Introduction
Octamer-binding transcription factor 4 (Oct-4), a
member of the family of POU-domain transcription factors, is
expressed in pluripotent embryonic stem (ES) and germ cells
(1,2).
Knocking out the Oct-4 gene in mice causes early lethality due to
the lack of inner cell mass formation, indicating that Oct-4 is
involved in the self-renewal of ES cells (3). Oct-4 activates transcription via the
octamer motif of an AGTCAAAT consensus sequence (4,5), and Oct-4
binding sites have been identified in various genes, including
fibroblast growth factor 4 and platelet-derived growth factor A
receptor (6,7). This suggests that Oct-4 functions as a
master switch during differentiation, by regulating the pluripotent
potential of stem cells, and that it is important during mammalian
development. Other studies have demonstrated that Oct-4 is
expressed in various types of human tumor, including gastric cancer
(8,9),
breast cancer (10), non-small cell
lung carcinoma (11), glioma
(12–14), esophageal squamous cell carcinoma
(15) and certain types of testicular
germ cell tumors (16,17). Furthermore, aberrant expression of
Oct-4 has been shown to be involved in maintaining self-renewal,
and the cancer stem cell-like, and chemoradioresistant properties
of lung cancer (18).
Colorectal cancer (CRC), one of the most common
types of malignant tumors, is the second leading cause of
cancer-related morbidity and mortality (19). Numerous studies have demonstrated that
only a small subpopulation of tumor cells in malignant tissues,
termed cancer stem cells (CSCs) or tumor initiating cancer cells
(TICs), has the capacity to regenerate the original tumor and to
maintain the heterogeneity of tumor tissues in animal models
(20,21). Studies conducted by O'Brien et
al (22) and Ricci-Vitian et
al (23), demonstrated that
CD133+ cells, but not CD133− cells, derived
from human colon carcinomas, initiated tumor development in
immunodeficient mice, and exhibited properties of CSCs. These
results led to novel research approaches aimed at improving
understanding of the development and treatment of CRC. CRC is known
to develop through a stepwise progression from benign polyps to
invasive adenocarcinoma and, ultimately, the occurrence of distant
metastasis (24). However, little is
known regarding the dynamic alteration of stem cells during the
evolution of CRC.
In the present study, the expression of Oct-4 in CRC
tissues, matched non-tumor tissues and benign polyp tissues was
measured, in order to evaluate the correlation between Oct-4
expression and the development of CRC. Clinicopathological analysis
was conducted to assess the association between Oct-4 expression
and certain clinicopathological parameters.
Patients and methods
Patients and specimens
CRC tissues, matched non-tumor tissues and benign
polyp tissues were obtained, which represented different steps in
the evolution of CRC. All specimens used in this study, including
primary tissue specimens and paraffin-embedded tissue specimens
were obtained from the No. 4 People's Hospital of Wuxi City,
Affiliated hospital of Jiangnan University (Wuxi, China)and the
study was approved by the ethic's committee of the same
institution. All patients voluntarily agreed to participate in the
study under the terms proposed by the ethic's committee. None of
the patients received preoperative treatment, such as radiotherapy
or chemotherapy.
Three groups of specimens were used for the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
flow cytometry (FCM) and immunohistochemistry (ICH) analysis that
were undertaken in the study. Primary CRC tissues and matched
non-tumor tissues were obtained from 33 patients with CRC, who were
undergoing radical resection. Benign polyp tissues were obtained
from 33 patients, who underwent endoscopy in 2010. All these
tissues were snap frozen and stored at 80°C, prior to RNA
extraction and RT-qPCR.
Ten further primary cancer tissues and matched
non-tumor tissues from patients with CRC, in addition to benign
polyp tissues, were also obtained in 2010 and cell suspensions were
prepared from these, for analysis by FCM.
A total of 158 paraffin-embedded CRC tissues and
matched non-tumor tissues from patients with CRC undergoing radical
resection, and 71 paraffin-embedded benign polyp tissues removed
from patients, who underwent endoscopy between August 2002 and
September 2003. Specimens were routinely fixed in 10% neutral
formalin and embedded in paraffin. The CRC tissues were obtained
from 158 patients, including 96 males and 62 females; age range,
36–76 years; median, 64 years; mean, 58.4 years. Of these, CRC
samples from 19 were grade 1, 87 were grade 2 and 52 were grade 3,
according to histological grading. In addition, 37 were stage I, 58
were stage II, 54 were stage III and 9 were stage IV, according to
the clinical TNM staging system, revised by the Union for
International Cancer Control in 2009. All patients were followed up
for survival. The follow-up period was defined from the date of
surgery to 30th September 2013, during which time 106 patients
died, while 52 survived (median survival time was 59 months).
