Introduction
Human defensins are a group of antimicrobial
peptides that defend hosts against bacterial, fungal and viral
infections (1). Human β-defensins
(hBDs) are a subfamily of defensins distributed at various mucosal
surfaces (2). Although multiple
gene duplications were observed in the human genome, only hBD-1,
hBD-2, hBD-3 and hBD-4 have been described in detail to date
(3). hBD-2 is the first member of
the human defensin family that has been observed to be highly
induced in inflamed epidermal cells, such as airway epithelia and
in psoriasis (4,5). Previous studies had demonstrated that
hBD-2 had high antimicrobial activity and broad spectrum activity
(6,7).
The epithelial surface of the lungs is exposed to a
large number of potentially pathogenic microorganisms. Several
defence mechanisms are required to protect against infection. The
production of antimicrobial peptides is one of the most important
and evolutionarily conserved mechanisms. hBD-1 and hBD-2 were found
to be locally expressed in the lung. Previous studies indicated
that hBD-2 expression was induced by inflammation, whereas hBD-1
serves as a defense mechanism in the absence of inflammation
(5,8). Furthermore, by measuring the
concentration of hBD-2 in the bronchoalveolar lavage fluid of
cystic fibrosis patients, the decrease of β-defensins in advanced
lung disease was found to lead to a secondary defect of the local
host defense (9). Hence, hBD-2 in
the lung is likely to provide an effective shield from microbial
infection.
hBD-2 exhibits particularly effective capacity
against Gram-negative bacteria and certain fungi (7,10).
Gram-negative bacteria, receiving notable concern in recent years,
show the feature of using a plethora of mechanisms against
antibiotics, which are due to gene cassettes and integrons
(11,12). This feature makes them more
resistant to antibiotics. While antibiotic resistance has become an
increasing global problem, the discovery and development of new
antibiotics has declined (13).
Infections due to antibiotic-resistant bacteria cause a major
public health dilemma, which may lead to future serious medical and
social problems. Moreover, hospital-acquired infections have become
a major threat to patient safety, and are associated with prolonged
hospital stays, higher healthcare costs and increased mortality
(14–16). The emergence of
antibiotic-resistant infections mainly caused by Gram-negative
bacteria aggravates the situation (14,15,17,18).
Thus, in this study, we intend to elevate endogenous hBD-2 protein
expression in order to meet the emerging clinical challenge.
Since the discovery of natural antimicrobial
peptides, much research has been carried out to improve the innate
immune response, including the study of many gene-based therapies
(19–24). However, to date, all these methods
are under preliminary investigation, and no new clinical
application has been proposed. In this study, we found that killed
vaccines, including the 23 valent pneumococcal polysaccharide
vaccine, Haemophilus influenzae vaccine and split influenza virus
vaccine, could serve as potential inducers of hBD-2. We propose
that these vaccines, which already have proven biosafety records,
may be used as a potential clinical approach.
Materials and methods
Primary cell culture and treatment
Human bronchial epithelia were surgically resected
from patients without lung infection or tumors. The research was
approved by the Ethics Committee of Tongji University, China.
Primary tissues were digested by 0.125% trypsin with 0.01% EDTA,
and cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture
F12 supplemented with 10% fetal bovine serum and 1%
penicillin-streptomycin. For treatment, cells were plated in 6-well
plates at a density of 1×105/well. Treatment with
23-valent pneumococcal polysaccharide vaccine, haemophilus
influenzae type b vaccine or split influenza virus vaccine was
performed after the cells reached ~80% confluence. Two independent
experiments were performed and each experiment consisted of at
least triplicate samples.
RNA extraction and semi-quantitative
RT-PCR
Following treatment, cells were lysed with 1 ml of
TRIzol reagent. Total RNA was extracted according to manufacturer’s
instructions and dissolved in 30 μl H2O with DEPC. Total
RNA (2 μg) was used for semi-quantitative RT-PCR according to the
manufacturer’s instructions. GAPDH was selected as an internal
control for normalization. For hBD-2 expression, the primers were
designed according to the cDNA sequences (GeneBank accession no.
NM_004942.2): the forward primer was 5′-CTT GTATCTCCTCTTCTCGTTCC-3′
and the reverse primer was 5′-TTTCTGAATCCGCATCAGC-3′. For GAPDH
expression, the forward primer was 5′-GTCGGTGTGAACGGATTT-3′ and the
reverse primer was 5′-ACTCCACGACGTACTC AGC-3′. cDNA (2 μl) was
amplified for hBD-2 and GAPDH. PCR was performed in a 20-μl final
volume containing 2 μl 10X reaction buffer, 0.2 mM dNTP, 2 mM
MgCl2 and 1.5 units TaqDNA polymerase. The cycle numbers
that generated approximately half maximal amplification were
determined by testing different cycle numbers of amplification for
each gene, and were found to be 40 cycles for hBD-2 and 30 cycles
for GAPDH. The PCR conditions were 94°C for 15 sec, annealing at
61°C for hBD-2 and 57°C for GAPDH for 20 sec and extension at 72°C
for 30 sec. The PCR product (5 μl) was checked on a 2.0% agarose
gel and visualized using ethidium bromide staining with UV
illumination. The intensity of each band was analyzed using the
BioSenSC300 system.
Enzyme-linked immunosorbent assay
The medium was collected at 6 and 12 h following
treatment. After centrifugation at 3000 g for 20 min, the
supernatant samples were removed. The manufacturer’s instructions
for enzyme-linked immunosorbent assay (ELISA) were followed.
