Introduction
As a novel cancer/testis antigen, cancer-associated
gene (CAGE) was originally isolated by serological analysis of a
cDNA expression library (SEREX) with serum from a patient with
gastric cancer (1). CAGE is known
to be widely expressed among cancer cell lines and cancer tissues,
yet restricted to testis tissue among normal tissues (2–4).
Cancer/testis antigens have the notable property of being
immunogenic in cancer patients and are considered promising targets
for cancer vaccines due to their restricted expression pattern
(5). Thus, anti-CAGE antibodies
are applicable for payload delivery. Particularly in oncology,
anti-CAGE antibodies may be combined with cytotoxic entities that
have limited selectivity.
Traditionally, monoclonal antibodies (mAbs) are
produced through rodent immunization using hybridoma technology.
However, this approach is laborious and poses difficulties in
generating antibodies against self-antigens. In recent years, in
vitro display techniques, including phage and ribosome display,
have become a platform technology for the design, selection and
production of reagents for targeted therapies in cancer (6). Ribosome display has a number of
advantages over phage display. A number of the limitations of phage
display including the inability to select antibodies under
conditions different from the cell environment, problems with the
selection of proteins that are toxic, cells circumventing selection
pressure and low transformation efficiency, may be circumvented
using in vitro ribosome display (7). Ribosome display enables rapid and
easy screening to isolate specific novel binders to target ligands.
Over the past decade, ribosome display has been widely used for the
selection of human antibodies for therapeutic intervention in
various cancers (8–10). In the present study, we describe
the construction of a single-chain variable fragment (scFv)
antibody library and selection of a fully human antibody fragment
from a gastric cancer patient-derived gene pool by in vitro
ribosome display technology.
Materials and methods
Construction of the scFv library
Peripheral blood lymphocytes were isolated from 10
gastric cancer patients who initially did not receive chemotherapy.
All patients provided a participant information statement and
informed consent. This study was approved by the Human Ethics
Committee of Nanchang University School of Medicine (Jiangxi,
China). Total RNA was isolated individually from peripheral blood
lymphocytes using an RNA purification kit for amplification of the
scFv library. Primers were designed as previously described
(11), with certain modifications
for the construction of ribosome display libraries (Table I). First-strand cDNA was
synthesized and V genes were amplified using suitable primers.
Agarose gel electrophoresis revealed bands of the expected sizes.
Splicing by splicing overlap extension polymerase chain reaction
(PCR) led to a full-length human scFv repertoire linking the heavy
and light chains. scFv antibody libraries in the format of
VH/κ and VH/Vλ-Cκ were constructed
for ribosome display.
 | Table IOligodeoxynucleotide primers used for
construction of the library. |
Table I
Oligodeoxynucleotide primers used for
construction of the library.
| A, Primers for
generating VH-linker fragments. |
|---|
| Upstream primer
sequence (5′-3′) |
|
HuVH1b-7a/reverse |
GAG(A/G)TGCAGCTGGTGCA(A/G)TCTGG |
|
HuVH1c/reverse |
(G/C)AGGTCCAGCTGGT(A/G)CAGTCTGG |
|
HuVH2b/reverse |
CAG(A/G)TCACCTTGAAGGAGTCTGG |
|
HuVH3b/reverse |
(G/C)AGGTGCAGCTGGTGGAGTCTGG |
|
HuVH3c/reverse |
GAGGTGCAGCTGGTGGAG(A/T)C(C/T)GG |
|
HuVH4b/reverse |