Total RNA extraction
Tissue sections were minced with scissors into small
fragments (1–2 mm3) and homogenized with TRIzol™ reagent
(Takara Bio, Inc., Otsu, Japan). Chloroform (200 µl; Sigma-Aldrich,
Santa Clara, CA, USA) was added to the TRIzol homogenate. The
preparations were then centrifuged at 12,000 × g for 15 min at 4°C,
and the upper aqueous layer was transferred to a clean Eppendorf
tube, containing an equal volume of isopropanol (Sigma-Aldrich).
The mixed suspensions were centrifuged at 12,000 × g for a further
15 min at 4°C. The precipitations were then collected. After
washing with 70% ethanol, total RNA was dissolved in RNase-free
water and the quality of RNA was evaluated by gel electrophoresis.
RNA concentrations were measured by optical density (260 nm, Q5000,
Quawell, San Jose, CA, USA) and the preparations stored at −80°C
for subsequent analysis.
RT-qPCR analysis
cDNA was reverse transcribed on the Bio-Rad S1000
Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using
oligo (dT) as primers. Briefly, the total RNA (1 µg) from each
sample was reverse transcribed in a 20 µl reaction volume,
containing 0.5 µg of oligo (dT) and 200 U M-MLV (MBI Fermentas,
Vilnius, Lithuania). All samples were amplified in triplicate under
the following conditions: 95°C for 2 min, 35 cycles of 95°C for 15
sec, 60°C for 30 sec and 72 C for 20 sec.
qPCR reaction was performed on the Bio-Rad C1000
Real-Time Fluorescence Thermal Cycler (Bio-Rad Laboratories), using
the following cycling conditions: Initiation at 95°C for 10 min;
amplification for 35 cycles, with denaturation at 95°C for 30 sec;
annealing at 60°C (Oct-4) or 56°C (GAPDH) for 30 sec; and
elongation at 72°C for 30 sec. A final extension at 72°C was
performed for 10 min. GAPDH mRNA level was used for normalization.
The following primers were used: Forward:
5′-CTGGAGAAGGAGAAGCTGGA-3′ and reverse:
5′-CAAATTGCTCGAGTTCTTTCTG-3′ for Oct-4 and forward:
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse: 5′-GAAGATGGTGATGGGATTTC-3′
for GAPDH. The expression level of Oct-4 mRNA relative to that of
GAPDH mRNA was calculated using the 2−ΔΔCt method.
Solid tissue disaggregation
Solid tissues, including matched normal, malignant
or benign polyp tissues, obtained from primary surgical specimens,
were mechanically and enzymatically disaggregated into single-cell
suspensions. Briefly, solid tissues were minced with scissors into
small (2-mm3) fragments and incubated for 15 min at room
temperature in 100 mM phosphate buffer (pH 7.0) with 6.5 mM DTT
(Sigma-Aldrich, Santa Clara, CA, USA), in order to remove mucus
contamination. Following gentle removal of the DTT solution, tissue
fragments were rinsed once with Hank's balanced salt solution
(Sigma-Aldrich), resuspended in serum-free RPMI 1640 medium with
200 units/ml Collagenase type III and 100 units/ml DNase I
(Invitrogen Life Technologies, Carlsbad, CA, USA), and incubated
for 2 h at 37°C for enzymatic disaggregation. Cells were then
resuspended by pipetting, and serially filtered using sterile gauze
with 70 µm and 40 µm nylon meshes. Contaminating erythrocytes were
removed by incubation in ammonium chloride potassium phosphate
hypotonic buffer for 5 min on ice.
FCM
For nuclear staining of cells from different
tissues, a Foxp3/Transcription Factor Staining kit was used
(eBioscience, San Diego, CA, USA). Cells prepared from matched
normal, neoplastic or benign polyp tissues, were initially fixed
with 1X fixation buffer, and subsequently permeabilized with 1X
permeabilization buffer. Cells were then incubated with anti Oct-4
antibody (SC-5279, Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). After washing with fixation buffer, cells were stained with
phycoerythrin-conjugated secondary antibody (Sigma-Aldrich). The
nuclear expression of Oct-4 was examined using FCM (Coulter Epics
XL Flow, Beckman Coulter, Inc., Brea, CA, USA).