Briefly, the microtiter plates were coated by adding 100 μl of the
samples. After 90 min incubation, the microtiter plates were
agitated to remove the samples. Biotin-labeled antibody (100 μl)
was added and the plates were then incubated at 37°C for 60 min.
After three washing steps with 0.01 M PBS, the plates were filled
with with 100 μl avidin-biotin-peroxidase complex at 37°C for 30
min. After extensive washing, the immunological reactions were
revealed by 30-min incubation in the dark with 90 μl TMB Microwell
substrate. TMB stop solution was added to stop the reaction. The
optical density results were converted into concentrations deduced
from a calibration curve.
Electrophoretic mobility shift assay
(EMSA)
According to the core recognition site of NF-κB, the
following oligonucleotides were synthesized for double-stranded
probes: forward (5′-AGT TGA GGG GAC TTT CCC AGG C-3′) and reverse
(3′-GCC TGG GAA AGT CCC CTC AAC T-5′). The annealed
oligonucleotides were labeled with DIG dUTP by terminal
transferase. The cellular extracts was incubated at room
temperature for 20 min with 1 μg Poly(dI-dC) in a binding buffer
containing 1X binding buffer, 2.5% glycerol, 0.05% NP-40, 100 mM
MgCl2 and 20 fM biotin end-labeled target DNA.
DNA-protein complexes were separated from free DNA by
electrophoresis in a non-denaturing polyacrylamide gel at 4°C and
electrophoretically transferred onto nylon membranes. The membranes
were then dried in a drier at 70°C for 1 h and blocked with
blocking buffer for 15 min. After extensive washing, the membranes
were incubated with substrate equilibration buffer for 5 min;
subsequently substrate working solution was added and the samples
were incubated for another 5 min. Finally, images of the membranes
were captured on film cassette following exposure to X-ray for 20
min.
Bacteriostatic experiment
The concentrated medium of cells treated with 23
valent pneumococcal polysaccharide vaccine, Haemophilus influenzae
vaccine and split influenza virus vaccine were collected. The round
filter paper was immersed in the medium, PBS and clean medium,
respectively. The spread plate method was used to measure the
antimicrobial activity.
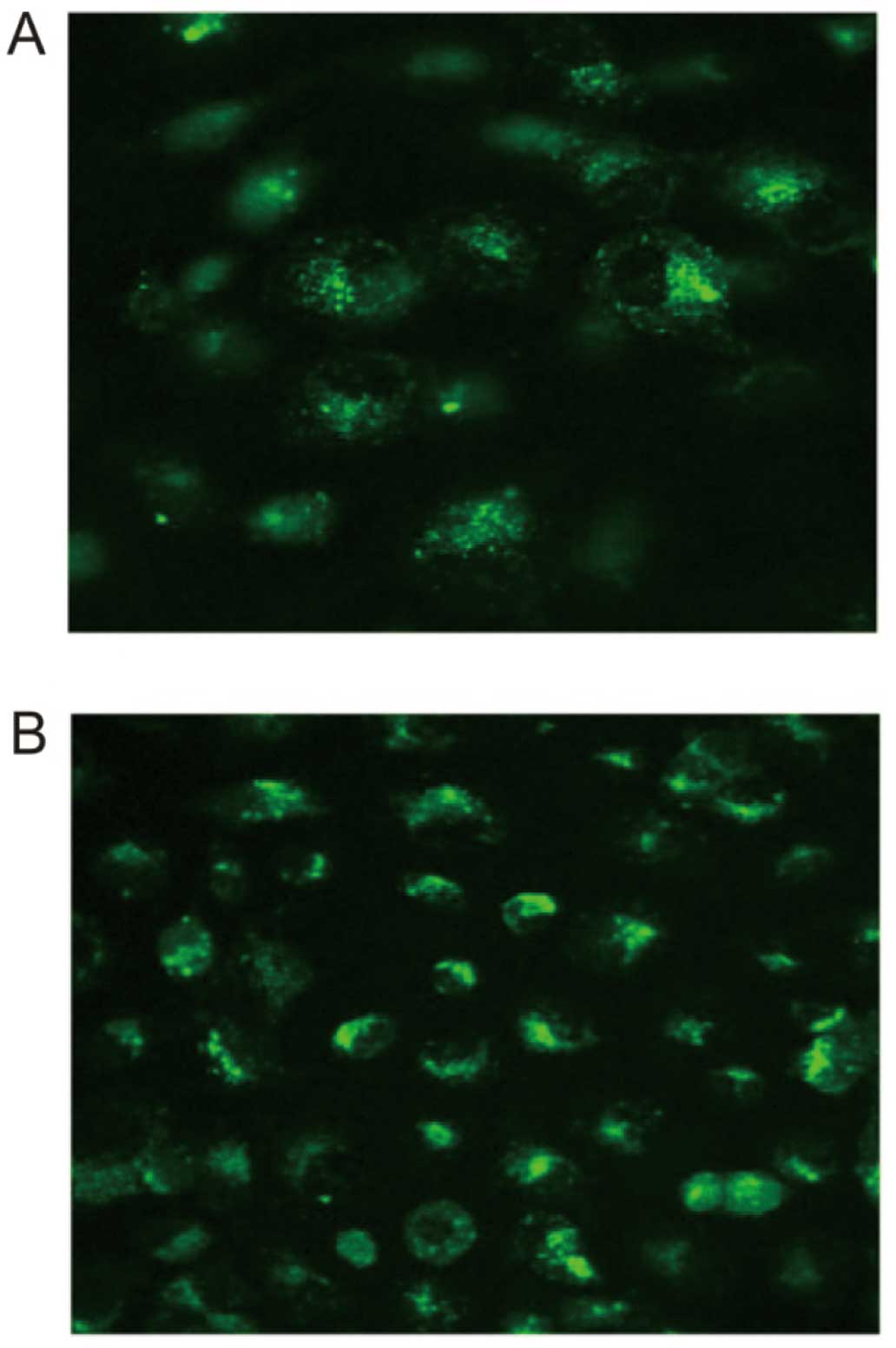
Measurement of the concentration of
Ca2+
Cells were plated in a 15×15-mm culture dish. After
the cells reached ~90% confluence, the medium was replaced with
D-Hanks buffer. An appropriate amount of Fluo-3/AM was added into
the buffer at a final concentration of 5–10 μmol/l and incubated
with the cells at 37°C in a 5% CO2 atmosphere for 60 min
in the dark. Cells were then washed twice by PBS and once by
D-Hanks buffer. Fluorescent images of the cells were visualized in
D-Hanks buffer under a laser scanning confocal microscope at an
excitation wavelength of 488 nm and an emission wavelength of 525
nm.
