Introduction
Lung cancer is the second most common cause of
cancer-related mortality worldwide in males and females (1). Non-small cell lung carcinoma (NSCLC),
which includes adenocarcinoma, squamous cell carcinoma and large
cell carcinoma accounts for >80% of lung cancer (2,3). The
majority of patients presenting with NSCLC have advanced disease,
which precludes curative treatment due to a lack of early diagnosis
detection (4). Early detection and
treatment may result in the identification of more patients with
early central lung cancer and improve survival rates (1). Thus, it is crucial to develop novel
molecular diagnostic markers and therapeutic targets for the
treatment of NSCLC.
A disintegrin and metalloprotease (ADAM) 17 is a
member of the disintegrin and metalloprotease gene family. This
family encodes proteins that mediate cellular responses to
environmental stress by interacting with a variety of cell surface
proteins and regulating diverse cellular processes, including
proliferation, extracellular matrix binding and ectodomain shedding
(5–7). Previously, ADAM17 has been analyzed
in various tumor entities and was revealed to be differentially
expressed, partially conveying prognostic information (8,9).
Previous studies demonstrated that ADAM17 is highly expressed in
NSCLC and highly expressed ADAM17 correlates with shortened
survival time, suggesting that ADAM17 may also become a useful
predictive biomarker for the selection of adjuvant chemotherapy
treatment of NSCLC (10). However,
little is known about whether the downregulation of ADAM17 effects
cell proliferation, cell invasion and the cell cycle in NSCLC, and
its underlying molecular mechanisms. Thus, the present study
assessed the feasibility of lentiviral vector-delivered small
hairpin RNA (shRNA) against ADAM17 for the treatment of NSCLC in
vitro and in vivo.
Materials and methods
Cell culture
The A549 human non-small cell lung cancer cell line
was obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, Shanghai Institute of Cell Biology
(Shanghai, China). A549 lung tumor cells were cultured in
Dulbecco’s modified Eagle’s medium containing 10% fetal bovine
serum (FBS), glutamine (2 mM), penicillin (100 U/ml), streptomycin
(100 mg/ml) and amphotericin B (250 mg/ml; all from Invitrogen Life
Technologies, Carlsbad, CA, USA) in 5% CO2 at 37°C.
Construction and transfection of
pGCSIL-GFP-ADAM17
In order to inhibit the expression of ADAM17, an
shRNA targeting the ADAM17 transcript was designed. The synthesized
oligonucleotides containing a specific target sequence, a loop, the
reverse complement of the target sequence, a stop codon for the U6
promoter and two sticky ends were cloned into the pGCSIL-GFP
lentivirus vector according to the manufacturer’s instructions
(Shanghai Gene Chem Co. Ltd., Shanghai, China). The target sequence
of ADAM17 in the oligonucleotide for suppressing shRNA was: Sense:
5′-GTGCCAGGAGGCGATTAAT-3′. The sequence for the scrambled negative
control (NC) for siRNA was: Sense: 5-′AATTCTCCGAACGTGTCACGT-3′.
This sequence did not target any gene product and had no
significant sequence similarity to human gene sequences, which is
essential for determining the effects of siRNA delivery. The
lentivirus carrying the ADAM17 siRNA was infected into A549 lung
cancer cells as previously described (11).