IHC analysis
Formalin-fixed paraffin-embedded tissue sections
(4-µm) were mounted on to APES-coated glass slides (Chenglin,
Shanghai, China). Slides were dewaxed in xylene (Sigma-Aldrich)
twice for 10 min and rehydrated through a graded ethanol series.
Antigen retrieval was performed in 0.01 mol/l citrate buffer (pH
6.0; GeneTech, Shanghai, China) by boiling for 10 min. Endogenous
peroxidase activity was suppressed with 3% hydrogen peroxide
(Sigma-Aldrich) for 10 min. After washing with phosphate-buffered
saline (PBS), the slides were blocked with 5% BSA (Sigma-Aldrich)
for 30 min at 37°C. Sections were incubated with primary mouse
monoclonal antibody to human Oct-4 (dilution, 1:200; clones
SC-5279; Santa Cruz Biotechnology, Inc.) at 4°C overnight, in a
humidified chamber. After washing three times with PBS, sections
were incubated for 30 min with the secondary antibody (peroxidase
goat anti-mouse IgG; dilution, 1:300; catalog no. 32230; Zymed, San
Diego, CA, USA). After washing three times in PBS,
3,3′-diaminobenzidine was used as the chromogen. Slides were
counter-stained with hematoxylin for 1 min. Sections not incubated
with the primary antibody were used as negative controls.
Immunohistochemically stained slides were reviewed
by two independent expert pathologists, who were blinded to the
clinical outcome, using an Olympus BX51 microscope, and images were
captured using an Olympus DP71 camera (Olympus Corporation, Tokyo,
Japan). Olympus BSW with DP Controller software version 2.2
(Olympus Corporation) was used for image acquisition and
storage.
Statistical analysis
Statistical analysis was performed using SPSS 14.0
computer software (SPSS, Inc., Chicago, IL, USA). Experiments were
conducted in duplicate or triplicate. Positivity rates and
differences in Oct-4 expression between these groups were estimated
using the χ2 test and the non-parametric Wilcoxon rank
sum test, respectively. Correlations between Oct-4 expression and
clinicopathological parameters were also statistically analyzed.
P<0.05 was considered to indicate a statistically significance
difference.
Results
Transcription of Oct-4 progressively
increased from normal tissues to malignant tissues
CRC tissues, benign polyp tissues and matched
non-tumor tissues were used to represent different steps during the
evolution of CRC. Oct-4 transcription in 33 colorectal tumor,
matched distant non-tumor and benign polyp tissue specimens were
analyzed using RT-qPCR, in order to evaluate changes in the
expression of Oct-4 during the development of CRC. The results
showed that Oct-4 was expressed in normal, benign and malignant
colorectal tissues (Fig. 1A).
Quantitative analysis demonstrated that there were significant
difference among matched normal, benign and malignant colorectal
tissue specimens, and a stepwise upregulation in the expression of
Oct-4 was observed (Fig. 1B).
 | Figure 1.Analysis of Oct-4 transcription in
normal, benign polyp and CRC tissues. (A) RT-PCR analysis of Oct-4
mRNA expression in normal (N1, N2), benign polyp (B1, B2) and CRC
tissues (M1, M2). (B) Comparison of the relative expression levels
of Oct-4 mRNA among three groups (n=33), as determined by qPCR.