Statistical analyses
Data were presented as the means ± SD. Statistical
analysis was performed using SPSS 11.0.0 (SPSS Inc., Chicago, IL,
USA). Data were analyzed by one-way ANOVA followed by Dunnett’s
multiple comparison test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of hBD-2 mRNA in epithelial
cells with various stimuli
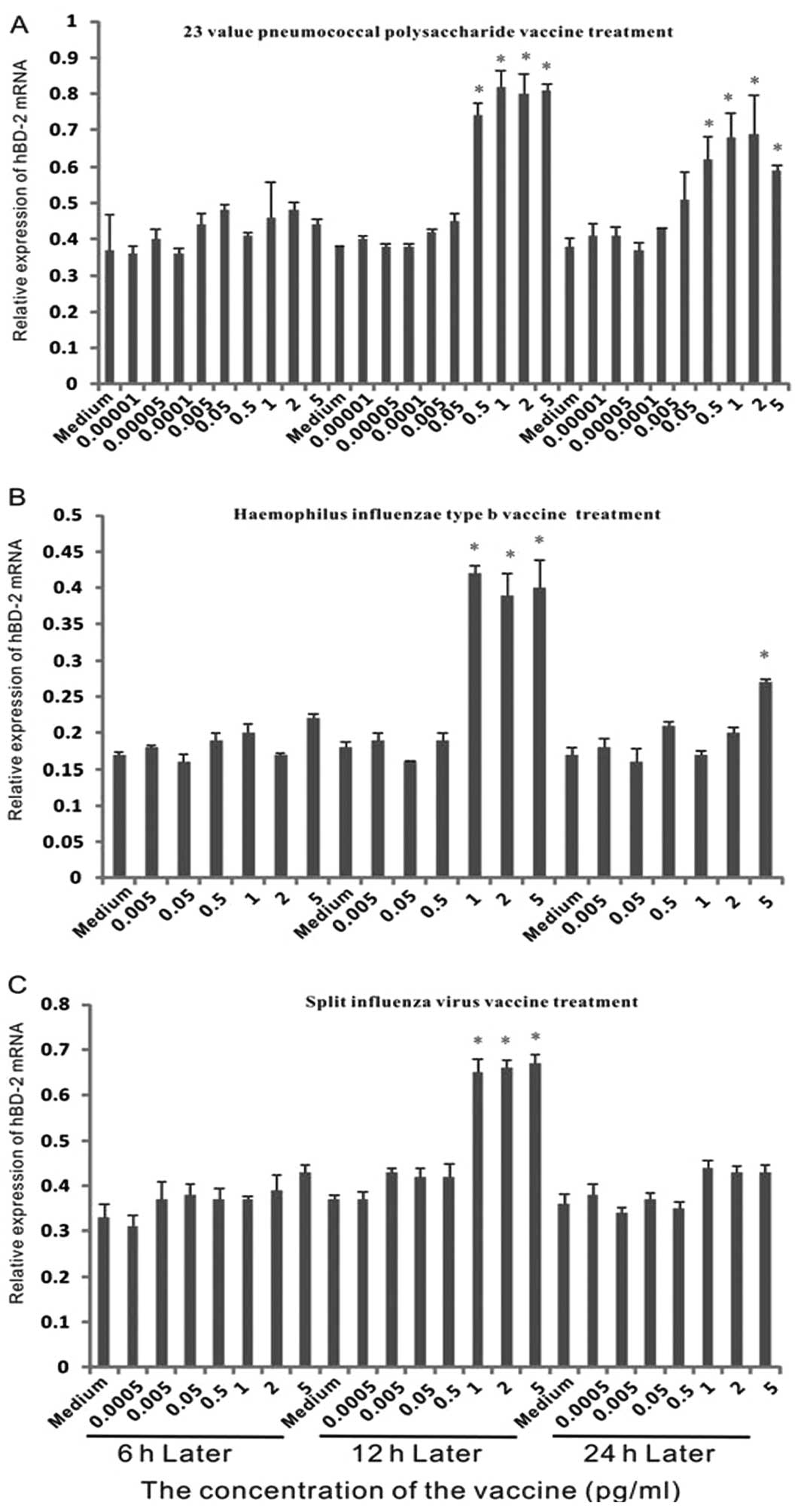
To investigate the expression of hBD-2 mRNA in
epithelial cells with different vaccine treatments, dose-effect
experiments were performed (Fig.