Quantitative polymerase chain reaction
(qPCR)
Total RNA was isolated from A549 cells using TRIzol
reagent (Invitrogen Life Technologies) according to the
manufacturer’s instructions. RNA was reverse-transcribed into cDNA
by a PrimeScript™ RT reagent kit according to the manufacturer’s
instructions (Takara Bio Inc., Shiga, Japan). qPCR was conducted
using the SYBR-Green fluorescent dye method and Rotor Gene 3000
real-time PCR apparatus (Qiagen, Hilden, Germany). The primer
sequences were as follows: Forward: 5′-ACTCTGAGGACAGTTAACCAAACC-3′
and reverse: 5′-AGTAAAAGGAGCCAATACCACAAG-3′ for ADAM17; and
forward: 5′-GATCATTGCTCCTCCTGAGC-3′ and reverse:
5′-ACTCCTGCTTGCTGATCCAC-3′ for β-actin. The PCR conditions were as
follows: A pre-denaturing at 95°C for 2 min, followed by 40 cycles
of denaturation at 95°C for 10 sec and annealing/extension at 55°C
for 20 sec. The amplification specificity was checked by melting
curve analysis. The 2−ΔΔCt method was used to calculate
the relative abundance of target gene expression generated by
Rotor-Gene Real-Time Analysis Software 6.1.81 (Qiagen). For each
cDNA, the target gene mRNA level was normalized to the β-actin mRNA
level.
Western blot analysis
Cultured cells were washed twice with
phosphate-buffered saline (PBS), then cells were lysed with Triton
X-100 in Hepes buffer [150 mM NaCl, 50 mM Hepes, 1.5 mM
MgCl2, 1% Triton X-100, 0.1% SDS and a protease
inhibitor cocktail (Sigma, St. Louis, MO, USA), 100 mM NaF and 100
mM Na3VO4] for 30 min. Cell lysates were
clarified by centrifugation (10,000 × g for 15 min), and protein
concentrations were determined using the Bradford reagent (Sigma).
The proteins were transferred onto nitrocellulose membranes
(Millipore, Bedford, MA, USA) incubated with specific primary
antibodies and then the corresponding horseradish
peroxidase-conjugated secondary antibodies were added. The specific
antibodies used in the western blot analysis were as follows:
Antibodies against ADAM17, β-actin, p21, cyclin D1 and cyclin D3
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and matrix
metalloproteinase (MMP)-9 and MMP-2 (Sigma). The secondary
antibodies used for immunodetection were as follows: Horseradish
peroxidase-conjugated goat anti-mouse IgG and goat anti-rabbit IgG
(Amersham Biosciences, Uppsala, Sweden). All immunoblots were
visualized by enhanced chemiluminescence (Pierce Biotechnology,
Inc., Rockford, IL, USA). All the assays using ADAM17 knockdown in
A549 cells were performed following the third day of siRNA
transfections.
Proliferation assays
To measure the effect of ADAM17 silencing on cell
proliferation, the transition of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
to formazan was used. Cells without any treatment, cells treated
only with negative siRNA and cells transfected with siRNAs were
seeded in 96-well plates and kept at 37°C for 24, 48 and 72 h. The
cells were then washed with PBS and incubated in 50 μl of 0.5 mg/ml
MTT in culture medium at 37°C for 4 h. Following the addition of
100 μl isopropanol, the absorbance was read at 595 nm by an ELISA
plate reader (Molecular Devices Corp., Sunnyvale, CA, USA). The
mean proliferation of cells without any treatment was expressed as
100%.
Cell cycle analysis
A549 cells were treated with siRNA for 24 h in DMEM
media containing 5% FBS. All cells were collected and
1×106 cells were centrifuged 700 × g for 5 min,
resuspended in ice-cold 70% ethanol and stored at −20°C until
analysis. Washed cells were stained with 0.1% Triton X-100 in 0.01
M PBS with 50 μg/ml propidium iodide (Sigma) and 1 mg/ml RNase A
(Invitrogen Life Technologies), and incubated at 37°C for 30 min in
the dark. The samples of cells were then analyzed for DNA content
by FACScan flow cytometry (Beckman Coulter, Inc., Miami, FL, USA)
and cell cycle phase distributions was analyzed with the Cell Quest
acquisition software (BD Biosciences, Franklin Lakes, NJ, USA).
Duplicates were performed in all experiments and experiments were
performed on three occasions.