*P<0.001. Oct-4, octamer-binding transcription factor 4; CRC,
colorectal cancer; RT-PCR, reverse transcription-polymerase chain
reaction; qPCR, quantitative PCR. |
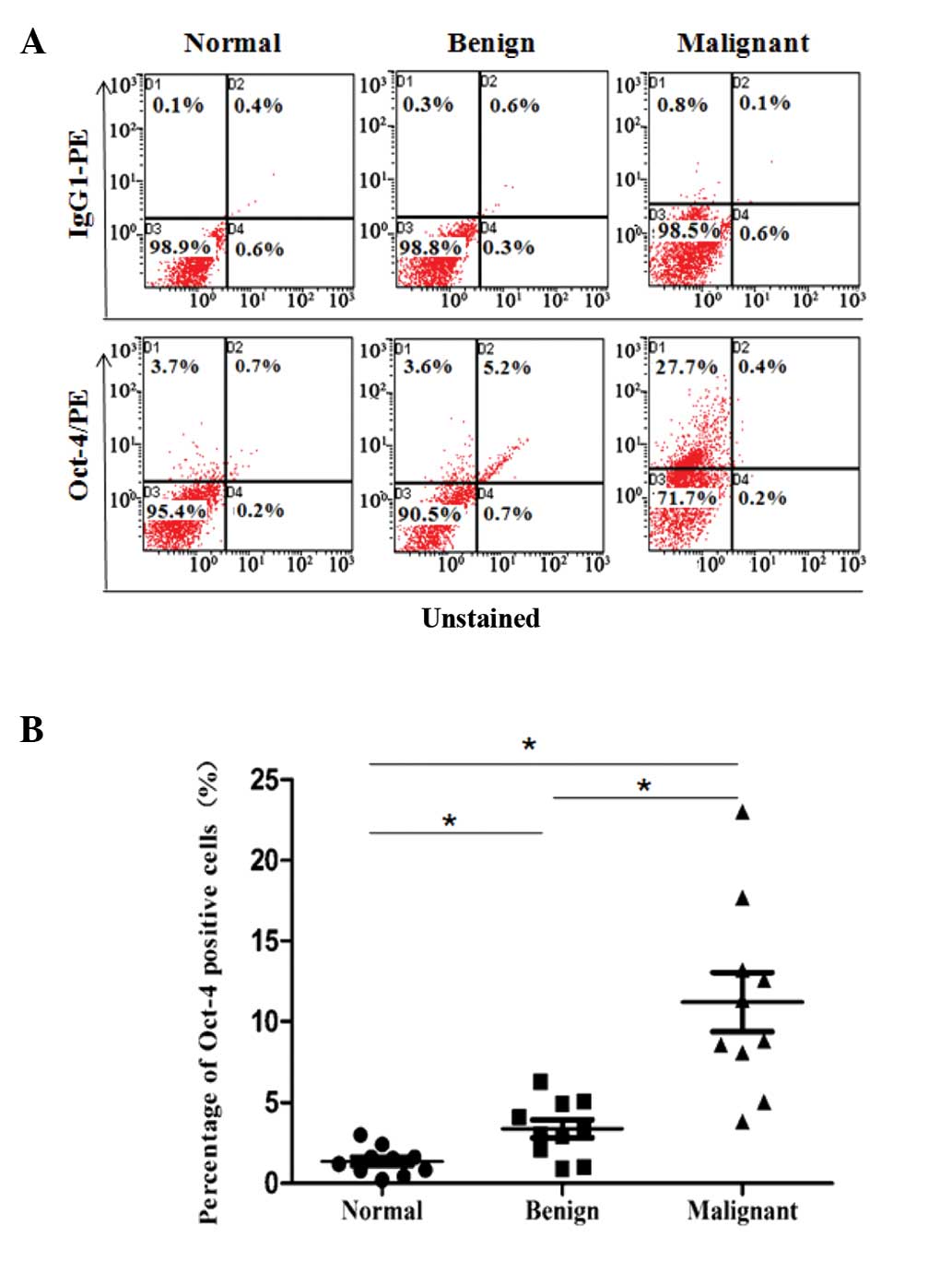
Oct-4+ cells increased from
normal tissues to malignant tissues
In order to analyze the number of Oct-4+
cells in different types of colorectal tissues, 10 primary CRC,
matched non-tumor and benign polyp tissues were mechanically and
enzymatically disaggregated into single-cell suspensions, and the
nuclear expression of Oct-4 in each specimen was determined by FCM.
Few Oct-4+ cells were detected in normal colorectal
tissues, while the number was significantly increased in benign
polyp tissues and was also significantly increased in CRC tissues,
compared with benign polyp tissues (Fig.
2A). The percentage of Oct-4+ cells in the three
types of colorectal tissues was 1.40±0.78, 2.91±1.57 and
11.37±6.32% respectively, and a significant difference was observed
among the three groups (Fig. 2B).
These results demonstrated that Oct-4 expression was also
upregulated at the protein level during the development of CRC.