1). As shown in Fig. 2, no
marked upregulation was observed following 6 h of treatment.
Another 6 h later, when the concentration of 23 valent pneumococcal
polysaccharide vaccine was above 0.5 μg/ml, it enhanced the
expression over 1.5-fold. The expression exhibited over 2-fold
induction when cells were treated with greater than 1 μg/ml
Haemophilus influenzae vaccine. For the split influenza virus
vaccine, when the concentration was above 1 μg/ml, it stimulated
the expression by greater than 1.5-fold.
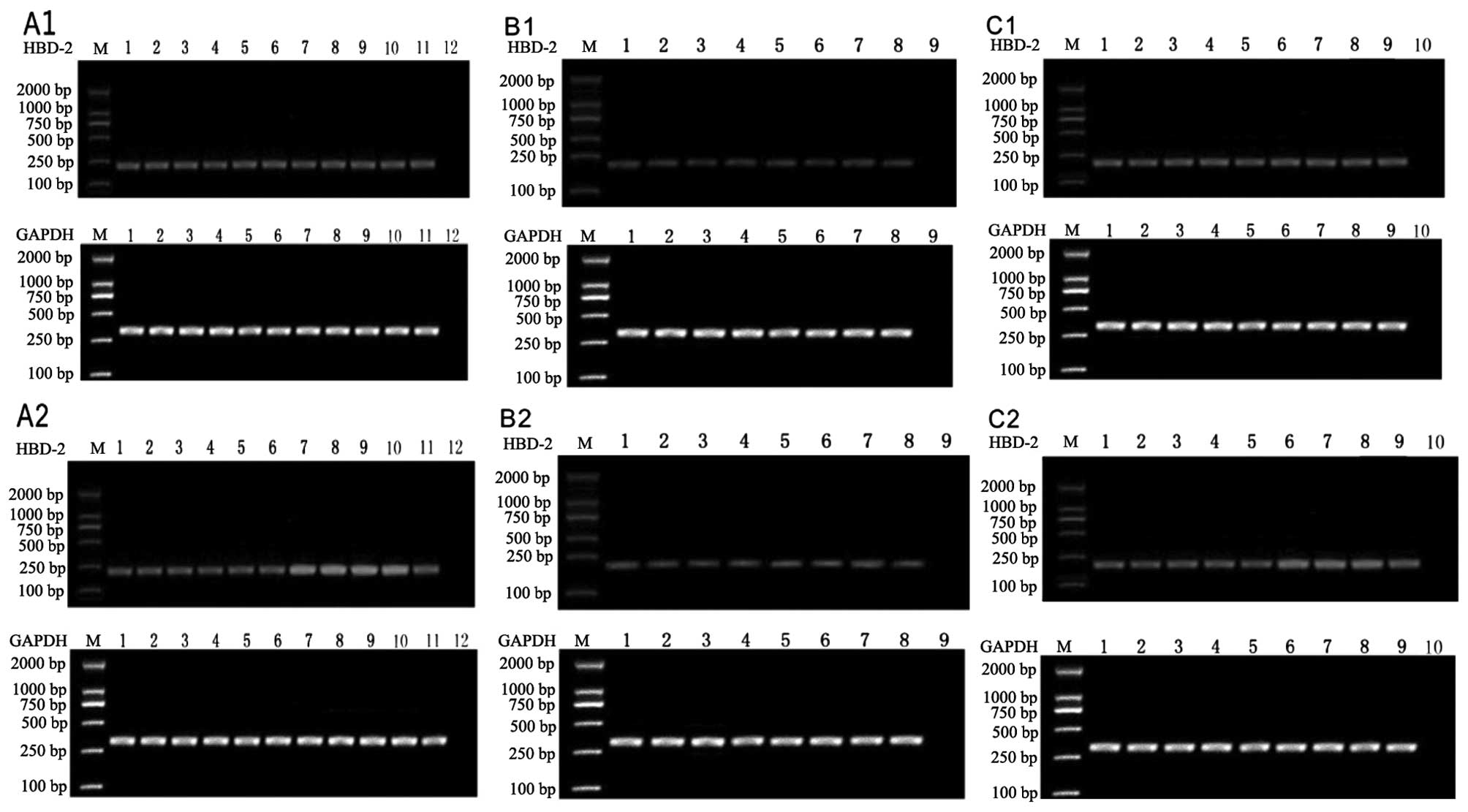 | Figure 1Analysis of mRNA expression for hBD-2
in epithelial cells treated with Streptococcus pneumoniae vaccine
for (A1) 6 h and (A2) 12 h, Hemophilus influenzae vaccine for (B1)
6 h and (B2) 12 h and influenza virus vaccine for (C1) 6 h and (C2)
12 h. Lanes 1–10: without the vaccine; 0.00001, 0.00005, 0.0001,
0.005, 0.05, 0.5, 1, 2, 5 μg/ml of the vaccine; lane 11, ATP; lane
12, PBS; M, marker. hBD-2, human β-defensin-2. |
At 24 h, the expression level of hBD-2 with 23
valent pneumococcal polysaccharide vaccine treatment was lower than
at 12 h. In the Haemophilus influenzae vaccine treatment group, a
significant difference was only observed at the concentration of 5
μg/ml. In the split influenza virus vaccine treatment group, no
significant difference was detected. This indicated that the effect
of vaccines on hBD-2 expression may be short-term.
