Introduction
Increasing evidence has confirmed that lung cancer,
predominantly non-small cell lung cancer (NSCLC), is the primary
cause of cancer-associated mortality worldwide, with a
significantly increased risk among males (1). It has been well established that lung
cancer is a complex disease that is highly associated with
environmental pollutants and smoking in the general population; in
addition, genetic factors are known to have an important role in
this disease. Therefore, the study of genetic variants relevant to
lung cancer may contribute to fully elucidating the pathogenesis of
this malignancy, including its formation and development, as well
as to predicting an individual's risk of developing NSCLC.
The p73 gene was identified at chromosome lp36.33 in
1997 (2), a site that has been
demonstrated to commonly undergo loss of heterozygosity (LOH) in
various types of cancer (2–4). LOH at the p73 gene has been reported to
occur in 62% of lung cancer patients (5). The p73 protein is a structural and
functional homolog of p53, a classic tumor suppressor (2). Numerous studies have demonstrated that
the p73 gene had a crucial role in the pathogenesis of various
types of cancer, including lung cancer (6–9).
Therefore, it was hypothesized that p73 may be a potential
susceptibility gene for lung cancer; in addition, it was suggested
that p73 gene polymorphisms may results in differences in
susceptibility to lung cancer (10).
The p73 gene contains a polymorphism in the 5′
untranslated region (UTR), which is located upstream of the
initiating AUG codon in exon 2 of p73 gene; in theory, this region
may have the ability to form a stem-loop structure and modulate
susceptibility to cancer (2,11,12). This
polymorphism involves two single-nucleotide polymorphisms (SNPs) at
positions 4 (G-A) and 14 (C-T) (2).
As these SNPs are in complete linage equilibrium, only three
genotypes are possible: GC/GC, GC/AT and AT/AT. Previous studies
have reported an association between the G4C14-to-A4T14
polymorphism and the risk of developing various types of cancer. Li
et al (13) revealed a marked
association in an American population between the AT allele or
genotype and the risk of developing squamous cell carcinoma of the
head and neck (13); in addition, a
study by Ryan et al (14)
indicated that AT/AT homozygotes were significantly uncommon in
esophageal cancer patients from an Irish population. Furthermore,
Hu et al (15) reported an
association between dinucleotide polymorphism of the p73 gene and a
reduced risk of lung cancer in a Chinese population and Li et
al (16) determined that the
variant GC/AT and AT/AT genotypes were associated with a
significantly increased risk of lung cancer (16). Associations between p73 gene
polymorphisms and the risk of lung cancer therefore illustrate that
the p73 gene has a critical role in lung cancer carcinogenesis
(15–18). However, there have been
inconsistencies in the data regarding these associations.
Therefore, the present study aimed to perform a case-control study
in order to examine whether the p73 G4C14-to-A4T14 polymorphism was
associated with NSCLC susceptibility and its effects on the
clinical characteristics of patients.
Materials and methods
Patients
A total of 184 consecutive lung cancer patients and
196 cancer-free controls were enrolled in the present study from
the Hunan Provincial Tumor Hospital (Changsha, China) between
January 2013 and June 2014. Patients included in the present study
had been newly diagnosed with NSCLC following histopathological
confirmation. At recruitment, each patient was interviewed to
collect personal information regarding demographic characteristics
factors, including age, gender, smoking status and histological
type (Table I). Written informed
consent was obtained from all participants and the study was
performed with the approval of the Changsha Clinical Research
Center Ethics Committee (Changsha, China).
 | Table I.Distributions of characteristics among
non-small cell lung cancer cases and controls. |
Table I.
Distributions of characteristics among
non-small cell lung cancer cases and controls.
|
| Cases (%) | Controls (%) |
|
|---|
|
|
|
|
|
|---|
| Variable | n=186 | n=198 | P-value |
|---|
| Age |
|
| 0.678 |
| ≤50 | 90 (48.3) | 100 (50.5) |
|
|
>50 | 96 (51.6) | 98 (49.5) |
|
| Gender |
|
| 0.837 |
| Male | 137 (73.7) | 144 (72.9) |
|
|
Female | 49 (26.3) | 54 (27.3) |
|
| Smoking status |
|
| <0.001 |
| No | 67 (36.0) | 125 (63.1) |
|
| Yes | 119 (64.0) | 73 (36.9) |
|
| Histological
type |
|
|
|
| Squamous
carcinomas | 116 (62.4) |
|
|
|
Adenocarcinomas | 58 (31.2) |
|
|
|
Others | 12 (6.4) |
|
|
Polymerase chain reaction (PCR)
analysis
Blood samples (400 µl) were collected from each
participant and genomic DNA was extracted using a DNA extraction
kit (Takara Biotechnology Co., Ltd., Dalian, China). Blood samples
and DNA samples were stored at −70°C. DNA samples were then
analyzed for the p73 G4C14-to-A4T14 polymorphism using PCR with
confronting two-pair primers (PCR-CTPP; Sangon Biotech Co., Ltd.,
Shanghai, China). The PCR primers used were as follows (19): Forward (F)1, 5′-CCA CGG ATG GGT CTG
ATCC-3′ and reverse (R)1, 5′-GGC CTC CAA GGG CAG CTT-3′; and F2,
5′-CCT TCC TTC CTG CAG AGCG-3′ and R2, 5′-TTAGCCCAGCGAAGGTGG-3′.