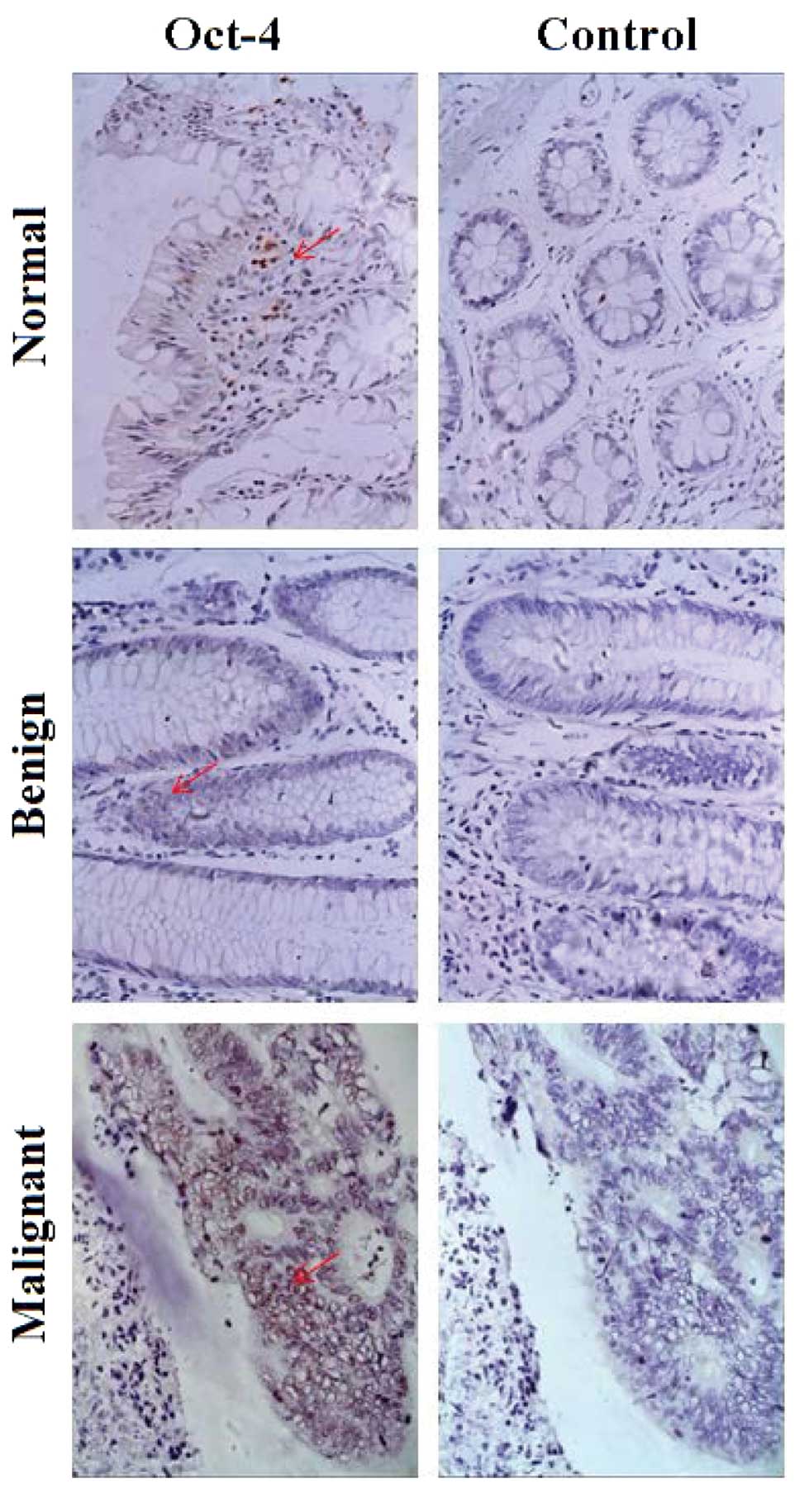
Expression of Oct-4 in colorectal
tumor, matched distant non-tumor and benign polyp tissues
Paraffin-embedded specimens, including 158 CRC
tissues, matched non-tumor tissues and 71 benign polyp tissues were
obtained, and IHC was performed in order to evaluate Oct-4
expression in the different types of colorectal tissues. Oct-4
protein was primarily located in the nuclei, and the expression
ratio was 4.43% (7/158) in normal tissues, 12.68% (9/71) in benign
polyp tissues and 42.41% (67/158) in CRC tissues (Fig. 3). No immunoreactivity was observed in
the negative controls.
Association of Oct-4 expression with
clinicopathological features
The correlations between various clinicopathological
features and Oct-4 expression in primary colorectal tumors are
summarized in Table I. A significant
positive correlation was observed between Oct-4 expression and
histological grade (P=0.007), lymph node metastasis (P=0.027),
distant metastasis (P=0.017) and TNM stage (P=0.041). Oct-4
expression was not associated with any other clinicopathological
factors, including gender, age and tumor size.
 | Table I.Association between Oct-4 expression
and clinicopathological factors in CRC. |
Table I.
