Introduction
Ulcerative colitis (UC) is precipitated by a complex
interaction of environmental, genetic and immunoregulatory factors
(1,2). Owing to the westernization of
lifestyle and dietary habits, the number of UC patients has
increased considerably in some Asian countries, including Japan and
China (3). UC has an impact on the
colon and rectum and usually involves the innermost lining mucosa,
appearing as continuous areas of inflammation, with no segments of
normal mucosa (4). Although the
pathogenesis of UC is only partially understood, previous studies
have indicated the significant role of the innate immune response
in the pathogenesis of UC (5,6). In
addition, a number of genes are associated with UC itself (7–10).
The biology of the innate immunity receptors is
crucial in the host response to the environment. Toll-like
receptors (TLRs) play a significant role in the gut innate immunity
and are involved in human inflammatory bowel diseases, including UC
(11). TLR activation initially
recruits the myeloid differentiation primary response gene (88)
(MyD88) protein, a common adaptor protein that is fundamental in
the innate immune response activation (12). MyD88 has an intermediate domain
that is crucial in TLR signaling as it interacts with interleukin-1
receptor-associated kinase. It has been reported that a
polymorphism in the 3′-untranslated region (UTR) of MyD88
(rs7744, *1244 A>G) is associated with susceptibility to Buerger
disease in Japanese individuals (13). MyD88 is located at
chromosome 3p22 and its length is ~4.5 kbp. There is no other
polymorphism excluding rs7744, with P>0.01 for Hardy-Weinberg
equilibrium and P>0.05 for the minor allele frequency, within 10
kbp around MyD88 and no polymorphism in LD with rs7744 is observed
within 20 kbp in the haplotype map of Japanese individuals
(HapMap-JPT). In addition, there are binding sites of several
microRNAs near *1244 A>G, including miR-29b-2-5p, miR-150-3p and
miR-1236. Polymorphisms near the miRNA binding site in mRNA 3′-UTR
may affect the binding of miRNAs to mRNA (14). Based on these results, we
hypothesized that rs7744 has an impact on the MyD88-dependent
pathway of the innate immune response via TLRs. It has been
indicated that the TLR-4-MyD88 pathway may play a significant role
on the pathogenesis of UC (15).
In the present study, the association of the MyD88
polymorphism rs7744 (*1244 A>G) with its susceptibility to UC
was investigated.
Materials and methods
Clinical samples
The studied population comprised 922 subjects,
including patients with UC (UC cases, n=200), who were enrolled at
the Fujita Health University Hospital or Kanazawa Medical
University Hospital (Kutsukake-cho or Uchinada-machi, Japan), and
subjects without UC (controls, n=722). The diagnosis of UC was
based on standard clinical, endoscopic, radiological and
histological criteria (16). The
control subjects had no lower abdominal symptoms, diarrhea or
hematochezia. Genomic DNA was isolated from peripheral blood using
a FlexiGene DNA kit (QIAGEN GmbH, Hilden, Germany).
The Ethics Committees of Fujita Health University
and Kanazawa Medical University approved the protocol, and written
informed consent was obtained from the participating subjects.
Sample size
First, we assessed the genotype of 200 UC cases.
Based on the frequency of the rs7744 minor allele in the UC cases
(43.5%) in the present study an assumption was made that a 20%
decrease in the prevalence of an allelic frequency would be of
clinical relevance. Assuming α=0.05 and power=0.80, at least 200 UC
cases and 400 controls would be sufficient to identify a clinically
relevant difference. Accordingly, 750 subjects without UC would be
of sufficiently clinical relevance for the study. A total of 722 of
750 subjects whose genotype was clearly determined were included as
controls.
Classification
According to their clinical courses, the UC cases
were classified into continuous and non-continuous disease
(relapsing and only one episode) (16). UC patients were also classified as
total colitis or non-total colitis (left sided, distal colitis and
proctitis) according to the location and extension of the
inflammatory lesions judged by the endoscopic findings. The cases
that need continuous intravenous or oral steroid therapy were
identified as steroid-dependent, and those that had one onset over
6 months or 2 onsets within one year were defined as refractory
cases.