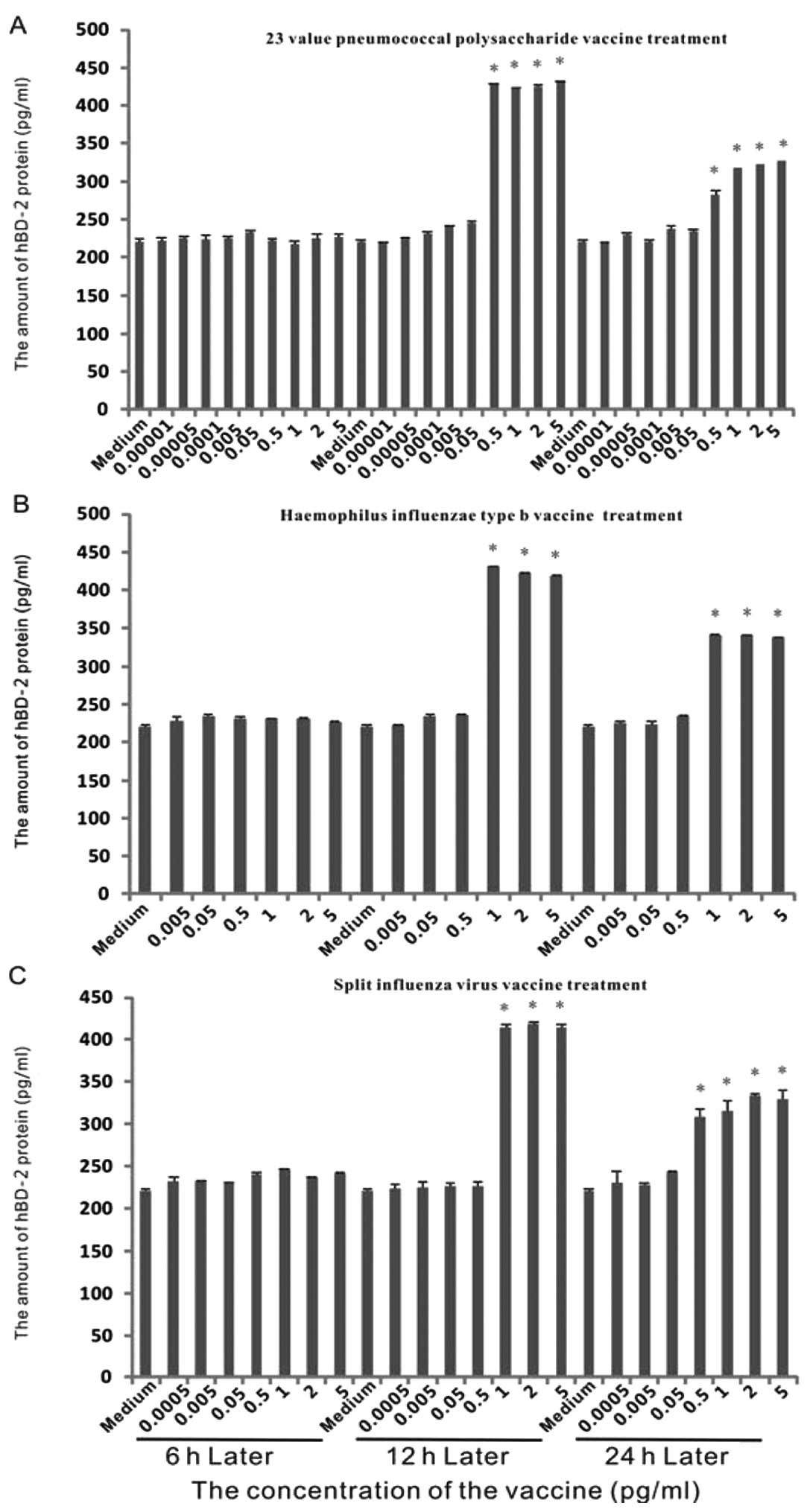
Expression of hBD-2 protein in epithelial
cells with various stimuli
To determine the concentration of protein, we
performed ELISA with the medium of the dose-effect experiments
mentioned previously (Fig. 3).
After the first 6 h, no marked increase of protein occurred.
Following 12 h of treatment, there was a ~2-fold induction by the
23 valent pneumococcal polysaccharide vaccine with concentrations
of above 0.5 μg/ml. Similarly, a ~2-fold induction was observed
when the Haemophilus influenzae vaccine or split influenza virus
vaccine was used with concentrations above 1 μg/ml. At 24 h, the
amount of hBD-2 protein was lower than at 12 h, but remained higher
than that in the control group.