CAGGTGCAGCTACAGCAGTGGGG |
|
HuVH4c/reverse |
CAG(G/C)TGCAGCTGCAGGAGTC(G/C)GG |
|
HuVH5b/reverse |
GA(A/G)GTGCAGCTGGTGCAGTCTGG |
|
HuVH6a/reverse |
CAGGTACAGCTGCAGCAGTCAGG |
| Downstream primer
sequence (5′-3′) |
| HuIgM
linker/for |
GGAGACGAGGGGGAAAAGGGTTGG |
|
| B, Primers for
generating Vκ light chain. |
|
| Upstream primer
sequence (5′-3′) |
| Hu
Vκ1b/reverse |
CTTTTCCCCCTCGTCTCCGACATCCAG(A/T)TGACCCAGTCTCC |
| Hu
Vκ2/reverse |
CTTTTCCCCCTCGTCTCCGATGTTGTGATGACTCAGTCTCC |
| Hu
Vκ3b/reverse |
CTTTTCCCCCTCGTCTCCGAAATTGTG(A/T)TGAC(A/G)CAGTCTCC |
| Hu
Vκ4b/reverse |
CTTTTCCCCCTCGTCTCCGATATTGTGATGACCCACACTCC |
| Hu
Vκ5/reverse |
CTTTTCCCCCTCGTCTCCGAAACGACACTCACGCAGTCTCC |
| Hu
Vκ6/reverse |
CTTTTCCCCCTCGTCTCCGAAATTGTGCTGACTCAGTCTCC |
| Downstream primer
sequence (5′-3′) |
| HuCκ/for | GCTCTAGA
ACACTCTCCCCTGTTGAAGCT |
|
| C, Primers for
generating Vλ fragment. |
|
| Upstream primer
sequence (5′-3′) |
| Hu
Vλ1a/reverse |
CTTTTCCCCCTCGTCTCCCAGTCTGTGCTGACTCAGCCACC |
| Hu
Vλ1b/reverse |
CTTTTCCCCCTCGTCTCCCAGTCTGTG(C/T)TGACGCAGCCCGCC |
| Hu
Vλ1c/reverse |
CTTTTCCCCCTCGTCTCCCAGTCTGTCGTGACGCAGCCGCC |
| Hu
Vλ2/reverse |
CTTTTCCCCCTCGTCTCCCA(A/G)TCTGCCCTGACTCAGCCT |
| Hu
Vλ3a/reverse |
CTTTTCCCCCTCGTCTCCTCCTATG(A/T)GCTGACTCAGCCACC |
| Hu
Vλ3b/reverse |
CTTTTCCCCCTCGTCTCCTCTTCTGAGCTGACTCAGGACCC |
| Hu
Vλ4/reverse |
CTTTTCCCCCTCGTCTCCCACGTTATACTGACTCAACCGCC |
| Hu
Vλ5/reverse |
CTTTTCCCCCTCGTCTCCCAGGCTGTGCTGACTCAGCCGTC |
| Hu
Vλ6/reverse |
CTTTTCCCCCTCGTCTCCAATTTTATGCTGACTCAGCCCCA |
| Hu
Vλ7–8/reverse |
CTTTTCCCCCTCGTCTCCCAG(A/G)TCGTGGTGAC(C/T)CAGGAGCC |
| Hu
Vλ9/reverse |
CTTTTCCCCCTCGTCTCCC(A/T)GCCTGTGCTGACTCAGCC(A/C)CC |
| Downstream primer
sequence (5′-3′) |
|
HuJλ/forward |
GCAAGATGGTGCAGCCACACCTA(A/G)(A/G)ACGGTGAGCTTGGTC |
|
| D, Primers for
generating full-length constructa. |
|
| Upstream primer
sequence (5′-3′) |
| T7Ab/reverse | GCAGC
TAATACGACTCACTATAGGAACAGACCACCATG
(C/G)AGGT(G/C)CA(G/C)CTCGAG (C/G)AGTCTGG |
| Downstream primer
sequence (5′-3′) |
| HuCκ/forward | GCTCTAGAACACTCTCCCCTGTTGAAGCT |
|
| E, Primers for
generating constant region (Cκ) of κ light chainb. |
|
| Upstream primer
sequence (5′-3′) |
| HuCκ/reverse |
ACTGTGGCTGCACCATCTG |
| Downstream primer
sequence (5′-3′) |
| HuCκ/forward | GCTCTAGAACACTCTCCCCTGTTGAAGCT |
Antibody-ribosome-mRNA (ARM) ribosome
display
The eukaryotic ribosome display was performed as
described previously (12), with
certain modifications. The PCR libraries were expressed in a
coupled rabbit reticulocyte lysate system to generate ARM
complexes. The reaction was performed in a volume of 50 μl
containing 40 μl TNT T7 Quick for PCR DNA, up to 5 μg PCR DNA, 1 μl
1 mM methionine, 0.5 μl 100 mM magnesium acetate and 3.5 μl
ddH2O. The mixture was incubated in a siliconized tube
at 30°C for 60 min. Forty units RNase-free DNase I were added to
the mixture together with 6 μl 10X DNase I digestion buffer and
ddH2O, yielding a final volume of 60 μl. The mixture was
then incubated at 30°C for an additional 15 min. Cold (4°C)
phosphate-buffered saline (PBS; 60 μl) was added prior to antigen
selection. Antigen-linked Dynabeads (2 μg) were then added and the
mixture was incubated at 4°C for 2 h with gentle agitation to
select a CAGE-specific antibody fragment. The beads were collected
magnetically and then washed three times with cold washing buffer
(PBS, 1% Tween-20, and 5 mM magnesium acetate), followed by washing
twice with cold ddH2O. The beads were collected by
magnetic particle clutch and resuspended in 10 μl nuclease-free
ddH2O for reverse transcription (RT)-PCR to recover
antigen-selected mRNA. The beads were stored at 4 or −20°C.