Cell migration assay
The migration assay was performed using a 12-well
Boyden Chamber (Neuro Probe, Gaithersburg, MD, USA) with an 8-μm
pore size. Approximately 1×105 cells were seeded into
the upper wells of the Boyden Chamber and incubated for 6 h at 37°C
in medium containing 1% FBS. DMEM medium with 10% FBS was used as a
chemoattractant in the bottom wells. Cells that did not migrate
through the pores of the Boyden chamber were manually removed with
a rubber swab. Cells that migrated to the lower side of the
membrane were stained with hematoxylin and eosin and images were
captured using an inverted microscope (Olympus, Tokyo, Japan).
Transwell invasion assay
The invasiveness of A549 cancer cells was assessed
using 24-well Transwell plates (Corning Inc., Lowell, MA, USA). In
brief, 2×105 cells in DMEM media with 0.5% FBS were
added to the upper chamber containing 8-mm pore polycarbonate
coated with 1 mg/ml matrigel. The lower chamber was filled with
media containing 5% FBS. Following 16 h of incubation, the upper
surface of the membrane was scrubbed with a cotton-tipped swab. The
invading cells on the lower surface of the membrane were fixed and
stained with 0.5% crystal violet. Images of the random fields (five
per membrane) were captured at ×40 magnification for calculating
the cell number. In addition, cells were quantified by measuring
the absorbance of dye extracts at 570 nm in 100 ml Sorenson’s
solution (9 mg trisodium citrate, 305 ml distilled water, 195 ml
0.1 NHCl and 500 ml of 90% ethanol). Triplicates were performed in
all experiments and experiments were performed on five
occasions.
Zymographic analysis
MMP-2 and MMP-9 enzymatic activities were measured
by gelatin zymography. Conditioned media from cells (cultured
without serum for 48 h) were collected and concentrated 20-fold
using a Centriprep YM-30 device (Millipore). Samples were then
mixed with Laemmli loading buffer and electrophoresed on a gelatin
containing 10% SDS-PAGE. Following electrophoresis, the gel was
washed twice with wash buffer (50 mM Tris-HCl, pH 7.5; 100 mM NaCl
and 2.5% Triton X-100), followed by a brief rinse in wash buffer
without Triton X-100. The gel was then placed in incubation buffer
(50 mM Tris-HCl, pH 7.5; 150 mM NaCl, 10 mM CaCl2, 0.02%
NaN3, 1 M ZnCl2) at 37°C for 24 h. The gel
was then stained with Coomassie Blue R-250 and destained with
destaining solution. A clear zone of gelatin digestion was
representative of the MMP activity.
Tumor xenograft model
All animal experiments were performed in accordance
with institutional guidelines, following a protocol approved by the
Ethics Committees of the Disease Model Research Center, The First
Hospital of Jilin University (Changchun, China). Male, nude mice
(age, ~6 -weeks) were maintained under specific pathogen-free (SPF)
conditions and provided with food and water ad libitum. All
the animals were fed with a normal pellet diet one week prior to
the experimentation. In vitro cultured A549 cells were
injected subcutaneously into mice. The tumor volume was calculated
using the following formula: Volume = length × width2 /
2. When tumors grew to an average volume of 75 mm3, the
mice were randomly divided into the siRNA group, control group
(untreated group), NC group (n=10 in each group) and treated by
administration of siRNA or NC plus PBS in a total volume of 20 μl
(10 μl of the virus plus 10 μl of PBS) once a week for 21 days,
respectively. When control mice started to succumb to their tumors,
mice in all treatment groups were anesthetized by sodium
pentobarbital (1.25 g/kg) after overnight fasting and decapitated..
The tumors were then removed and directly embedded in an optimal
cutting temperature compound in a deep freezer at −80°C.
Statistical analysis
Statistical analysis between two samples was
performed using Student’s t-test. Statistical comparison of more
than two groups was performed using one-way analysis of variance
followed by Tukey’s post hoc test. All data are expressed as the
mean ± standard error of the mean. The SPSS 13.0 software (SPSS
Inc., Chicago, IL, USA) for Windows was used for statistical
analyses. P<0.05 or P<0.01 were considered to indicate a
statistically significant difference.