Genotyping of polymorphisms
Polymorphism was genotyped by the PCR single-strand
conformation polymorphism (SSCP) method as reported previously
(17,18). The primers used to detect rs7744
A>G were: MyD88 forward, 5′-ccttttctcttgtgt ccctactcattg-3′ and
reverse, 5′-cagctctcttcctctctctgtgcttc-3′. A PCR reaction was
carried out in a volume of 20 μl containing 0.1 μg genomic DNA. The
DNA was denatured at 95°C for 3 min, followed by 35 cycles at 95°C
for 30 sec, 52°C for 40 sec and 72°C for 45 sec, with a final
extension at 72°C for 5 min. Thereafter, 2 μl PCR product was
denatured with 10 μl formamide (Sigma-Aldrich Co., St. Louis, MO,
USA) at 90°C for 5 min. SSCP was carried out at 18°C using a
GenePhor DNA separation system with GeneGel Excel 12.5/24 (GE
Healthcare Japan, Tokyo, Japan), after which the denatured
single-strand DNA bands were detected using a DNA silver staining
kit (Amersham Biosciences Corp.).
Statistical analysis
The data of the age was expressed as the mean ± SD.
The mean ages between the two groups were compared by the Student’s
t-test. Allelic and genotype frequencies were calculated by direct
counting. The allele counts and distribution of genotype were
compared between the cases and the controls by a 2×2 table using
Fisher’s exact test. Furthermore, the strength of the association
between allele frequencies and the disease was assessed by
calculating the odds ratio (OR) and 95% confidence intervals (CIs)
by logistic multivariate regression analysis. For all the analyses,
P<0.05 was used to indicate a statistically significant
difference.
Results
Characteristics of subjects and the
frequencies of genotypes
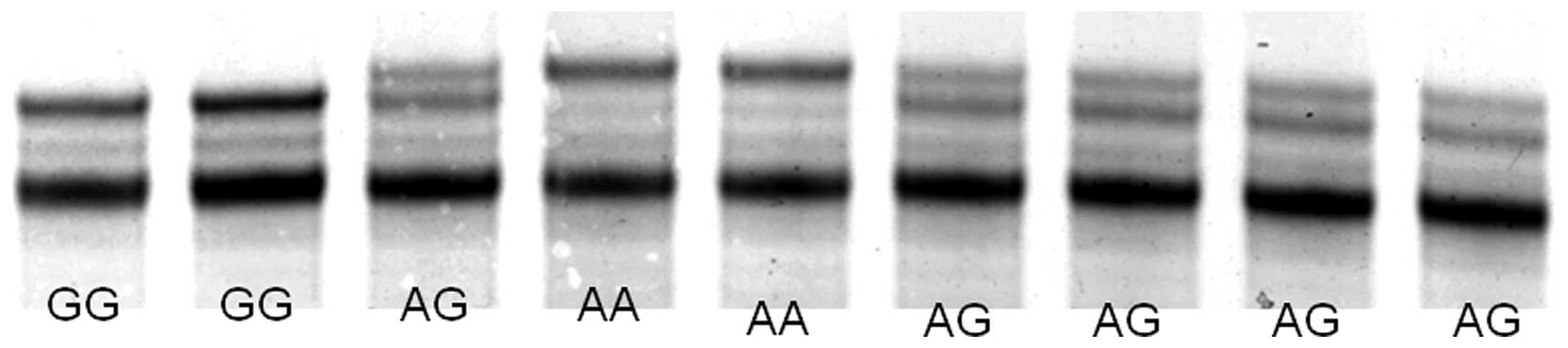
As shown in Fig. 1,
single-stranded DNAs were clearly separated by SSCP. The allele
frequency of MyD88 rs7744 in the controls was in
Hardy-Weinberg equilibrium (P=0.31). The mean age in the controls
was significantly higher compared with that in the UC cases
(Table I). The genotype frequency
of the rs7744 AA homozygote was significantly lower and that of the
GG homozygote was significantly higher in the UC cases compared
with the controls (P=0.0012 and 0.0012, respectively). The minor
allele frequency of rs7744 was 32.8 and 43.5% in the controls and
UC cases, respectively (P<0.0001).
 | Table ICharacteristics of the subjects and
allelic frequency. |
Table I
Characteristics of the subjects and
allelic frequency.