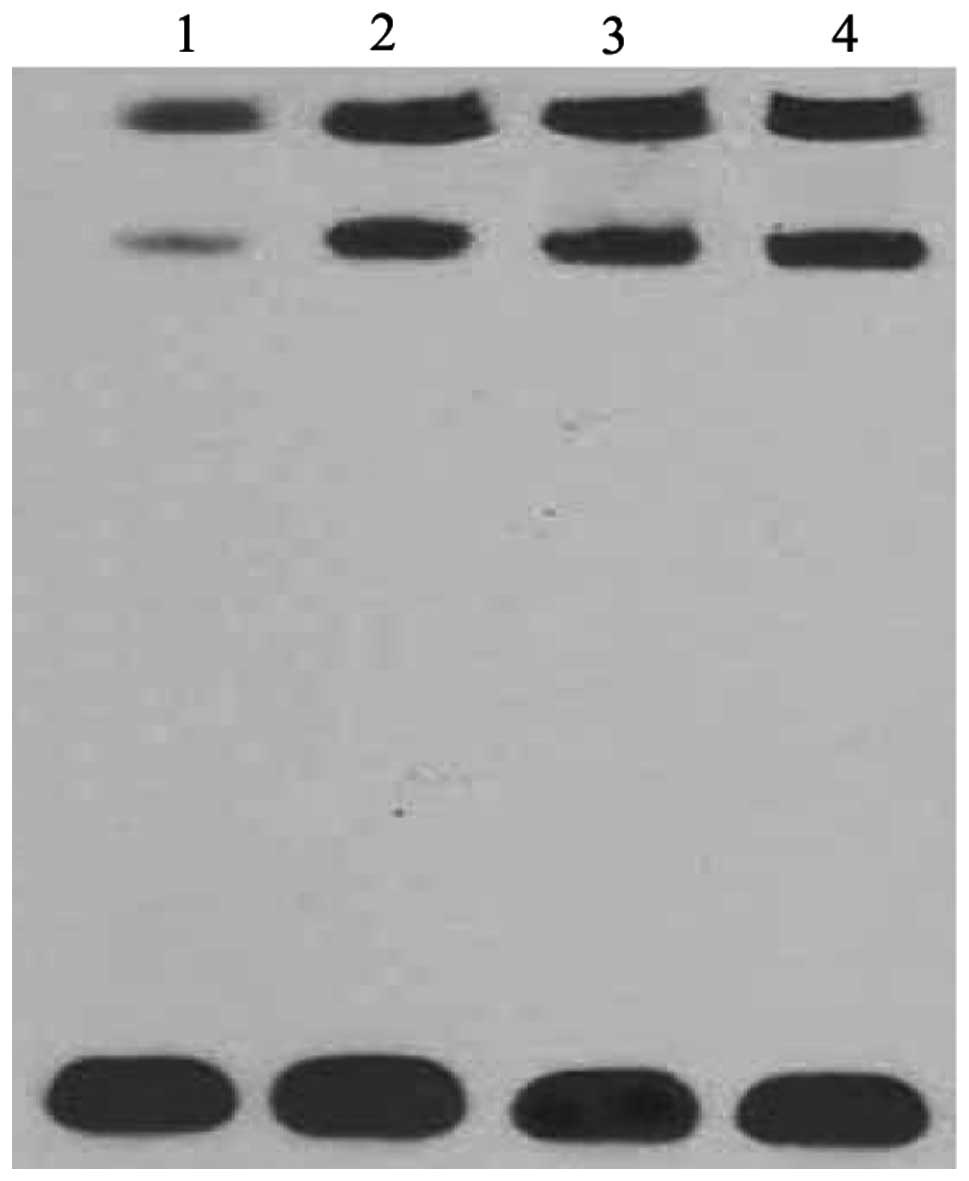
NF-κB involved in the signal pathway of
vaccine-stimulation
Since NF-κB is a well-known mediator of induction of
hBD-2 in epithelial cells, we tested the activity of the NF-κB
subunit in normal epithelial cells and epithelial cells treated
with the vaccines by EMSA (Fig.
4). Treatment with vaccines markedly increased the activity of
the NF-κB subunit, while the activity of the NF-κB subunit was
barely detected in normal epithelial cells.
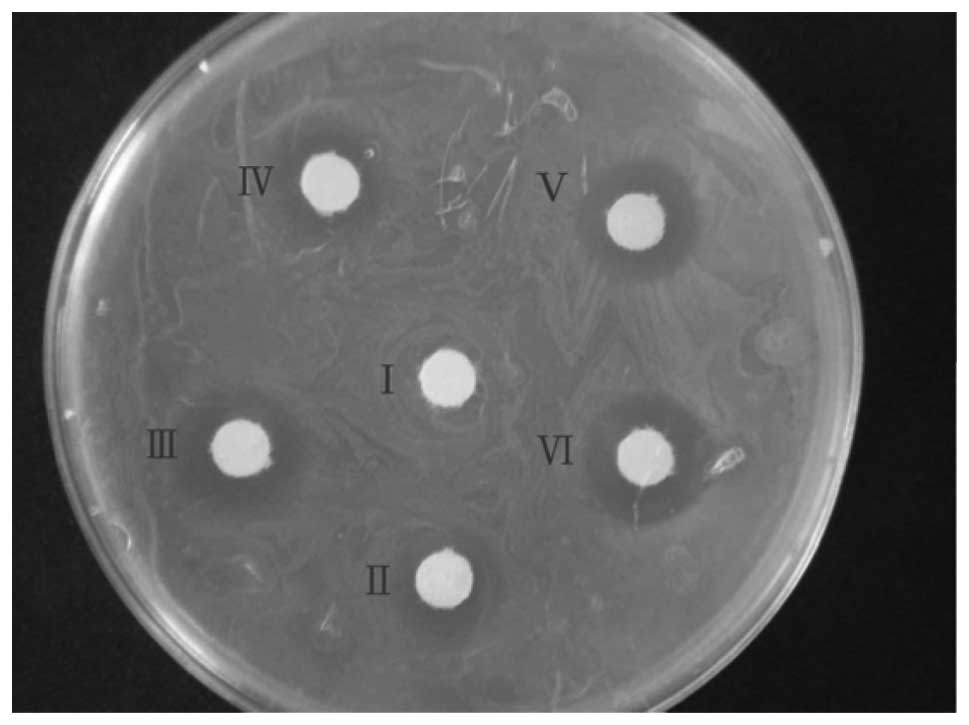
Comparison of antimicrobial ability
To analyze the antimicrobial ability, concentrated
medium was used for the bacteriostatic experiment (Fig. 5). The spots of inhibition were not
clear for PBS and normal medium. The spot diameter was 12 mm with
medium treated with 23 valent pneumococcal polysaccharide vaccine,
10 mm with medium treated with Haemophilus influenzae vaccine and
19 mm with medium treated with split influenza virus vaccine.