Results
ADAM17 silencing is detected at the mRNA
and protein levels
The lentivirus carrying the ADAM17 siRNA or scramble
siRNA were transfected into A549 cells. ADAM17 gene expression was
markedly decreased in A549 cells treated with siRNA at the mRNA and
protein level (Fig. 1) compared
with the controls (untreated cell) and scrambled-treated cells.
qPCR analysis demonstrated a downregulation of ADAM17 expression in
RNAi-mediated knockdown A549 cells when compared with the control
cells and scrambled-treated cells (Fig. 1A). At the protein level, similar
results were obtained by western blot analysis using an anti-ADAM17
antibody (Fig. 1B).
ADAM17 silencing affects A549 cell
proliferation and apoptosis
To investigate whether siRNA affects A549 cell
proliferation and apoptosis, A549 cells were treated with siRNA and
negative siRNA for 24, 48 and 72 h. The antiproliferative and
apoptotic effects of siRNA on A549 cells were examined by MTT and
acridine orange assays, respectively. It was revealed that ADAM17
silencing was able to significantly inhibit the proliferation of
A549 cells when compared with scrambled-treated cells and control
cells (Fig. 2A). In addition, to
determine the effects of A549 cell cycle progression following
ADAM17 silencing, flow cytometry was performed in the present
study. The percentage of cells in the siRNA therapy groups
significantly increased at the G0/G1 phase compared with
scrambled-treated cells and control cells. These results suggest
that ADAM17 silencing is able to induce cell cycle arrest at the
G0/G1 phase in A549 cells (Fig.
2B).
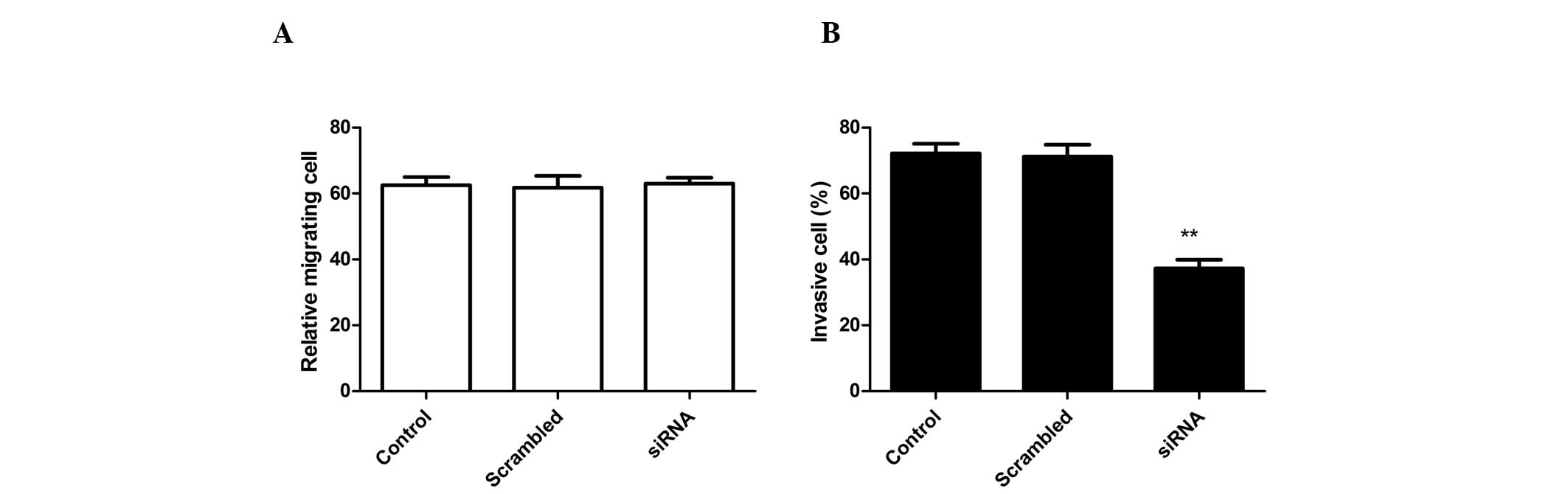
ADAM17 silencing inhibits A549 cell
invasion without affecting cell migration
To analyze whether siRNA effects A549 cell
migration, migration assays were performed using Boyden chambers.