Association between Oct-4 expression
and clinicopathological factors in CRC.
|
| Oct-4 |
|
|---|
|
|
|
|
|---|
| Variables | + | – | P-value |
|---|
| Gender |
|
| 0.484 |
| Male | 41 | 55 |
|
| Female | 23 | 39 |
|
| Age (years) |
|
| 0.383 |
| <45 | 12 | 15 |
|
| 45–60 | 24 | 31 |
|
| ≥60 | 28 | 48 |
|
| Tumor size
(cm) |
|
| 0.330 |
| <5 | 40 | 66 |
|
| 5–10 | 21 | 24 |
|
| ≥10 | 3 | 4 |
|
| Histological
grade |
|
| 0.007a |
| 1 | 4 | 15 |
|
| 2 | 32 | 55 |
|
| 3 | 28 | 24 |
|
| Lymph node
status |
|
| 0.027a |
| N0 | 31 | 60 |
|
| N1 | 20 | 26 |
|
| N2 | 13 | 8 |
|
| Distant
metastasis |
|
| 0.017a |
| Negative | 51 | 89 |
|
| Positive | 13 | 5 |
|
| TNM stage |
|
| 0.041a |
| I | 9 | 28 |
|
| II | 25 | 33 |
|
| III | 26 | 28 |
|
| IV | 4 | 5 |
|
Prognostic implication of Oct-4
expression in CRC
Follow-up information was available for 158 patients
over a minimum period of 10 years. Kaplan-Meier survival curves and
the log-rank test showed that Oct-4+ cases had a
significantly shorter median survival time (37.0 months) compared
with Oct-4− cases (76.0 months; P=0.001; Fig 4).
Discussion
CRC is one of the most common malignant tumors
worldwide (18,25). Recently, its incidence has markedly
increased, and it is currently a significant public health problem
in China (26). Despite considerable
improvements in diagnosis and therapy protocols, including surgical
resection, chemotherapy and radiotherapy, the clinical outcome for
patients with CRC remains unsatisfactory. Therefore, it is
necessary to achieve greater understanding of the development of
this disease, in order to establish novel strategies for the
treatment and assessment of prognosis in CRC.
Recently, heterogeneity within tumors has been
demonstrated. Only a small subpopulation of tumor cells, termed
CSCs or TICs, which had the capacity to generate the original tumor
and to maintain the heterogeneity of tumor tissues in animal
models, were identified in malignant tissues. Two research groups
initially identified CD133+ colorectal tumor cells as
colorectal tumor stem cells (22,23). The
studies conducted by these groups demonstrated that CD133
expression was markedly increased in colorectal carcinoma tissues
compared with that in normal colorectal tissues, and that
CD133+ cells effectively generated new tumors in
NOD/SCID mice. While there are uncertainties regarding the
phenotype of colorectal CSCs (27,28), the
identification of colorectal CSCs provided novel directions for
research into CRC.
Oct-4, part of the family of POU-domain
transcription factors, was originally shown to be expressed in
pluripotent embryonic stem (ES) and germ cells (1,2). Numerous
studies have shown that Oct-4 activates transcription via the
octamer motif of an AGTCAAAT consensus sequence, and affects
mammalian development by regulating the pluripotent potential of
stem cells (3–7). Subsequent studies have demonstrated that
Oct-4 is also expressed in a number of types of tumor cells, and
that it has potential as a biomarker for the diagnosis and
prognosis of malignant tumors (8–18). Further
studies have indicated that Oct-4 may be important in cancer cell
survival, and that it exerts multiple functions in tumor cells.
Overexpression of Oct-4 was hypothesized to lead to the
inappropriate activation of growth factors, promotion of cellular
proliferation and, ultimately, malignant transformation (29). Dai et al (30) reported that Oct-4 regulates
epithelial-mesenchymal transition and contributes to CRC cell
migration and invasion. Wang et al (31) showed that Oct-4 is significantly
associated with an unfavorable clinical outcome in human esophageal
squamous cell carcinoma.
In recent years, it has been shown that Oct-4 is
expressed in CSCs, and that it is an important molecule with which
to identify and research properties of CSCs. Chen et al
(18) proposed that Oct-4 expression
maintains cancer stem-like properties in lung cancer-derived
CD133+ cells. Cortes-Dericks et al (32) proposed that high expression of Oct-4
is involved in the initiation of lung adenocarcinoma and results in
a reduction in disease-free survival. The evolution of CRC is a
stepwise process. Little is known regarding the changes that occur
in stem cells during this process. Therefore Oct-4, as an important
functional molecule, requires further investigation in the context
of colorectal carcinogenesis.