Discussion
In this study, we have used vaccines, including 23
valent pneumococcal polysaccharide vaccine, Haemophilus influenzae
vaccine and influenza virus vaccine, to elevate the expression of
hBD-2. The expression level of hBD-2 is markedly low in the absence
of inflammation (8). In
vivo, the vaccines may significantly induce the expression of
hBD-2 within the first 12 h at an appropriate concentration. After
24 h, the expression of hBD-2 declined, but it remained
significantly higher than the control group in the 23 valent
pneumococcal polysaccharide vaccine treatment group and in the
Haemophilus influenzae vaccine treatment group. This indicated that
the effect of vaccines may be short-term.
Several microorganisms are capable of inducing the
expression of certain defensins (25–27).
Besides microorganisms, LTB4, L12, L23, L27 and certain other
immune factors also induce the expression of hBD-2 (24,28).
However, using these microorganisms to study the regulation and
function in vivo and in vitro often leads to the
threat of a biological hazard. Our results showed that vaccines
could also serve as inducers of hBD-2, but with improved biosafety,
simplicity and inexpensiveness. Hence, vaccines may be an
alternative to the use of microorganisms and may facilitate the
study of hBD-2.
Functional binding sites for NF-κB and AP-1 were
identified in the promoter of hBD-2 (29–31).
Besides these, little was known about the mechanism of induction of
hBD-2. Multiple signaling pathways are involved in the inducible
expression of hBD-2 (26). Hence,
our study set up a biosecure model that facilitates the study of
the mechanism of inducible expression in epithelial cells. As the
NF-κB subunit was activated by the vaccines in our model, this
indicated that our model could simulate the in vivo defense
mechanism against microorganisms. Moreover, our preliminary results
showed that vaccines could increase the concentration of
Ca2+ within epithelial cells, which is in line with a
previous study that proposes that the increase of Ca2+
is necessary for the activation of NF-κB (Fig. 6) (32).
During our investigation, the medium of split
influenza virus vaccine treatment was revealed to have the highest
antimicrobial activity with the least amount of hBD-2. Certain
other antimicrobial peptides induced by the specific vaccine may
contribute to the variation of the antimicrobial activity. The 23
valent pneumococcal polysaccharide vaccine was more sensitive to
stimulating hBD-2 expression. Further research is required to
explain the variation in innate immune response to exogenous
stimulation.
Although the pharmaceutical industry endeavors to
develop effective therapies against Gram-negative bacteria, it is
difficult to meet the clinical challenge (33). hBD-2, with high antimicrobial
activity against Gram-negative bacteria, is regarded as an approach
to solve the problem. Thus, the effectiveness of vaccine-induced
hBD-2 should be investigated in future studies.
Acknowledgements
This study was financially supported by the National
Natural Science Foundation of China (No. 30670932), the Social
Development Bureau Fund of Shanghai Municipal Pudong New District
(No. pw2005A-10) and the Medicine Leading Talent Project Fund of
Shanghai Municipal Pudong New District (CX2004004).
References
|
1
|
Lehrer RI: Primate defensins. Nat Rev
Microbiol. 2:727–738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pazgier M, Hoover DM, Yang D, Lu W and
Lubkowski J: Human beta-defensins. Cell Mol Life Sci. 63:1294–1313.
2006. View Article : Google Scholar
|
|
3
|
Rodriguez-Jimenez FJ, Krause A, Schulz S,
et al: Distribution of new human beta-defensin genes clustered on
chromosome 20 in functionally different segments of epididymis.
Genomics. 81:175–183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schroder JM and Harder J: Human
beta-defensin-2. Int J Biochem Cell Biol. 31:645–651. 1999.
View Article : Google Scholar
|
|
5
|
Liu L, Wang L, Jia HP, et al: Structure
and mapping of the human beta-defensin HBD-2 gene and its
expression at sites of inflammation. Gene. 222:237–244. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hiratsuka T, Nakazato M, Date Y, et al:
Identification of human beta-defensin-2 in respiratory tract and
plasma and its increase in bacterial pneumonia. Biochem Biophys Res
Commun. 249:943–947. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bals R, Wang X, Wu Z, et al: Human
beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in
human lung. J Clin Invest. 102:874–880. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh PK, Jia HP, Wiles K, et al:
Production of beta-defensins by human airway epithelia. Proc Natl
Acad Sci USA. 95:14961–14966. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen CI, Schaller-Bals S, Paul KP, Wahn U
and Bals R: Beta-defensins and LL-37 in bronchoalveolar lavage
fluid of patients with cystic fibrosis. J Cyst Fibros. 3:45–50.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harder J, Bartels J, Christophers E and
Schröder JM: A peptide antibiotic from human skin. Nature.