RNA-mediated ADAM17 silencing had no effect on A549 cell migration
when compared with control cells (scrambled-treated cells and
untreated cell; Fig. 3A). By
contrast, RNAi-mediated ADAM17 knockdown in A549 cells markedly
inhibited invasion in vitro when compared with
scrambled-treated cells and control cells (Fig. 3B). Control cells (untransfected
cell) and NC transfected cells remained invasive and no
statistically significant differences were identified.
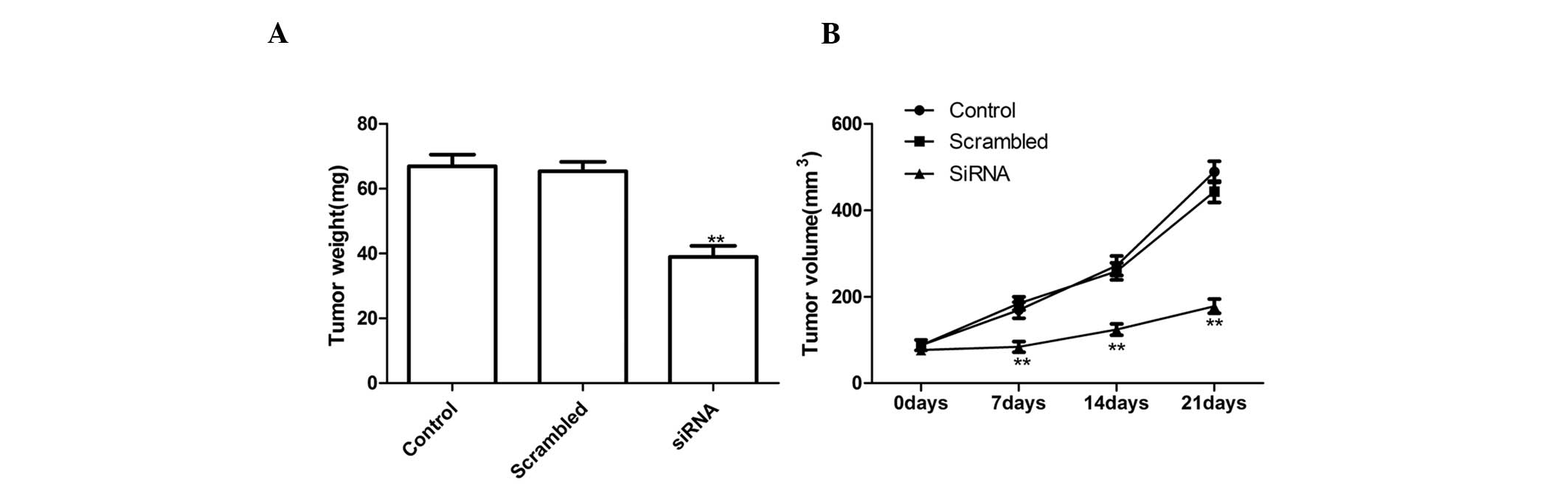
Silencing ADAM17 suppressed tumor growth
in vivo
To investigate the effects of ADAM17 on tumor growth
and metastasis in a nude mouse model, a xenograft assay was
conducted by injecting the NC cells and ADAM17-silenced tumor cells
subcutaneously into mice and comparing the growth rate of the solid
tumors. It was revealed that tumor volume and weight was
significantly slower for ADAM17 tumor cells compared with the
control cells and NC cells following ADAM17 silencing (Fig. 4). These results indicated that the
suppression of ADAM17 expression in lung tumor cells markedly
suppressed their tumorigenicity in mice.
