |
|
| 4.9 | 2.64 |
| BAbS19_I02050 | D-amino acid
oxidase family protein |
| D-amino-acid
oxidase activity [GO:0003884]; FAD binding [GO:0071949] | D-amino acid
metabolic process [GO:0046416] |
| 2.3 | 3.37 |
| BAbS19_I19580 | Phosphoenolpyruvate
carboxykinase [ATP] | cytoplasm
[GO:0005737] | ATP binding
[GO:0005524]; kinase activity [GO:0016301]; metal ion binding
[GO:0046872]; phosphoenolpyruvate carboxykinase (ATP) activity
[GO:0004612] | gluconeogenesis
[GO:0006094] |
| 3.5 | 4.59 |
| BAbS19_I17580 | Nitrilase/cyanide
hydratase/apolipoprotein N-acyltransferase |
| hydrolase activity,
acting on carbon-nitrogen (but not peptide) bonds [GO:0016810];
transferase activity, transferring acyl groups [GO:0016746] | nitrogen compound
metabolic process [GO:0006807] |
| 5.3 | 2.02 |
| BAbS19_I14530 | MaoC-like
dehydratase |
|
|
| Butanoate | 6.5 | 1.58 |
| BAbS19_I01700 | CysG, siroheme
synthase |
| NAD binding
[GO:0051287]; precorrin-2 dehydrogenase activity [GO:0043115];
sirohydrochlorin ferrochelatase activity [GO:0051266];
uroporphyrin-III C-methyltransferase activity [GO:0004851] | cobalamin
biosynthetic process [GO:0009236]; siroheme biosynthetic process
[GO:0019354] | metabolism
[path:ko00650] | 5.7 | 2.07 |
Discussion
Bacteria can produce an extracellular matrix that
helps them adhere to inert or biological surfaces. Bacteria that
colonize different surfaces and invade susceptible hosts to cause
infections predominantly grow in biofilms (28). Biofilm formation is a developmental
process characterized by altered expression of structural and
regulatory genes (28). Most
bacteria grow in biofilms, and only a small portion grow in
planktonic mode (29). There have
been previous studies regarding the differences between biofilms
and planktonic cells for bacteria, including Lactobacillus
plantarum (30), swine
Brodetella bronchiseptica (31), Porphyromonas gingivalis
(32) and Clostridium
perfringens (33).
Nevertheless, the majority of the studies focus only one aspect,
either proteomic analysis or transcriptomic analysis. In the
present study, the differences in both protein and gene expression
levels in B. abortus cultured under either biofilm or
planktonic conditions were analyzed. A total of 9 downregulated
proteins under conditions of biofilm growth were identified by
proteomics, and 14 upregulated genes were identified in biofilm via
high-throughput sequencing.
Bacteria in biofilms exhibit persistence in spite of
sustained host defense (8);
however, little is known regarding the host immune response to
biofilm infections. The protein expression in biofilms grown in
vivo is difficult to study due to the difficulty of extracting
bacterial proteins from in vivo biofilms (10). The present study separated proteins
via 2-D electrophoresis and analyzed with MALDI-TOF/TOF-MS to
identify the differentially expressed protein spots, which included
elongation factor Tu, enolase, chaperone protein DnaJ and
periplasmic binding protein.
Among the differentially expressed protein spots,
elongation factor Tu had a higher fold change. It was also found to
have the greatest fold change in mRNA expression, which suggested
the potential role of elongation factor Tu in B. abortus.
Elongation factor Tu is one of the most abundant proteins in
bacterial cells, involved in critical steps in protein biosynthesis
and forming structural filaments in vitro (34). It has been observed on the surface
of several pathogenic bacteria, including Burkholderia
pseudomallei and the closely-related Pseudomonas
aeruginosa (35,36). It has also been demonstrated that
elongation factor Tu may play a role as a bacterial virulence
factor. Barel et al (37)
reported that elongation factor Tu can facilitate invasion of host
cells by Francisella tularensis via interaction with
nucleolin. Notably, it also acts as a biofilm component in
Serratia aureus (38).
Thus, it was hypothesized that elongation factor Tu may be
associated with virulence in B. abortus biofilm.