The selected mRNA was reverse transcribed to cDNA
using a PrimeScript® One Step RT-PCR Kit (ver. 2) for 30
min at 50°C for the reverse transcription and then for 2 min at
94°C for deactivation of the reverse transcriptase. The obtained
cDNA was amplified in a 50 μl PCR mixture for 30 cycles (30 sec at
94°C, 30 sec at 55°C and 1 min at 72°C) with T7A1
(5′-GCAGCTAATACGACTCACTATAGAACA GACCACCATG-3′) and Hu1
(5′-GCTCAGCGTCAGGGT GCTGCT-3′). For nested PCR, different
downstream primers for subsequent cycles were used to enrich
production: Hu2 (5′-CTCTCCTGGGAGTTACC-3′) in the second cycle and
Hu3 (5′-GAAGACAGATGGTGCAGC-3′) in the third cycle. Purified
products were applied to the subsequent selection round.
Cloning and expression
The DNA generated using the above-described steps
was digested with XhoI and XbaI and ligated into the
vector pBluescript SK+ for periplasmic expression of the
antibody fragment. Escherichia coli (E. coli) BL21
containing the expression plasmid was grown in 20 ml 2X
tryptone-yeast extract (TY; 16 g tryptone; 10 g yeast extract, 5 g
NaCl) medium containing 100 μg/ml ampicillin and 0.5% glucose at
37°C overnight. Following centrifugation at 300 × g for 10 min, the
bacterial pellet was induced by resuspension in 50 ml fresh 2X TY
containing 100 μg/ml ampicillin and 0.5 mM
isopropylthio-β-galactoside (IPTG) and then grown at room
temperature (20–25°C) for 5–7 h. The bacteria were then harvested
by centrifugation at 300 × g for 10 min and the pellet was either
stored at −20°C or used immediately. The pellet was first washed
with 1 ml 20% (w/v) sucrose, 0.3 M Tris-HCl (pH 8.0) and 1 mM
ethylenediaminetetraacetic acid (EDTA) to prepare a periplasmic
extract. Following centrifugation, the bacteria were subjected to
osmotic shock by rapidly mixing with 1 ml ice-cold 0.5 mM
MgCl2 and leaving on ice for 10 min with periodic
agitation. The supernatant periplasmic fraction containing the
soluble scFv was recovered by centrifugation at 13,000 × g for 10
min at room temperature. Recombinant protein was further purified
by affinity chromatography using Ni-NTA spin kits. Following
binding of the protein to Ni-NTA agarose, the column was washed
with increasing concentrations of imidazole. The expressed protein
was eluted with 200 mM imidazole and dialyzed overnight in PBS at
4°C.
Immunoblot analysis
The purified periplasmic extracts from selected
anti-CAGE-producing clones were subjected to sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) on 12% polyacrylamide
gel. Following SDS-PAGE, the gel was transferred to a
nitrocellulose membrane [2% (w/v) skimmed milk in PBS]. The
transblotted membrane was blocked for 1 h at room temperature with
blocking solution and then incubated for 1 h at room temperature
with peroxidase-conjugated mouse anti-His tag (1/1000 dilution with
blocking solution). 4-Chloro-1-naphthol was used as a peroxidase
substrate to visualize the immunoreactivity. Protein concentration
was determined using the BCA protein assay kit and purity was
assessed with SDS-PAGE analysis.