387:8611997. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hall RM and Collis CM: Antibiotic
resistance in gram-negative bacteria: the role of gene cassettes
and integrons. Drug Resist Updat. 1:109–119. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Partridge SR: Analysis of antibiotic
resistance regions in Gram-negative bacteria. FEMS Microbiol Rev.
35:820–855. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boucher HW, Talbot GH, Bradley JS, et al:
Bad bugs, no drugs: no ESKAPE! An update from the Infectious
Diseases Society of America. Clin Infect Dis. 48:1–12. 2009.
|
|
14
|
Slama TG: Gram-negative antibiotic
resistance: there is a price to pay. Crit Care. 12(Suppl 4):
S42008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peleg AY and Hooper DC: Hospital-acquired
infections due to gram-negative bacteria. N Engl J Med.
362:1804–1813. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mauldin PD, Salgado CD, Hansen IS, Durup
DT and Bosso JA: Attributable hospital cost and length of stay
associated with health care-associated infections caused by
antibiotic-resistant gram-negative bacteria. Antimicrob Agents
Chemother. 54:109–115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Siegel RE: Emerging gram-negative
antibiotic resistance: daunting challenges, declining
sensitivities, and dire consequences. Respir Care. 53:471–479.
2008.PubMed/NCBI
|
|
18
|
Chopra I, Schofield C, Everett M, et al:
Treatment of health-care-associated infections caused by
Gram-negative bacteria: a consensus statement. Lancet Infect Dis.
8:133–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu B, Dong CY, Zhang F, Lin YM, Wu KF and
Ma XT: Synergistic antileukemia effect of combinational gene
therapy using murine beta-defensin 2 and IL-18 in L1210 murine
leukemia model. Gene Ther. 14:1181–1187. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yin C, Dang HN, Gazor F and Huang GT:
Mouse salivary glands and human beta-defensin-2 as a study model
for antimicrobial gene therapy: technical considerations. Int J
Antimicrob Agents. 28:352–360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harvey SA, Romanowski EG, Yates KA and
Gordon YJ: Adenovirus-directed ocular innate immunity: the role of
conjunctival defensin-like chemokines (IP-10, I-TAC) and phagocytic
human defensin-alpha. Invest Ophthalmol Vis Sci. 46:3657–3665.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meyerholz DK, Grubor B, Gallup JM, et al:
Adenovirus-mediated gene therapy enhances parainfluenza virus 3
infection in neonatal lambs. J Clin Microbiol. 42:4780–4787. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bals R, Weiner DJ, Moscioni AD, Meegalla
RL and Wilson JM: Augmentation of innate host defense by expression
of a cathelicidin antimicrobial peptide. Infect Immun.
67:6084–6089. 1999.PubMed/NCBI
|
|
24
|
Gaudreault E and Gosselin J: Leukotriene
B4 induces release of antimicrobial peptides in lungs of virally
infected mice. J Immunol. 180:6211–6221. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alekseeva L, Huet D, Femenia F, et al:
Inducible expression of beta defensins by human respiratory
epithelial cells exposed to Aspergillus fumigatus organisms.
BMC Microbiol. 9:332009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krisanaprakornkit S, Kimball JR, Weinberg
A, Darveau RP, Bainbridge BW and Dale DA: Inducible expression of
human beta-defensin 2 by Fusobacterium nucleatum in oral
epithelial cells: multiple signaling pathways and role of commensal
bacteria in innate immunity and the epithelial barrier. Infect
Immun. 68:2907–2915. 2000.PubMed/NCBI
|
|
27
|
Liu AY, Destoumieux D, Wong AV, et al:
Human beta-defensin-2 production in keratinocytes is regulated by
interleukin-1, bacteria, and the state of differentiation. J Invest
Dermatol. 118:275–281. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda N, Tada Y, Shimizu T and Watanabe S:
IL-12, IL-23, and IL-27 enhance human beta-defensin-2 production in
human keratinocytes. J Invest Dermatol. 38:1287–1296.
2008.PubMed/NCBI
|
|
29
|
Tsutsumi-Ishii Y and Nagaoka I: NF-kappa
B-mediated transcriptional regulation of human beta-defensin-2 gene
following lipopolysaccharide stimulation. J Leukoc Biol.
71:154–162. 2002.PubMed/NCBI
|
|
30
|
Yoon YM, Lee JY, Yoo D, et al: Bacteroides
fragilis enterotoxin induces human beta-defensin-2 expression in
intestinal epithelial cells via a mitogen-activated protein
kinase/I kappa B kinase/NF-kappa B-dependent pathway. Infect Immun.
78:2024–2033. 2010. View Article : Google Scholar
|
|
31
|
Wehkamp J, Harder J, Wehkamp K, et al:
NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in
intestinal epithelial cells by Escherichia coli Nissle 1917:
a novel effect of a probiotic bacterium. Infect Immun.
72:5750–5758. 2004.PubMed/NCBI
|
|
32
|
Palkowitsch L, Marienfeld U, Brunner C,
Eitelhuber A, Krappmann D and Marienfeld RB: The
Ca2+-dependent phosphatase calcineurin controls the
formation of the Carma1-Bcl10-Malt1 complex during T cell
receptor-induced NF-kappaB activation. J Biol Chem. 286:7522–7534.
2011.PubMed/NCBI
|
|
33
|
Lavigne JP, Brunel JM, Chevalier J and
Pages JM: Squalamine, an original chemosensitizer to combat
antibiotic-resistant gram-negative bacteria. J Antimicrob
Chemother. 65:799–801. 2010. View Article : Google Scholar : PubMed/NCBI
|