Enolase is an enzyme involved in the glycolytic
pathway, catalyzing the reversible conversion of 2-phosphoglycerate
to phosphoenolpyruvate (39).
Enolase was previously regarded as a soluble glycolytic enzyme,
present in cytosol exclusively (39). Enolase has been found to be a
multifaceted protein with sub-cellular localizations and diverse
biological functions (40,41). It acts as a plasminogen receptor on
the cell surface of various microorganisms and pathogens, and
serves an important role in bacterial colonization, persistence and
host tissue invasion (42,43). It has been reported that in
Candida albicans, enolase expresses on the surface of
biofilm-forming cells and contributes to the adhesion of Candida
albicans to different substrates with potential implications
for biofilm adhesion and formation (44). Importantly, enolase has been cloned
in B. abortus A19 and exhibits critical roles in the
colonization and invasion of this pathogen (45). Specially, Han et al
(45) also find that enolase
protein can bind to B. abortus-positive sera, indicating
that enolase may serve as a useful diagnostic marker for
brucellosis.
DnaJ chaperone is a prototypical member of the Hsp40
family, which is important for numerous cellular functions, such as
membrane lipid composition and cell division (46,47).
Grudniak et al (48)
demonstrated that this chaperone was involved in the biofilm
formation of Escherichia coli. Periplasmic binding proteins
have been introduced into bacteria to function in synthetic signal
transduction pathways that respond to extracellular ligands and act
as biologically active enzymes (49). As for the other differentially
expressed proteins, their roles in biofilm formation have not been
reported to the best of the authors' knowledge. Given their
differential expression between biofilm and planktonic cells, it
can be hypothesized that they may be associated with the biofilm
characteristics of B. abortus.
From high-throughput sequencing, 14 function- and
pathway-associated genes were identified. For instance,
BAbS19_I19580 was enriched in functions related to
phosphoenolpyruvate carboxykinase activity [GO:0004612] and
gluconeogenesis [GO:0006094]. In most organisms,
phosphoenolpyruvate carboxykinase can catalyze the formation of
phosphoenolpyruvate via the phosphorylation and decarboxylation of
oxaloacetate, which is the first step in the gluconeogenic pathway
(50). A previous study by Li
et al (51) suggested that
gluconeogenesis serves a key role in the development of
Saccharomyces cerevisiae biofilms. It was found that during
the attachment period of biofilms, the expression of
gluconeogenesis pathway-associated genes was upregulated, which was
consistent with the findings of the present study. Viadas et
al (52) found that the
expression level of phosphoenolpyruvate carboxykinase gene was
increased in B. abortus with bvrR mutant compared with wild
type cells. Notably, phosphoenolpyruvate carboxykinase has been
reported to be an acid-induced virulence factor in Agrobacterium
tumefaciens (53). Therefore,
it was proposed that phosphoenolpyruvate carboxykinase serves an
important role in the virulence of B. abortus through the
functions described above.
Binding protein-dependent transport systems have
been found to be closely associated with structure, organization,
mechanism and evolutionary origin (54). Numerous binding protein-dependent
transport systems have been identified in Gram-negative bacteria
(54). The major role of the
binding protein systems is to recapture substrates that leak from
the cell and retain them near the cell (55). In the present study, BAbS19_I14970
(binding-protein-dependent transport systems inner membrane
component) was differentially expressed between biofilm and
planktonic bacteria and enriched in an integral component of
membrane (GO:0016021), indicating an important role in biofilm
function.
Despite the identification of differentially
expressed proteins and genes between biofilm and planktonic cells,
no common protein or gene was identified, which may be attributed
to several reasons. First, the detectability and abundance of
proteomic and genomic analyses were different. Second, only 9
differentially expressed proteins were validated in proteomics,
which may affect the consistency between proteomic and genomic
analyses. Third, the thresholds used in proteomic and genomic
analyses were different. Of note, there were certain common genes
between proteomic and genomic analyses when significant differences
were disregarded. Finally, there were differences in expression
between transcriptional and protein levels due to
post-transcriptional and posttranslational modifications.