Determination of scFv titer and affinity
constant
The equilibrium dissociation constant
(KD) value of scFv antibodies was determined in the
solution phase by enzyme-linked immunosorbent assay (ELISA)
(13,14). A 96-well plate was first coated
with 5 μg/ml CAGE protein and then serially diluted scFv protein in
2% skimmed milk was added at 37°C for 1 h. The bound antibodies
were detected with normal ELISA. The concentration of scFv
antibodies presenting 50% of the maximum antigen-binding activity
was used in competitive ELISA, which was performed on immobilized
CAGE as described above, with the exception that CAGE at various
concentrations was mixed with a constant amount of antibody in 100
mM PBS (pH 7.4) supplemented with 10 mg/ml bovine serum albumin
(BSA). The antibody concentration was derived using ELISA as
mentioned above. After overnight incubation at room temperature, 50
μl each mixture was transferred into the well, coated with CAGE and
incubated for 1 h at 37°C. After washing, the bound scFv antibodies
were detected as above in triplicate and their affinity was
calculated using the Scatchard analysis equation.
Sequencing analysis
Plasmid DNA from anti-CAGE-producing clones was
isolated from E. coli BL21. The scFv DNA was sequenced using
the dideoxy method with the pBluescript SK+ sequence
primer set in an ABI Prism automated sequencing machine system.
Results
Antibody library construction
In this study, a large non-immune ribosome display
antibody library was established for routine isolation of a
high-affinity human scFv antibody to the CAGE antigen. Purified
lymphocytes from blood samples were obtained from 10 gastric cancer
patients for amplification of the scFv library. Following total RNA
isolation, first-strand cDNA synthesis and amplification of V genes
with their corresponding primers were performed. Agarose gel
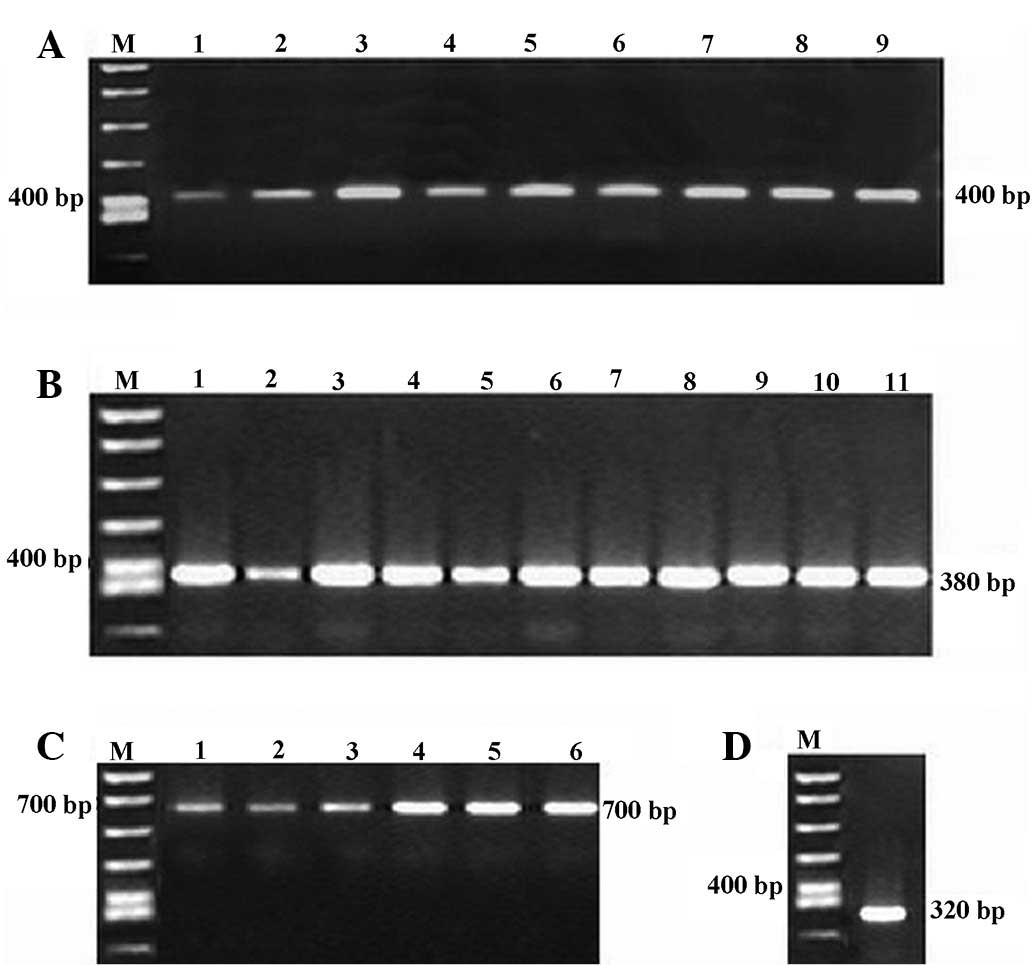
electrophoresis revealed bands of the expected sizes (Fig. 1). Splicing by overlap extension PCR
was then used to generate full-length scFv gene templates, i.e.,
VH/κ and VH/Vλ-Cκ, which were
detected by gel electrophoresis (Fig.