| Characteristics | Controls | UC cases | P-value |
|---|
| Number of
samples | 722 | 200 | |
| Mean age ± SD (age of
onset) | 54.5±16.4 | 40.0±13.6
(32.7±13.3) | P<0.0001 |
| Male:female | 424:298 | 113:87 | NS |
| rs7744 A>G |
| AA | 320 | 63 | P=0.0012 |
| AG | 331 | 100 | |
| GG | 71 | 37 | P=0.0012 |
| G allele
freqency | 32.8% | 43.5% | P<0.0001 |
Association between the rs7744 and
UC
The rs7744 minor allele was significantly associated
with an increased risk for UC using the dominant and recessive
genetic models (OR, 1.64; 95% CIs, 1.15–2.34; P=0.0063 and OR,
1.87; 95% CIs, 1.17–3.00 and P=0.0094, respectively, Table II). In the analysis using the
number of minor alleles as a co-variate, the significant
association of the rs7744 minor allele was also observed (OR, 1.52;
95% CIs, 1.18–1.95 and P=0.0010). This association was observed in
the male subjects in the same manner (dominant genetic model: OR,
1.90; 95% CIs, 1.19–3.04 and P=0.0071, recessive genetic model: OR,
1.93; 95% CIs, 1.01–3.68 and P=0.045, and the number of minor
allele: OR, 1.66; 95% CIs, 1.19–2.33 and P=0.0029), but was not
observed in the female individuals.
 | Table IIAssociation between rs7744 and UC. |
Table II
Association between rs7744 and UC.
| Variables | Genotype, n | AG + GG vs. AA | GG vs. AA + AG | No. of G
allelea |
|---|
|
|
|
|
|
|---|
| AA | AG | GG | OR (95% CI) | OR (95% CI) | OR (95% CI) |
|---|
| Overall |
| Controls (722) | 320 | 331 | 71 | Reference | Reference | Reference |
| UC cases (200) | 63 | 100 | 37 | 1.64
(1.15–2.34)b | 1.87
(1.17–3.00)c | 1.52
(1.18–1.95)d |
| Male |
| Controls (424) | 191 | 198 | 35 | Reference | Reference | Reference |
| UC cases (113) | 33 | 61 | 19 | 1.90
(1.19–3.04)e | 1.93
(1.01–3.68)f | 1.66
(1.19–2.33)g |
| Female |
| Controls
(298) | 129 | 133 | 36 | Reference | Reference | Reference |
| UC cases (87) | 30 | 39 | 18 | 1.35
(0.78–2.34) | 1.83
(0.90–3.72) | 1.37
(0.931–2.00) |
Association between the rs7744 and
phenotypes of UC
The rs7744 polymorphism was associated with UC cases
with onset after 31 years of age by all the genetic models
(Table III). In addition, this
polymorphism was significantly associated with non-continuous
disease and non-total colitis by all the genetic models. In the
recessive genetic model, the rs7744 polymorphism was associated
with non-total and total colitis. On the other hand, this
association of rs7744 appeared not to have a difference because of
disease severity or hospitalization. Moreover, the rs7744
polymorphism was significantly associated with steroid-dependent or
-unused cases in all the genetic models, whereas this polymorphism
was associated with steroid-dependent cases only in the recessive
genetic model.
 | Table IIIAssociation between rs7744 and the
phenotype of UC. |
Table III
Association between rs7744 and the
phenotype of UC.
| Genotype, n | AG + GG vs. AA | GG vs. AA + AG | No. of G
allele |
|---|
|
|
|
|
|
|---|
| Variables | AA | AG | GG | OR (95% CI) | OR (95% CI) | OR (95% CI) |
|---|
| Controls (722) | 320 | 331 | 71 | Reference | Reference | Reference |
| Age of onset |
| ≤30 (92) | 33 | 42 | 17 | 1.19
(0.717–1.98) | 1.51
(0.766–2.96) | 1.22
(0.852–1.74) |
| ≥31 (93) | 27 | 49 | 17 | 1.90
(1.18–3.05)a | 2.03
(1.13–3.66)b | 1.67
(1.21–2.31)c |
| Clinical type |
| Not continuous
(111) | 31 | 58 | 22 | 1.91
(1.21–3.02)d | 2.02
(1.15–3.54)e | 1.67
(1.23–2.28)f |
| Continuous
(82) | 31 | 38 | 13 | 1.23
(0.754–2.00) | 1.58
(0.807–3.11) | 1.25
(0.881–1.79) |
| Extension |
| Non-total colitis
(108) | 31 | 57 | 20 | 1.82
(1.15–2.88)g | 1.89
(1.06–3.38)h | 1.61
(1.17–2.20)i |
| Total colitis
(89) | 29 | 43 | 17 | 1.55
(0.955–2.52) | 1.92
(1.04–3.56)j | 1.49
(1.06–2.09)k |
| Max. UCDAI
score |
| ≤8 (105) | 32 | 56 | 17 | 1.74
(1.11–2.74)l | 1.68
(0.927–3.06) | 1.52
(1.11–2.09)m |
| ≥9 (88) | 30 | 40 | 18 | 1.37
(0.838–2.25) | 2.01