Furthermore, there is a limitation in the present study in that
only one isolate was used for the analysis. Further isolates of
B. abortus will be applied in future studies to further
investigate the differentially expressed proteins and genes between
the two culture conditions.
In conclusion, differential expression analysis at
the protein and genomic levels suggested that the proteins and
genes differentially expressed in B. abortus biofilms may
serve important roles in bacterial defense, colonization, invasion,
and virulence.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Priority
Academic Program Development of Jiangsu Higher Education
Institutions (grant no. PAPD), the Science Foundation of General
Administration of Quality Supervision, Inspection and Quarantine of
the People's Republic China (grant no. 2008IK004) and National
Basic Research Program of China (973 Program; grant no.
2012CB518801).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TT, CL and YJ participated in the design of this
study. TT, MW, LZ an AG performed the experiments. TT, GC, YX and
CZ analyzed the data. CL, LZ and AG obtained funding. TT and YX
drafted the manuscript. GC and CZ participated in revision of
manuscript for important intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Byndloss MX and Tsolis RM: Brucella spp.
virulence factors and immunity. Annu Rev Anim Biosci. 4:111–127.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hasanjani Roushan MR and Ebrahimpour S:
Human brucellosis: An overview. Caspian J Intern Med. 6:46–47.
2015.PubMed/NCBI
|
|
3
|
Atluri VL, Xavier MN, de Jong MF, den
Hartigh AB and Tsolis RM: Interactions of the human pathogenic
Brucella species with their hosts. Annu Rev Microbiol. 65:523–541.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Halling SM, Peterson-Burch BD, Bricker BJ,
Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP and Olsen SC: Completion
of the genome sequence of Brucella abortus and comparison to
the highly similar genomes of Brucella melitensis and
Brucella suis. J Bacteriol. 187:2715–2726. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Doganay G and Doganay M: Brucella as a
potential agent of bioterrorism. Recent Pat Antiinfect Drug Discov.
8:27–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dang H and Lovell CR: Microbial surface
colonization and biofilm development in marine environments.
Microbiol Mol Biol Rev. 80:91–138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu H, Moser C, Wang HZ, Høiby N and Song
ZJ: Strategies for combating bacterial biofilm infections. Int J
Oral Sci. 7:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burmølle M, Thomsen TR, Fazli M, Dige I,
Christensen L, Homøe P, Tvede M, Nyvad B, Tolker-Nielsen T, Givskov
M, et al: Biofilms in chronic infections a matter of opportunity
monospecies biofilms in multispecies infections. FEMS Immunol Med
Microbiol. 59:324–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parsek MR and Singh PK: Bacterial
biofilms: An emerging link to disease pathogenesis. Annu Rev
Microbiol. 57:677–701. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Yi L, Wu Z, Shao J, Liu G, Fan H,
Zhang W and Lu C: Comparative proteomic analysis of streptococcus
suis biofilms and planktonic cells that identified biofilm
infection-related immunogenic proteins. PLoS One. 7:e333712012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumon H, Ono N, Iida M and Nickel JC:
Combination effect of fosfomycin and ofloxacin against Pseudomonas
aeruginosa growing in a biofilm. Antimicrob Agents Chemother.
39:1038–1044. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilson M: Susceptibility of oral bacterial
biofilms to antimicrobial agents. J Med Microbiol. 44:79–87. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Li Y, Wang L, Zhu M, Zhu X, Qian C
and Li W: Responses of biofilm microorganisms from moving bed
biofilm reactor to antibiotics exposure: Protective role of
extracellular polymeric substances. Bioresour Technol. 254:268–277.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Flury D, Behrend H, Sendi P, von Kietzell
M and Strahm C: Brucella melitensis prosthetic joint
infection. J Bone Jt Infect. 2:136–142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Almirón MA, Roset MS and Sanjuan N: The
aggregation of Brucella abortus occurs under microaerobic
conditions and promotes desiccation tolerance and biofilm
formation. Open Microbiol J. 7:87–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conde-Álvarez R, Arce-Gorvel V, Iriarte M,
Manček-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C,
Chaves-Olarte E, Martirosyan A, von Bargen K, et al: The
lipopolysaccharide core of Brucella abortus acts as a shield
against innate immunity recognition. PLoS Pathog. 8:e10026752012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spera JM, Ugalde JE, Mucci J, Comerci DJ
and Ugalde RA: A B lymphocyte mitogen is a Brucella abortus
virulence factor required for persistent infection. Proc Natl Acad
Sci USA. 103:16514–16519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Manterola L, Guzmán-Verri C, Chaves-Olarte
E, Barquero-Calvo E, de Miguel MJ, Moriyón I, Grilló MJ, López-Goñi
I and Moreno E: BvrR/BvrS-controlled outer membrane proteins Omp3a
and Omp3b are not essential for Brucella abortus virulence.