2). Although variable regions in the light and heavy chains of
the scFv genes were different, their upstream primer, downstream
primer and linker were the same. Thus, these scFvs had an identical
sequential pattern graph. DNA sequencing of randomly selected
clones confirmed in-frame cloned V genes of human origin derived
from various V-gene families (data not shown).
ARM ribosome display
The scFv library was expressed in vitro in
the rabbit reticulocyte coupled transcription/translation system to
generate ternary RNA-ribosome-scFv complexes. The ribosome
complexes were enriched by the binding of scFv antibody to
recombinant CAGE protein-conjugated Dynabeads. Following RT-PCR
recovery of the selected RNA in the complexes, the small DNA band
was detected following three rounds of subtractive panning, as
shown by the results of agarose gel electrophoresis (Fig. 3). For nested PCR, we used the
downstream primer Hu2 in the second cycle and Hu3 in the third
cycle in order that the recovered DNA became progressively shorter
in each cycle. The shortening only affected the constant domain of
the κ-light chain.
Western blot analysis
After three rounds of panning, the CAGE-specific
scFv antibody genes were digested and ligated into the expression
vector. The expressed scFv antibodies were collected from the
periplasm of the bacteria and purified by affinity chromatography
using Ni-NTA spin kits. The amount of the best-expressed scFv
anti-CAGE proteins expressed in E. coli BL21 was ~30% of the
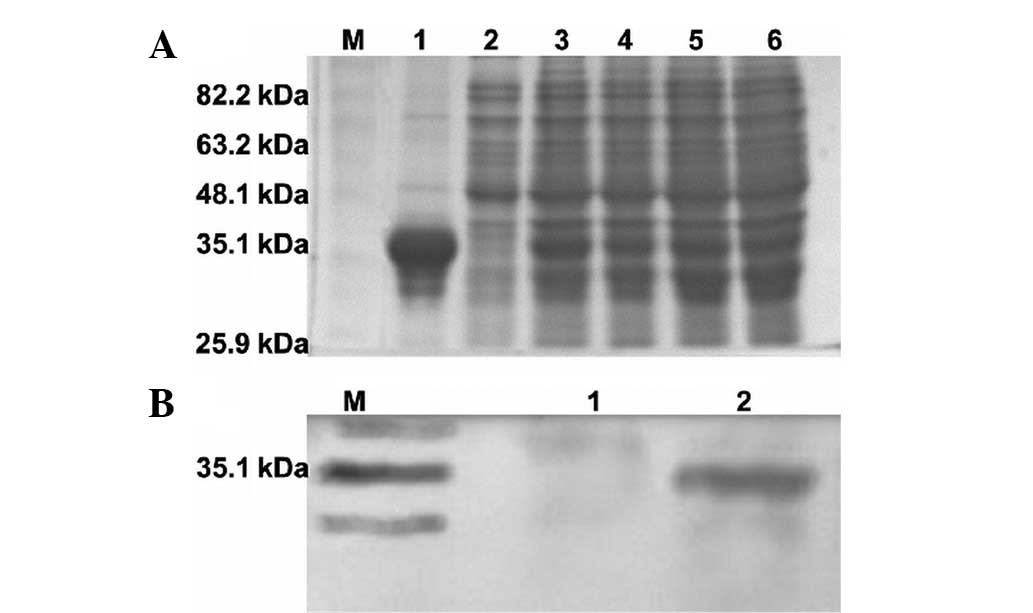
total bacterial proteins. SDS-PAGE results for the full-length
fusion proteins are shown in Fig.