(1.07–3.75)n | 1.42
(1.01–2.01)o |
|
Hospitalization |
| None (121) | 35 | 67 | 19 | 1.87
(1.21–2.88)p | 1.63
(0.919–2.90) | 1.57
(1.16–2.12)q |
| More than once
(69) | 26 | 29 | 14 | 1.19
(0.696–2.03) | 2.05
(1.04–4.04)r | 1.33
(0.912–1.95) |
| Response to
treatment |
| Steroid-dependent
(42) | 17 | 14 | 11 | 1.03
(0.532–1.98) | 2.71
(1.25–5.85)s | 1.37
(0.864–2.18) |
| Steroid-refractory
(56) | 18 | 27 | 11 | 1.46
(0.796–2.67) | 1.88
(0.886–3.97) | 1.44
(0.948–2.19) |
| Steroid
independent or unused (155) | 41 | 70 | 24 | 1.76
(1.17–2.66)t | 1.85
(1.08–3.16)u | 1.57
(1.18–2.09)v |
Discussion
In the present study, the association between
polymorphism rs7744 (*1244 A>G) in the 3′-UTR of MyD88
and the risk for development of UC was evaluated. The rs7744 minor
allele variant was significantly associated with an increased risk
for UC, in particular in the male subjects. In addition, this
polymorphism was associated with cases with an older age of onset,
non-continuous disease and steroid-independent or -unused cases in
the dominant and recessive genetic models. On the other hand, the
rs7744 GA + GG genotype was associated with cases without
hospitalization and cases with a UCDAI score <8, whereas the GG
homozygote was associated with cases with hospitalization, with a
UDCAI score >9 and steroid-dependent cases. These results
indicate that the rs7744 minor allele variant may be associated
with the development of UC and the minor allele homozygote with the
severity of UC. In the present study, sample selection may have
affected the outcome as the controls included unhealthy individuals
who presented at hospital for treatment for problems excluding
diarrhea, bloody feces and lower abdominal discomfort. Although a
comparison of allele frequencies between the UC cases and controls
have an adequately statistical power (1-β=0.971), the effect of
type II error cannot be excluded by relatively small sample sizes
in other comparisons. Another limitation of the present study was
that the mean age was different among the controls and UC cases.
Therefore, an analyis was performed using a logistic multivariate
regression analysis following adjustment for age. However, it
appears that this is not an obstacle to the analysis, as UC
develops at a relatively young age.
It is well known that a major inducer of the
inflammation response to Gram-negative bacteria is
lipopolysaccharide, derived from the outer envelopes of these
microorganisms (19).
Lipopolysaccharide signaling is mainly mediated through the cell
surface TLRs, which have been shown to be extremely important to
gut homeostasis during host-microbial interactions (20,21).
TLR-4 plays a significant role in gut innate immunity, protection
and is involved in human inflammatory bowel diseases (IBDs),
including UC (22,23). Cantó et al reported that a
marked increase of TNF-α response to TLR2 ligands correlated with a
higher TLR2 expression in Crohn’s disease and UC patients,
indicating that an abnormal mechanism may provide an excess of
inflammatory mediators during the active phases of IBDs (24). This TLR signaling acts through a
downstream regulator, MyD88, which initiates a signal transduction
cascade leading to the induction of NF-κB (12). Although TLRs signal transmit
through the MyD88-dependent or -independent pathway, previous
studies have indicated a significant role of the TLR-MyD88 pathway
on IBD (15,25). Aoyagi et al have reported
that mRNA levels of MyD88, TLR-4 and NF-κB p65 are significantly
increased in colonic mucosa of UC patients (26). It has also been reported that the
Myd88 protein level is increased in the colonic mucosa of
non-treated UC and this increase is suppressed by azathioprine
treatment (15). Therefore, MyD88
is considered one of the key molecules in the pathogenesis of UC
and an overexpression of MyD88 may promote the development of
UC.