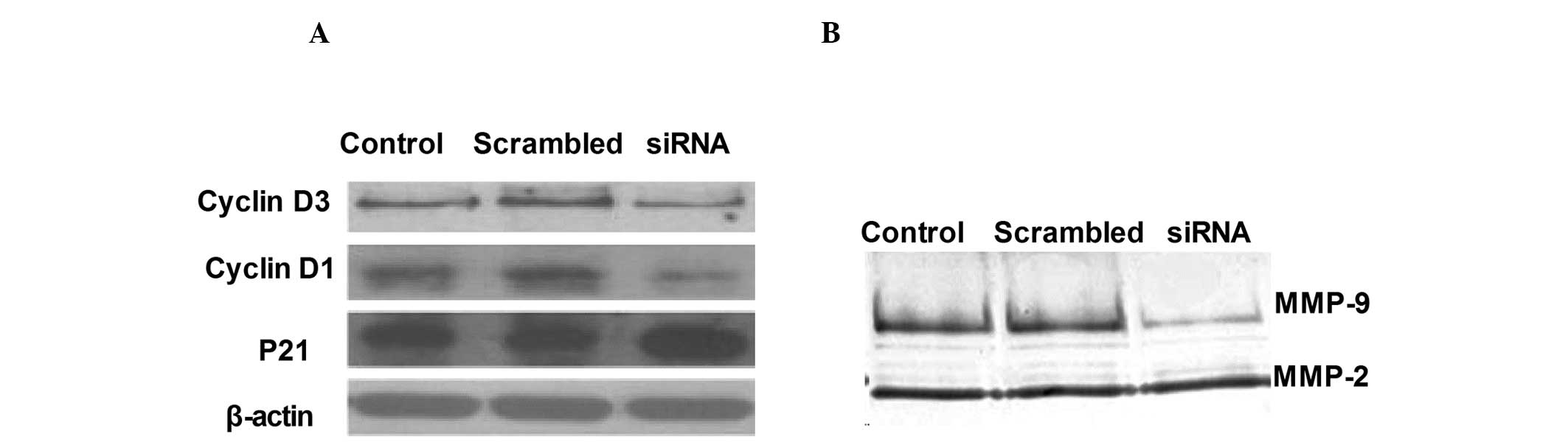
Preliminary mechanisms involved in the
regulation of cell proliferation and invasion by ADAM17
To investigate the preliminary mechanisms involved
in the regulation of cell proliferation and invasion by ADAM17
in vitro, the activation of cyclin D1, cyclin D3 and p21 in
ADAM17 silencing were analyzed by western blot analysis and
compared with that of control cells transfected with empty vectors.
As shown in Fig. 5A, ADAM17
silencing significantly inhibited cyclin D1 and cyclin D3
expression and increased p21 expression in the A549 tumor
cells.
In order to investigate the mechanisms involved in
the inhibition of the invasion ability of RNAi-mediated ADAM17
silencing of A549 cells, zymography assays were performed to
evaluate the activity of MMP-2 and MMP-9. Data demonstrated that
ADAM17 silencing exhibited much lower MMP-9 activity compared with
its control cells as determined by zymography assays (Fig. 5B).
Discussion
ADAMs are known as ectodomain sheddases, a function
of their metalloprotease domains and is a member of the superfamily
of Zn-dependent metalloproteinases (12,13).
ADAMs are therefore important in the remodeling or processing of
cell membrane proteins. Several of the substrates processed by
ADAMs, particularly by ADAM10 and ADAM17, have been implicated in
the pathogenesis or progression of cancer (14,15).
Previous studies have demonstrated that ADAM17 is involved in
invasion and proliferation (16,17).
The results of the present study are in line with this theory and
demonstrate that ADAM17 silencing suppresses A549 tumor
proliferation and invasion in vitro and tumor growth in
vivo. The data further demonstrated that ADAM17 is important in
tumor progression.