In the present study, CRC, benign polyp, and distant
non-tumor tissues were obtained in order to represent different
steps in the development of CRC, and the expression of Oct-4 was
measured in these tissues. The results of RT-qPCR showed a
progressive upregulation of the transcription of Oct-4 from normal
tissues to malignant tissues. Oct-4 expression in cells from the
various types of tissues was further investigated using FCM. As
hypothesized, the percentage of Oct-4+ cells in these
tissues increased in a stepwise manner, from normal to benign polyp
tissues, and from benign polyps to CRC tissues. Subsequently, IHC
was performed in order to confirm the variation in expression of
Oct-4 in the three types of colorectal tissues. The results
demonstrated that the Oct-4 protein was primarily located in the
nuclei, and that the expression ratios in normal tissues, polyp
tissues and CRC tissues were 4.43, 12.68 and 42.41%, respectively.
These results indicated that aberrant expression of Oct-4 may
contribute to carcinogenesis within colorectal tissues. As Oct-4 is
known to act as a functional molecule for stem cells, the present
results suggested that abnormal biological behavior may occur in
stem cells during the development of CRC, and that aberrant
expressed of Oct-4 may contribute to the functional alteration of
colorectal stem cells. Statistical analysis showed that Oct-4
expression in CRC was significantly correlated with histological
grade (P=0.007), lymph node metastasis (P=0.027), distant
metastasis (P=0.017) and TNM stage (P=0.041). Kaplan-Meier survival
curves and the log-rank test showed that Oct-4+ cases
had a significantly shorter median survival time (37.0 months)
compared with Oct-4− cases. These results suggested that
Oct-4 may also be a useful biomarker with which to assess prognosis
in CRC.
To the best of our knowledge, the present study is
the first to demonstrate the dynamic expression of Oct-4 during the
evolution of CRC. In conclusion, the current findings suggest that
aberrant expression of Oct-4 may be involved in the development of
CRC. Oct-4 may function as a novel oncogene, and has potential for
use as a biomarker for the prediction, and assessment of prognosis
and survival in patients with CRC. Furthermore, Oct-4 is implicated
in the de-differentiation of cells and is a marker for stem cell
populations. Overexpression of Oct-4 may result in the
amplification of resident colorectal stem cell populations which
subsequently leads to the initiation, progression and
differentiation of human CRC. Further investigation into these
processes is required. However, Oct-4 may eventually be a novel
therapeutic target for CRC.
Acknowledgements
This study was supported by the Social Development
Foundation of Wuxi city (grant no. CSE31N1313) and the Scientific
Research Program of Wuxi Hospital Administration Centre (grant no.
YGZXQ1305).
Glossary
Abbreviations
Abbreviations:
|
Oct-4
|
octamer-binding transcription factor
4
|
|
CRC
|
colorectal cancers
|
|
CSCs
|
cancer stem cells
|
|
TICs
|
tumor initiating cancer cells
|
References
|
1
|
Burdon T, Smith A and Savatier P:
Signalling, cell cycle and pluripotency in embryonic stem cells.
Trends Cell Biol. 12:432–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosner MH, Vigano MA, Ozato K, Timmons PM,
Poirier F, Rigby PW and Staudt LM: A POU-domain transcription
factor in early stem cells and germ cells of the mammalian embryo.
Nature. 345:686–692. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pesce M and Schöler HR: Oct-4: Gatekeeper
in the beginnings of mammalian development. Stem Cells. 19:271–278.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scholer HR: Octamania: The POU factors in
murine development. Trends Genet. 7:323–329. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kraft HJ, Mosselman S, Smits HA,
Hohenstein P, Piek E, Chen Q, Artzt K and van Zoelen EJ: Oct-4
regulates alternative platelet-derived growth factor alpha receptor
gene promoter in human embryonal carcinoma cells. J Biol Chem.
271:12873–12878. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lamb KA and Rizzino A: Effects of
differentiation on the transcriptional regulation of the FGF-4
gene: Critical roles played by a distal enhancer. Mol Reprod Dev.
51:218–224. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Xu WR, Qian H, Zhu W, Bu XF, Wang
S, Yan YM, Mao F, Gu HB, Cao HL, et al: Oct4, a novel marker for
human gastric cancer. J Surg Oncol. 99:414–419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Zhang X, Wang X, Gan L, Yu G,
Chen Y, Liu K, Li P, Pan J, Wang J, et al: Inhibition of LDH-A by
lentivirus-mediated small interfering RNA suppresses
intestinal-type gastric cancer tumorigenicity through the
downregulation of Oct4. Cancer Lett. 321:45–54. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim RJ and Nam JS: OCT4 expression
enhances features of cancer stem cells in a mouse model of breast
cancer. Lab Anim Res. 27:147–152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iida H, Suzuki M, Goitsuka R and Ueno H:
Hypoxia induces CD133 expression in human lung cancer cells by
up-regulation of OCT3/4 and SOX2. Int J Oncol. 40:71–79.