MyD88 is located on chromosome 3p22, a
position that is not included in UC susceptible loci (27). However, rs7744, which is the only
polymorphism in MyD88, may be associated with the
susceptibility or pathogenesis of UC, if the expression and
function of MyD88 undergo certain effects by the polymorphisms in
MyD88. To the best of our knowledge, there is only one study
regarding the investigation of an association between MyD88
polymorphism and human disorders. Although Chen et al have
reported that rs7744 is associated with Buerger disease in Japanese
individuals (131 cases and 270 controls) (13), the function of rs7744 has not been
identified. In addition, there is no evidence regarding the
function of rs7744. However, there may be a possibility of
overexpressing the MyD88 protein in the rs7744 minor allele
variants, considering overexpression of mRNA and protein levels of
MyD88 in UC patients (15,26). There are binding sites of
miR-150-3p and miR-1236 of 100 bp around rs7744. miRNAs bind to the
RNA-induced silencing complex, making it non-symmetrical which then
binds to the target mRNA to regulate its expression (28). In the rs7744 minor allele variants,
cleavage of mRNA or repression of protein synthesis by miRNAs may
not be well-regulated.
In the present study, MyD88 rs7744 was
associated with non-continuous phenotypes of UC and steroid
unused/independent UC. In addition, this minor allele homozygote
was associated with the disease severity of UC, hospitalization and
response to steroid treatment. Potter et al have reported
that rs7744 is nominally associated with a response to anti-TNF
therapy for rheumatoid arthritis (29). This evidence indicates that rs7744
may be associated with a response to treatment for UC. The reason
for this genotype being associated with specific phenotypes or the
male cases of UC remains to be determined. UC is a multi-factorial
disorder including genetic and environmental factors, and is
considered a complex genetic disorder predicted to involve multiple
genes of relatively low penetrance (30). In fact, Fisher et al have
reported that several regions of the male-specific linkage were
identified in the susceptibility to inflammatory bowel disease
(31). It may not be surprising
that the MyD88 polymorphism is more closely associated with
the specific phenotypes of UC. Future studies should be conducted
to clarify how the MyD88 polymorphism affects the
susceptibility to UC.
In conclusion, the minor allele variant of rs7744,
which is located in MyD88 3′-UTR, is significantly
associated with the susceptibility to UC, in particular in the
Japanese male subjects. This polymorphism may also be associated
with the response to treatment for UC.
References
|
1
|
Podolsky DK: Inflammatory bowel disease. N
Engl J Med. 347:417–429. 2002. View Article : Google Scholar
|
|
2
|
Chamaillard M, Philpott D, Girardin SE, et
al: Gene-environment interaction modulated by allelic heterogeneity
in inflammatory diseases. Proc Natl Acad Sci USA. 100:3455–3460.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakamoto N, Kono S, Wakai K, et al;
Epidemiology Group of the Research Committee on Inflammatory Bowel
Disease in Japan. Dietary risk factors for inflammatory bowel
disease: a multicenter case-control study in Japan. Inflamm Bowel
Dis. 11:154–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Head KA and Jurenka JS: Inflammatory bowel
disease part 1. Ulcerative colitis - pathophysiology and
conventional and alternative treatment options. Altern Med Rev.
8:247–283. 2003.PubMed/NCBI
|
|
5
|
Lakatos PL, Fischer S, Lakatos L, Gal I
and Papp J: Current concept on the pathogenesis of inflammatory
bowel disease-crosstalk between genetic and microbial factors:
pathogenic bacteria and altered bacterial sensing or changes in
mucosal integrity take ‘toll’? World J Gastroenterol. 12:1829–1841.
2006.
|
|
6
|
Gersemann M, Wehkamp J and Stange EF:
Innate immune dysfunction in inflammatory bowel disease. J Intern
Med. 271:421–428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
UK IBD Genetics Consortium. Barrett JC,
Lee JC, Lees CW, et al: Genomewide association study of ulcerative
colitis identifies three new susceptibility loci, including the
HNF4A region. Nat Genet. 41:1330–1334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Silverberg MS, Cho JH, Rioux JD, et al:
Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by
genome-wide association study. Nat Genet. 41:216–220. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGovern DP, Gardet A, Törkvist L, et al:
Genome-wide association identifies multiple ulcerative colitis
susceptibility loci. Nat Genet. 42:332–337. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Franke A, Balschun T, Sina C, et al:
Genome-wide association study for ulcerative colitis identifies
risk loci at 7q22 and 22q13 (IL17REL). Nat Genet. 42:292–294. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fukata M and Abreu MT: TLR4 signaling in
the intestine in health and disease. Biochem Soc Trans.