4A. The expression of scFv was confirmed by western blot
analysis using anti-His tag antibodies (Fig. 4B). The selected scFv antibody
presented a single band measuring ~35 kDa in size.
Dissociation constant of CAGE-specific
scFvs and sequence analysis
According to the Scatchard analysis equation
[A0/(A0 - A) =
KD/Ag +1, where
A0 is the absorbance when the antibody was
incubated without any antigen, A is the absorbance
corresponding to free antibody following incubation with antigen
and Ag is the free antigen concentration that is
equal to the antigen obtained for experimentation considering a
pseudo-first-order reaction], the KD of scFv for
CAGE was 7.6×10−8 M. Fig.
5 shows the RT-PCR product sequence obtained following the
third screening cycle and its deduced amino acid sequence.
Complementarity determining regions (CDRs) are marked as per the
Kabat definition.
Discussion
Gastric cancer is one of the most common
malignancies in the digestive system. Chemotherapy remains the
mainstay of treatment for advanced gastric cancer; however, its
efficacy is modest. An understanding of the molecular biological
mechanisms underlying the formation, progression and metastasis of
advanced gastric cancer has enabled the use of this new approach to
treat this disease in clinical practice. In recent years,
molecular-targeted therapies have emerged as a novel approach to
the treatment of gastric cancer (15). mAb-based cancer immunotherapy has
attracted significant research interest. Immunoconjugate agents are
obtained by coupling radioactive materials, chemotherapeutic agents
and biotoxins with mAbs (16–18).
The coupled agents produce antitumor effects by directing mAbs to
the cell surface that expresses the corresponding antigen. The
coupled agents also reduce the damage to normal tissues.
At present, therapeutic mAbs are one of the most
rapidly growing components of the biopharmaceutical industry.
Investigators have created large numbers of mouse mAbs to treat or
diagnose human diseases. However, the availability of mouse mAbs
has been greatly limited by their high immunogenicity in humans and
rapid clearance due to the human anti-mouse antibody reactions that
occur in patients (19). Thus,
mouse mAbs have exhibited significantly limited and inefficient
effector functions in clinical trials. To reduce the immunogenicity
of mouse mAbs, an attempt has been made to establish methods to
generate human-derived mAbs. Several methods for generating human
mAbs have been established, including the phage and ribosome
display methods, as well as transchromosome mice technology, in
which human Ig genes are expressed (20). Ribosome display is considered to be
one of the most promising biotechnologies among the next generation
of display technologies (21). It
is a technique used to perform in vitro protein evolution to
create proteins that bind to a desired ligand (22). The process results in translated
proteins associated with their mRNA progenitor, which is used as a
complex to bind to an immobilized ligand in a selection step.
scFv-ribosome-mRNA complexes bind to their specific antigens and
non-specific complexes are removed by extensive washing. Eluted
mRNAs from the remaining complexes are reverse transcribed to cDNA
and reamplified by PCR. The outcome is a nucleotide sequence that
may be used to create tightly binding proteins. In addition,
ribosome display has advantages in selection from larger libraries
and is capable of affinity maturation without any need for
additional ligation, transformation or construction of a second
library (23).
Together with the progression of genome medical
science, an increasing number of target molecules in various
diseases have been revealed. Cancer testis antigens are a unique
class of tumor antigens expressed in a variety of cancer tissues
but silent in normal tissues, with the exception of the testis. Due
to their restricted gene expression in the testis and various
malignancies, cancer testis antigens are potential defined targets
for antigen-based vaccination and antigen-directed immunotherapy
for the regulation of cancer growth (24). CAGE is a typical cancer/testis
antigen that is overexpressed in gastric cancer tissues, whereas
its expression in normal tissues is limited to the testis (25). Studies have shown that anti-CAGE
IgG antibodies are present in the sera of patients with various
cancers (26). Therefore, CAGE may
be an ideal target for delivering cytotoxic agents to tumors.
In this study, we used ribosome display to select a
fully human anti-CAGE scFv. Small antibody fragments, including Fab
and scFv, exhibit applicable pharmacokinetics for tissue
penetration and full binding specificity as the antigen-binding
surface is unaltered (27). A
single-chain antibody library in the format of VH/κ and
VH/Vλ-Cκ was constructed using a PCR-based
recombination method. Using this DNA library, we performed
CAGE-specific scFv selection and enrichment by ribosome display.