Infect Immun. 75:4867–4874. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Harro JM, Peters BM, O'May GA, Archer N,
Kerns P, Prabhakara R and Shirtliff ME: Vaccine development in
Staphylococcus aureus: Taking the biofilm phenotype into
consideration. FEMS Immunol Med Microbiol. 59:306–323. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yi L, Wang Y, Ma Z, Zhang H, Li Y, Zheng
JX, Yang YC, Fan HJ and Lu CP: Biofilm formation of Streptococcus
equi ssp. zooepidemicus and comparative proteomic analysis of
biofilm and planktonic cells. Curr Microbiol. 69:227–233. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang L, Wang S and Li W: RSeQC: Quality
control of RNA-seq experiments. Bioinformatics. 28:2184–2185. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Yi L, Zhang Z, Fan H, Cheng X and
Lu C: Biofilm formation, host-cell adherence, and virulence genes
regulation of Streptococcus suis in response to autoinducer-2
signaling. Curr Microbiol. 68:575–580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Steinberg D: Studying plaque biofilms on
various dental surfaces. Handbook of bacterial adhesion Springer.
353–370. 2000. View Article : Google Scholar
|
|
29
|
Costerton JW, Stewart PS and Greenberg E:
Bacterial biofilms: A common cause of persistent infections.
Science. 284:1318–1322. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Angelis M, Siragusa S, Campanella D, Di
Cagno R and Gobbetti M: Comparative proteomic analysis of biofilm
and planktonic cells of Lactobacillus plantarum DB200. Proteomics.
15:2244–2257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ning J, Gao X, Xiao M, et al: Comparative
proteomic analysis between biofilm-forming cells and planktonic
cells of swine Brodetella bronchiseptica. J Agric
Biotechnol. 26:159–166. 2018.[In Chinese].
|
|
32
|
Romero-Lastra P, Sánchez MC, Ribeiro-Vidal
H, Llama-Palacios A, Figuero E, Herrera D and Sanz M: Comparative
gene expression analysis of Porphyromonas gingivalis ATCC
33277 in planktonic and biofilms states. PLoS One. 12:e01746692017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Charlebois A, Jacques M and Archambault M:
Comparative transcriptomic analysis of Clostridium perfringens
biofilms and planktonic cells. Avian Pathol. 45:593–601. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Furano AV: Content of elongation factor Tu
in Escherichia coli. Proc Natl Acad Sci USA. 72:4780–4784.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Harding SV, Sarkar-Tyson M, Smither SJ,
Atkins TP, Oyston PC, Brown KA, Liu Y, Wait R and Titball RW: The
identification of surface proteins of Burkholderia pseudomallei.