ADAM17 sheds a variety of important cell surface
molecules, including cytokines and adhesion molecules (18), which are important in cell
proliferation and invasion. It has been demonstrated that
ADAM17-mediated epidermal growth factor receptor ligand cleavage
enhances the proliferation and survival of squamous cell carcinoma
cells as well as lung cancer cells (19,20).
The present study employed an A549 human lung cancer cell line to
investigate the effect of ADAM17 on lung cancer proliferation, cell
cycle progression and invasion in vitro. The downregulation
of ADAM17 expression was established by siRNA transfection. To
assess A549 cell viability and proliferation, an MTT assay was
employed. The data demonstrated that a reduction of ADAM17 by shRNA
significantly decreased tumor cell proliferation when compared with
the control. In addition, the present study also demonstrated that
ADAM17 knockdown induces cell cycle arrest at the G0/G1 transition
in NSCLC cells. To investigate the exact mechanism, cell
cycle-related proteins were analyzed, after silencing ADAM17, by
western blot analysis. The p21 protein is a widely accepted cell
cycle regulator, as a cyclin dependent kinase (CDK) inhibitor and a
negative regulator in the G1/S transition (21). In the present study, compared with
the control cells transfected with empty vectors, p21 expression
was markedly increased after silencing ADAM17. Whereas, cyclin D1
or cyclin D3 expression decreased following silencing. These data
demonstrated that ADAM17 downregulation was able to effect p21,
cyclin D3 and cyclin D1 levels to reduce lung carcinoma cell
proliferation.
The progression of malignant tumors results from the
invasion of the primary tumor to a secondary site, causing
metastasis in a multistep process that requires cell-cell and
cell-matrix interactions within the host tissue. These interactions
lead to the production, release and activation of a variety of
cytokines and growth factors and subsequent generation of signals
to directly or indirectly promote tumor growth and survival
(22). Different proteases have
been implicated in these processes, including MMPs, ADAMs and a
disintegrin and metalloprotease with thrombospondin motifs
(23,24). Due to the strong involvement of
ADAM17 in the metastatic process, the present study demonstrated
that ADAM17 silencing significantly inhibited the invasion capacity
of A549 human lung cancer cells using a matrigel invasion assay,
which suggests that this protein is important in cell invasion.
Furthermore, it was revealed that ADAM17 silencing was able to
inhibit MMP-2 and MMP-9 expression by a zymographic assay
indicating reduced cell invasion.
In conclusion, the present study demonstrated that
ADAM17 silencing significantly suppressed cell proliferation and
invasion in vitro, and tumor growth in vivo. ADAM17
is an important regulator of the tumorigenic properties of human
NSCLC and may be used as a potential anticancer therapeutic target
in NSCLC. Considering the significance of cell invasion in
metastatic progression, ADAM17 may be a key target for the design
of drugs involved in the treatment or prevention of NSCLC
cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Askoxylakis V, Thieke C, Pleger ST, Most
P, Tanner J, Lindel K, Katus HA, Debus J and Bischof M: Long-term
survival of cancer patients compared to heart failure and stroke: a
systematic review. BMC Cancer. 10:1052010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schaake EE, Kappers I, Codrington HE,
Valdés Olmos RA, Teertstra HJ, van Pel R, Burgers JA, van Tinteren
H and Klomp HM: Tumor response and toxicity of neoadjuvant
erlotinib in patients with early-stage non-small-cell lung cancer.
J Clin Oncol. 30:2731–2738. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shepherd FA, Rodrigues Pereira J, Ciuleanu
T, et al: Erlotinib in previously treated non-small-cell lung
cancer. N Engl J Med. 353:123–132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fischer OM, Hart S, Gschwind A, Prenzel N
and Ullrich A: Oxidative and osmotic stress signaling in tumor
cells is mediated by ADAM proteases and heparin-binding epidermal
growth factor. Mol Cell Biol. 24:5172–5183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mahimkar RM, Visaya O, Pollock AS and
Lovett DH: The disintegrin domain of ADAM9: a ligand for multiple
beta1 renal integrins. Biochem J. 385:461–468. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh B, Schneider M, Knyazev P and
Ullrich A: UV-induced EGFR signal transactivation is dependent on
proligand shedding by activated metalloproteases in skin cancer
cell lines. Int J Cancer. 124:531–539. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kenny PA and Bissell MJ: Targeting
TACE-dependent EGFR ligand shedding in breast cancer. J Clin
Invest. 117:337–345. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGowan PM, Ryan BM, Hill AD, McDermott E,
O’Higgins N and Duffy MJ: ADAM-17 expression in breast cancer
correlates with variables of tumor progression. Clin Cancer Res.