2012.PubMed/NCBI
|
|
12
|
Fang XF, Zhang WY, Zhao N, Yu W, Ding D,
Hong X, Li LS, Zhang HR, Zheng S and Lin BY: Genome-wide analysis
of OCT4 binding sites in glioblastoma cancer cells. J Zhejiang Univ
Sci B. 12:812–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing
L, Zhang Y, Ling EA, Gao J and Hao A: Expression profile of
embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human
gliomas. Histopathology. 59:763–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ikushima H, Todo T, Ino Y, Takahashi M,
Saito N, Miyazawa K and Miyazono K: Glioma-initiating cells retain
their tumorigenicity through integration of the Sox axis and Oct4
protein. J Biol Chem. 286:41434–41441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He W, Li K, Wang F, Qin YR and Fan QX:
Expression of OCT4 in human esophageal squamous cell carcinoma is
significantly associated with poorer prognosis. World J
Gastroenterol. 18:712–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng L, Sung MT, Cossu-Rocca P, Jones TD,
MacLennan GT, De Jong J, Lopez-Beltran A, Montironi R and Looijenga
LH: OCT4: Biological functions and clinical applications as a
marker of germ cell neoplasia. J Pathol. 211:1–9. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jones TD, Ulbright TM, Eble JN and Cheng
L: OCT4: A sensitive and specific biomarker for intratubular germ
cell neoplasia of the testis. Clin Cancer Res. 10:8544–8547. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen YC, Hsu HS, Chen YW, Tsai TH, How CK,
Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, et al: Oct-4
expression maintained cancer stem-like properties in lung
cancer-derived CD133-positive cells. PLoS One. 3:e26372008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alison MR, Islam S and Wright NA: Stem
cells in cancer: Instigators and propagators? J Cell Sci.
123:2357–2368. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y and Laterra J: Cancer stem cells:
Distinct entities or dynamically regulated phenotypes? Cancer Res.
72:576–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu
K, Yu J and Sung JJ: MicroRNA in colorectal cancer: From benchtop
to bedside. Carcinogenesis. 32:247–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Compton CC: Colorectal carcinoma:
Diagnostic, prognostic and molecular features. Mod Pathol.
16:376–388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
You WC, Jin F, Devesa S, Gridley G,
Schatzkin A, Yang G, Rosenberg P, Xiang YB, Hu YR and Li Q: Rapid
increase in colorectal cancer rates in urban Shanghai, 1972–97, in
relation to dietary changes. J Cancer Epidemiol Prev. 7:143–146.
2002.PubMed/NCBI
|
|
27
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shmelkov SV, Butler JM, Hooper AT, Hormigo
A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et
al: CD133 expression is not restricted to stem cells and both
CD133+ and CD133-metastatic colon cancer cells initiate tumors. J
Clin Invest. 118:2111–2120. 2008.PubMed/NCBI
|
|
29
|
Hochedlinger K, Yamada Y, Beard C and
Jaenisch R: Ectopic expression of Oct-4 blocks progenitor-cell
differentiation and causes dysplasia in epithelial tissues. Cell.
121:465–477. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dai X, Ge J, Wang X, Qian X, Zhang C and
Li X: OCT4 regulates epithelial-mesenchymal transition and its
knockdown inhibits colorectal cancer cell migration and invasion.
Oncol Rep. 29:155–160. 2013.PubMed/NCBI
|
|
31
|
Wang Q, He W, Lu C, Wang Z, Wang J,
Giercksky KE, Nesland JM and Suo Z: Oct3/4 and Sox2 are
significantly associated with an unfavorable clinical outcome in
human esophageal squamous cell carcinoma. Anticancer Res.
29:1233–1241. 2009.PubMed/NCBI
|
|
32
|
Cortes-Dericks L, Galetta D, Spaggiari L,
Schmid RA and Karoubi G: High expression of octamer-binding
transcription factor 4A, prominin-1 and aldehyde dehydrogenase
strongly indicates involvement in the initiation of lung
adenocarcinoma resulting in shorter disease-free intervals. Eur J
Cardiothorac Surg. 41:e173–e181. 2012. View Article : Google Scholar : PubMed/NCBI
|