After three rounds of selection, anti-CAGE scFv was cloned into an
expression vector.
In vitro display techniques have certain
potential advantages over in vivo display methods. These
include ease of generating larger libraries, fewer biases than
would be caused by cell expression and more facile application of
round-by-round mutagenesis (28).
In this study, we performed ribosome display, an in vitro
display method, using an in vitro transcription and
translation process. As the library size of a ribosome display is
not limited by transformation, anti-CAGE scFv is more easily
obtained from DNA libraries using ribosome display than by in
vivo display methods. We used a rabbit reticulocyte lysate
system since such a system has a lower level of RNase activity
compared with the E. coli ribosome display system, rendering
the selection conditions less complex. Moreover, eukaryotic
conditions may improve the translation and/or folding efficiency of
certain proteins. Therefore, our study is valuable as an example of
antibody selection from an antibody library by ribosome display
using a eukaryotic translation system.
Although we observed antigen-specific gene recovery
by RT-PCR and monitored the enrichment rate during the selection
procedure, our results suggest that three rounds are not sufficient
for complete selection. One reason may be that stringent selection
conditions are essential for the enrichment of specific binders
(29). Low temperature and
RNase-free conditions are required to maintain intact
scFv-ribosome-RNA complexes. With the aim of developing more
efficient selection in ribosome display using eukaryotic
translation, we are currently studying the factors involved in
maintaining ARM complexes. To the best of our knowledge, this is
the first time that this technique has been used successfully for
the direct generation of human scFv against CAGE. As the scFv
originates from humans, it is expected to be less immunogenic.
In summary, a new human anti-CAGE scFv was isolated
using in vitro ribosome display technology. The
technological challenge of the ribosome display methodology is
reflected in the screening process. Nevertheless, it successfully
isolates a fully human antibody construct with specificity to a
clinically relevant target antigen. It may provide the basis for
the development of anti-scFv-drug conjugates for the proliferation
and metastasis of gastric cancer cells.
Acknowledgements
The authors thank Dr Bing-Ya Liu and Wen-Tao Liu
(Shanghai Institute of Digestive Surgery, Ruijin Hospital, Shanghai
Jiao Tong University School of Medicine) for their technical
assistance. This study was supported by the National Natural
Science Foundation of China (no. 30960442).
References
|
1
|
Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim
WH, Yang H, Bang YJ and Jeoung DI: Identification and
characterization of a novel cancer/testis antigen gene CAGE.
Biochem Biophys Res Commun. 292:715–726. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim Y and Jeoung D: Role of CAGE, a novel
cancer/testis antigen, in various cellular processes, including
tumorigenesis, cytolytic T lymphocyte induction, and cell motility.
J Microbiol Biotechnol. 18:600–610. 2008.
|
|
3
|
Kim Y, Park H, Park D, Lee YS, Choe J,
Hahn JH, Lee H, Kim YM and Jeoung D: Cancer/testis antigen CAGE
exerts negative regulation on p53 expression through HDAC2 and
confers resistance to anti-cancer drugs. J Biol Chem.
285:25957–25968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Por E, Byun HJ, Lee EJ, Lim JH, Jung SY,
Park I, Kim YM, Jeoung DI and Lee H: The cancer/testis antigen CAGE
with oncogenic potential stimulates cell proliferation by
up-regulating cyclins D1 and E in an AP-1- and E2F-dependent
manner. J Biol Chem. 285:14475–14485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Caballero OL and Chen YT: Cancer/testis
(CT) antigens: potential targets for immunotherapy. Cancer Sci.
100:2014–2021. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Filpula D: Antibody engineering and
modification technologies. Biomol Eng. 24:201–215. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dreier B and Plückthun A: Ribosome
display: a technology for selecting and evolving proteins from
large libraries. Methods Mol Biol. 687:283–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rothe A, Nathanielsz A, Hosse RJ,
Oberhäuser F, Strandmann EP, Engert A, Hudson PJ and Power BE:
Selection of human anti-CD28 scFvs from a T-NHL related scFv
library using ribosome display. J Biotechnol. 130:448–454. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rothe A, Nathanielsz A, Oberhäuser F, von
Pogge SE, Engert A, Hudson PJ and Power BE: Ribosome display and
selection of human anti-cD22 scFvs derived from an acute
lymphocytic leukemia patient. Biol Chem. 389:433–439. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li F, Su P, Lin C, Li H, Cheng J and Shi
D: Ribosome display and selection of human anti-placental growth
factor scFv derived from ovarian cancer patients. Protein Pept
Lett. 17:585–590. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He M, Cooley N, Jackson A and Taussig MJ:
Production of human single-chain antibodies by ribosome display.