13:2335–2343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ni SS, Zhang J, Zhao WL, Dong XC and Wang
JL: ADAM17 is overexpressed in non-small cell lung cancer and its
expression correlates with poor patient survival. Tumour Biol.
34:1813–1818. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang WY, Chen DH, Ning L and Wang LW:
siRNA mediated silencing of NIN1/RPN12 binding protein 1 homolog
inhibits proliferation and growth of breast cancer cells. Asian Pac
J Cancer Prev. 13:1823–1827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Black RA: Tumor necrosis factor-alpha
converting enzyme. Int J Biochem Cell Biol. 34:1–5. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Primakoff P and Myles DG: The ADAM gene
family: surface proteins with adhesion and protease activity.
Trends Genet. 16:83–87. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duffy MJ, McKiernan E, O’Donovan N and
McGowan P: Role of ADAMs in cancer formation and progression. Clin
Cancer Res. 15:1140–1144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duffy MJ, McKiernan E, O’Donovan N and
McGowan P: The role of ADAMs in disease pathophysiology. Clin Chim
Acta. 403:31–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takamune Y, Ikebe T, Nagano O and
Shinohara M: Involvement of NF-kappaB-mediated maturation of
ADAM-17 in the invasion of oral squamous cell carcinoma. Biochem
Biophys Res Commun. 365:393–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng X, Jiang F, Katakowski M, Zhang ZG,
Lu QE and Chopp M: ADAM17 promotes breast cancer cell malignant
phenotype through EGFR-PI3K-AKT activation. Cancer Biol Ther.
8:1045–1105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding X, Yang LY, Huang GW, Wang W and Lu
WQ: ADAM17 mRNA expression and pathological features of
hepatocellular carcinoma. World J Gastroenterol. 10:2735–2739.
2004.PubMed/NCBI
|
|
19
|
Gschwind A, Hart S, Fischer OM and Ullrich
A: TACE cleavage of proamphiregulin regulates GPCR-induced
proliferation and motility of cancer cells. EMBO J. 22:2411–2421.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hart S, Fischer OM and Ullrich A:
Cannabinoids induce cancer cell proliferation via tumor necrosis
factor α-converting enzyme (TACE/ADAM17)-mediated transactivation
of the epidermal growth factor receptor. Cancer Res. 64:1943–1950.
2004.PubMed/NCBI
|
|
21
|
Niculescu AB III, Chen X, Smeets M, Hengst
L, Prives C and Reed SI: Effects of p21(Cip1/Waf1) at both the G1/S
and the G2/M cell cycle transitions: pRb is a critical determinant
in blocking DNA replication and in preventing endoreduplication.
Mol Cell Biol. 18:629–643. 1998.PubMed/NCBI
|
|
22
|
Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL,
Chang YC, Chiou GY, Chou MY and Chiou SH: miR145 targets the
SOX9/ADAM17 axis to inhibit tumor-initiating cells and
IL-6-mediated paracrine effects in head and neck cancer. Cancer
Res. 73:3425–3440. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mochizuki S and Okada Y: ADAMs in cancer
cell proliferation and progression. Cancer Sci. 98:621–628. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rocks N, Paulissen G, El Hour M, Quesada
F, Crahay C, Gueders M, Foidart JM, Noel A and Cataldo D: Emerging
roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie.
90:369–379. 2008. View Article : Google Scholar : PubMed/NCBI
|