Vaccine. 25:2664–2672. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kunert A, Losse J, Gruszin C, Hühn M,
Kaendler K, Mikkat S, Volke D, Hoffmann R, Jokiranta TS, Seeberger
H, et al: Immune evasion of the human pathogen Pseudomonas
aeruginosa: Elongation factor Tuf is a factor H and plasminogen
binding protein. J Immunol. 179:2979–2988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barel M, Hovanessian AG, Meibom K, Briand
JP, Dupuis M and Charbit A: A novel receptor-ligand pathway for
entry of Francisella tularensis in monocyte-like THP-1 cells:
Interaction between surface nucleolin and bacterial elongation
factor Tu. BMC Microbiol. 8:1452008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Foulston L, Elsholz AK, DeFrancesco AS and
Losick R: The extracellular matrix of Staphylococcus aureus
biofilms comprises cytoplasmic proteins that associate with the
cell surface in response to decreasing pH. MBio. 5:e01667–e01614.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Entelis N, Brandina I, Kamenski P,
Krasheninnikov IA, Martin RP and Tarassov I: A glycolytic enzyme,
enolase, is recruited as a cofactor of tRNA targeting toward
mitochondria in Saccharomyces cerevisiae. Genes Dev. 20:1609–1620.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pancholi V: Multifunctional a-enolase: Its
role in diseases. Cell Mol Life Sci. 58:902–920. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sriram G, Martinez JA, McCabe ER, Liao JC
and Dipple KM: Single-gene disorders: What role could moonlighting
enzymes play? Am J Hum Genet. 76:911–924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kolberg J, Aase A, Bergmann S, Herstad TK,
Rødal G, Frank R, Rohde M and Hammerschmidt S: Streptococcus
pneumoniae enolase is important for plasminogen binding despite low
abundance of enolase protein on the bacterial cell surface.
Microbiology. 152:1307–1317. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pancholi V and Fischetti VA:
alpha-enolase, a novel strong plasmin(ogen) binding protein on the
surface of pathogenic streptococci. J Biol Chem. 273:14503–14515.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Silva RC, Padovan AC, Pimenta DC, Ferreira
RC, da Silva CV and Briones MR: Extracellular enolase of Candida
albicans is involved in colonization of mammalian intestinal
epithelium. Front Cell Infect Microbiol. 4:662014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han X, Ding C, Chen H, Hu Q and Yu S:
Enzymatic and biological characteristics of enolase in Brucella
abortus A19. Mol Biol Rep. 39:2705–2711. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sieńczyk J, Skłodowska A, Grudniak A and
Wolska KI: Influence of DnaK and DnaJ chaperones on Escherichia
coli membrane lipid composition. Pol J Microbiol. 53:121–123.
2004.PubMed/NCBI
|
|
47
|
McCarty JS and Walker GC: DnaK mutants
defective in ATPase activity are defective in negative regulation
of the heat shock response: Expression of mutant DnaK proteins
results in filamentation. J Bacteriol. 176:764–780. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Grudniak AM, Włodkowska J and Wolska KI:
Chaperone DnaJ influences the formation of biofilm by Escherichia
coli. Pol J Microbiol. 64:279–283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dwyer MA and Hellinga HW: Periplasmic
binding proteins: A versatile superfamily for protein engineering.
Curr Opin Struct Biol. 14:495–504. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Inui M, Nakata K, Roh JH, Zahn K and
Yukawa H: Molecular and functional characterization of the
Rhodopseudomonas palustris no. 7 phosphoenolpyruvate carboxykinase
gene. J Bacteriol. 181:2689–2696. 1999.PubMed/NCBI
|
|
51
|
Li Z, Chen Y, Liu D, Zhao N, Cheng H, Ren
H, Guo T, Niu H, Zhuang W, Wu J and Ying H: Involvement of
glycolysis/gluconeogenesis and signaling regulatory pathways in
Saccharomyces cerevisiae biofilms during fermentation. Front
Microbiol. 6:1392015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Viadas C, Rodríguez MC, Sangari FJ, Gorvel
JP, García-Lobo JM and López-Goñi I: Transcriptome analysis of the
Brucella abortus BvrR/BvrS two-component regulatory system.
PLoS One. 5:e102162010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu P, Wood D and Nester EW:
Phosphoenolpyruvate carboxykinase is an acid-induced, chromosomally
encoded virulence factor in Agrobacterium tumefaciens. J Bacteriol.
187:6039–6045. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Higgins C, Hyde S, Mimmack M, Gileadi U,
Gill D and Gallagher M: Binding protein-dependent transport
systems. J Bioenerg Biomembr. 22:571–592. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Stirling D, Hulton C, Waddell L, Park SF,
Stewart GS, Booth IR and Higgins CF: Molecular characterization of
the proU loci of Salmonella typhimurium and Escherichia coli
encoding osmoregulated glycine betaine transport systems. Mol
Microbiol. 3:1025–1038. 1989. View Article : Google Scholar : PubMed/NCBI
|