35:1473–1478. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arancibia SA, Béltran CJ, Aguirre IM, et
al: Toll-like receptors are key participants in innate immune
responses. Biol Res. 40:97–112. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Z, Nakajima T, Inoue Y, et al: A
single nucleotide polymorphism in the 3′-untranslated region of
MyD88 gene is associated with Buerger disease but not with Takayasu
arteritis in Japanese. J Hum Genet. 56:545–547. 2011.
|
|
14
|
Mishra PJ and Bertino JR: MicroRNA
polymorphisms: the future of pharmacogenomics, molecular
epidemiology and individualized medicine. Pharmacogenomics.
10:399–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siddique I and Khan I: Mechanism of
regulation of Na-H Exchanger in inflammatory bowel disease: role of
TLR-4 signaling mechanism. Dig Dis Sci. 56:1656–1662. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Podolsky DK: Inflammatory bowel disease
(2). N Engl J Med. 325:1008–1016. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arisawa T, Tahara T, Shibata T, et al: A
polymorphism of microRNA 27a genome region is associated with the
development of gastric mucosal atrophy in Japanese male subjects.
Dig Dis Sci. 52:1691–1697. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arisawa T, Tahara T, Shiroeda H, et al:
NFKB1 polymorphism is associated with age-related gene methylation
in Helicobacter pylori-infected subjects. Int J Mol Med.
30:255–262. 2012.PubMed/NCBI
|
|
19
|
Ruemmele FM, Beaulieu JF, Dionne S, et al:
Lipopolysaccharide modulation of normal enterocyte turnover by
toll-like receptors is mediated by endogenously produced tumour
necrosis factor alpha. Gut. 51:842–848. 2002. View Article : Google Scholar
|
|
20
|
Boone DL and Ma A: Connecting the dots
from Toll-like receptors to innate immune cells and inflammatory
bowel disease. J Clin Invest. 111:1284–1286. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pierik M, Joossens S, Van Steen K, et al:
Toll-like receptor-1, -2, and -6 polymorphisms influence disease
extension in inflammatory bowel diseases. Inflamm Bowel Dis.
12:1–8. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lubbad A, Oriowo MA and Khan I: Curcumin
attenuates inflammation through inhibition of TLR-4 receptor in
experimental colitis. Mol Cell Biochem. 322:127–135. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sepúlveda SE, Beltrán CJ, Peralta A, et
al: Inflammatory bowel diseases: an immunological approach. Rev Med
Chil. 136:367–375. 2008.(In Spanish).
|
|
24
|
Cantó E, Ricart E, Monfort D, et al: TNF
alpha production to TLR2 ligands in active IBD patients. Clin
Immunol. 119:156–165. 2006.PubMed/NCBI
|
|
25
|
Tomita T, Kanai T, Fujii T, et al:
MyD88-dependent pathway in T cells directly modulates the expansion
of colitogenic CD4+ T cells in chronic colitis. J
Immunol. 180:5291–5299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aoyagi Y, Nagata S, Kudo T, et al:
Peroxisome proliferator- activated receptor γ 2 mutation may cause
a subset of ulcerative colitis. Pediatr Int. 52:729–734. 2010.
|
|
27
|
Hugot JP and Thomas G: Genome-wide
scanning in inflammatory bowel diseases. Dig Dis. 16:364–369. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Potter C, Cordell HJ, Barton A, et al;
Biologics in Rheumatoid Arthritis Genetics and Genomics Study
Syndicate (BRAGGSS). Association between anti-tumour necrosis
factor treatment response and genetic variants within the TLR and
NF{kappa}B signalling pathways. Ann Rheum Dis. 69:1315–1320.
2010.PubMed/NCBI
|
|
30
|
Cho JH and Brant SR: Genetics and genetic
markers in inflammatory bowel disease. Curr Opin Gastroenterol.
14:283–288. 1998. View Article : Google Scholar
|
|
31
|
Fisher SA, Hampe J, Macpherson AJ, et al:
Sex stratification of an inflammatory bowel disease genome search
shows male-specific linkage to the HLA region of chromosome 6. Eur
J Hum Genet. 10:259–265. 2002. View Article : Google Scholar : PubMed/NCBI
|