Methods Mol Biol. 248:177–189. 2004.PubMed/NCBI
|
|
12
|
He M and Taussig MJ:
Antibody-ribosome-mRNA (ARM) complexes as efficient selection
particles for in vitro display and evolution of antibody combining
sites. Nucleic Acids Res. 25:5132–5134. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Friguet B, Chaffotte AF, Djavadi-Ohaniance
L and Goldberg ME: Measurements of the true affinity constant in
solution of antigen-antibody complexes by enzyme-linked
immunosorbent assay. J Immunol Methods. 77:305–319. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan XH and Xu ZR: Production of human
single-chain variable fragment (scFv) antibody specific for digoxin
by ribosome display. Indian J Biochem Biophys. 42:350–357.
2005.PubMed/NCBI
|
|
15
|
Yoong J, Michael M and Leong T: Targeted
therapies for gastric cancer: current status. Drugs. 71:1367–1384.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Andratschke M, Luebbers CW, Johannson V,
Schmitt B, Mack B, Zeidler R, Lang S, Wollenberg B and Gildehaus
FJ: Biodistribution of 131I-labeled anti-CK8 monoclonal antibody in
HNSCC in xenotransplanted SCID mice. Anticancer Res. 31:3315–3321.
2011.PubMed/NCBI
|
|
17
|
Kreitman RJ and Pastan I: Antibody fusion
proteins: anti-CD22 recombinant immunotoxin moxetumomab pasudotox.
Clin Cancer Res. 17:6398–6405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ricart AD: Antibody-drug conjugates of
calicheamicin derivative: gemtuzumab ozogamicin and inotuzumab
ozogamicin. Clin Cancer Res. 17:6417–6427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bernett MJ, Karki S, Moore GL, et al:
Engineering fully human monoclonal antibodies from murine variable
regions. J Mol Biol. 396:1474–1490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamashita M, Katakura Y and Shirahata S:
Recent advances in the generation of human monoclonal antibody.
Cytotechnology. 55:55–60. 2007. View Article : Google Scholar
|
|
21
|
He M and Khan F: Ribosome display:
next-generation display technologies for production of antibodies
in vitro. Expert Rev Proteomics. 2:421–430. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zahnd C, Amstutz P and Plückthun A:
Ribosome display: selecting and evolving proteins in vitro that
specifically bind to a target. Nat Methods. 4:269–279. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He M and Taussig MJ: Selection of
recombinant antibodies by eukaryotic ribosome display. Methods Mol
Biol. 484:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suri A: Cancer testis antigens - their
importance in immunotherapy and in the early detection of cancer.
Expert Opin Biol Ther. 6:379–389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cho B, Lee H, Jeong S, Bang YJ, Lee HJ,
Hwang KS, Kim HY, Lee YS, Kang GH and Jeoung DI: Promoter
hypomethylation of a novel cancer/testis antigen gene CAGE is
correlated with its aberrant expression and is seen in premalignant
stage of gastric carcinoma. Biochem Biophys Res Commun. 307:52–63.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iwata T, Fujita T, Hirao N, et al:
Frequent immune responses to a cancer/testis antigen, CAGE, in
patients with microsatellite instability-positive endometrial
cancer. Clin Cancer Res. 11:3949–3957. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Accardi L and Di Bonito P: Antibodies in
single-chain format against tumour-associated antigens: present and
future applications. Curr Med Chem. 17:1730–1755. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ullman CG, Frigotto L and Cooley RN: In
vitro methods for peptide display and their applications. Brief
Funct Genomics. 10:125–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee MS, Kwon MH, Kim KH, Shin HJ, Park S
and Kim HI: Selection of scFvs specific for HBV DNA polymerase
using ribosome display. J Immunol Methods. 284:147–157. 2004.
View Article : Google Scholar : PubMed/NCBI
|