PCR was performed with a 15 µl reaction mixture, containing 7.5 µl
Taq PCR Master Mix (Sangon Biotech Co., Ltd.), 2.5 µl
double-distilled H20, 1 µl each of four primers and 1 µl
DNA template. PCR was performed using a Gene Amp PCR System 9700
(Applied Biosystems, Waltham, MA, USA) as follows: Initial
denaturation at 95°C for 5 min; 35 cycles of 95°C for 40 sec, 60°C
for 40 sec and 72°C for 40 sec; and a final extension at 72°C for 5
min (19). The PCR products were
separated on a 2% agarose gel (Sangon Biotech Co., Ltd.) with
ethidium bromide staining (Sinopharm Chemical Reagent Co., Ltd.,
Shanghai, China) and visualized using the Vilber Fusion FX7
gel-imaging system (11200238; Vilber Lourmat Deutchland,
Eberharedzell, Germany).
Statistical analysis
Gene counting was used to determine the genotype and
allele frequencies for p73 G4C14-to-A4T14 polymorphism using the
χ2-test. Comparisons of age, sex and smoking status
distributions between NSCLC cases and controls were performed using
the χ2-test. The association between the p73 genotypes
and NSCLC risk was assessed using the logistic regression model,
while odds ratios (ORs), 95% confidence intervals (95% CIs) and
P-values were also calculated. The association between p73
polymorphic variants and clinicopathological characteristics was
determined using the χ2-test. P<0.05 was considered
to indicate a statistically significant difference between values.
All statistical analyses were performed using SPSS 17.0 software
(SPSS Inc., Chicago, IL, USA).
Results
Characteristics of study subjects
In total, 186 NSCLC cases and 198 control subjects
were recruited into the present study. The frequency distribution
of the basic characteristics of the patients and controls is
summarized in Table I. The
distributions of age and sex seemed to be matched fully and there
was no significant difference between cases and controls. However,
there were a significantly increased number of smokers in the NSCLC
group compared with the control group (64.0 vs. 36.9%,
respectively; P<0.001). This therefore indicated that smoking
was a high risk factor associated with NSCLC in patients enrolled
in the present study.
Distribution of p73 polymorphisms
The association between p73 polymorphisms and
susceptibility to NSCLC were explored using PCR-CTPP; as the two
SNPs were in complete linage equilibrium, only three genotypes were
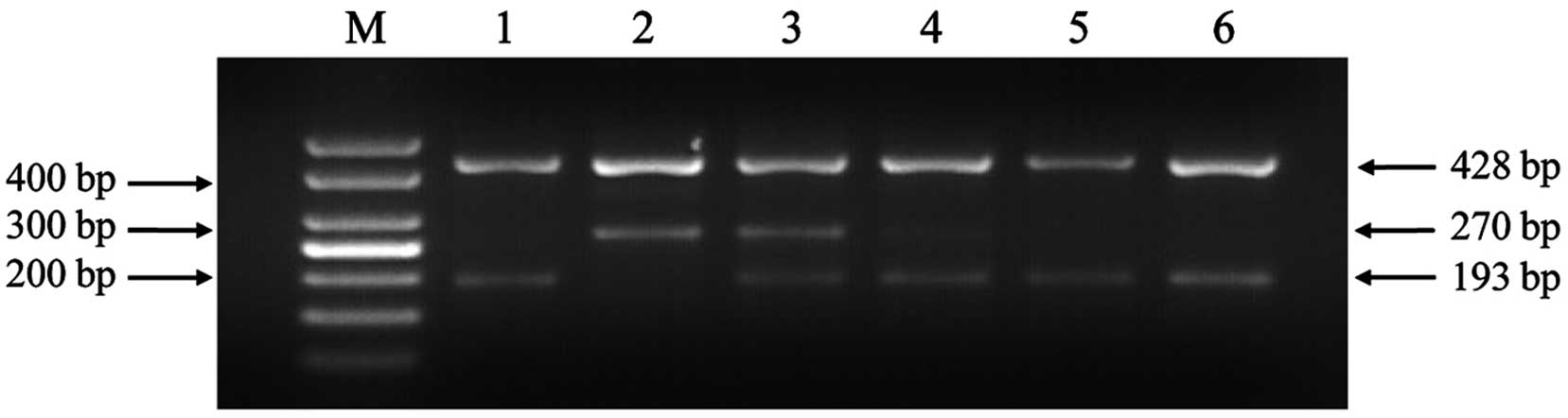
possible: GC/GC, GC/AT and AT/AT. The electrophoresis results are
shown in Fig. 1 and the sequencing
results of PCR products are shown in Fig.
2. As shown in Fig. 1, the GC
allele yielded two bands of 428 and 193 bp, the AT allele yielded
two bands of 428 and 270 bp and the GC/AT heterozygosity yielded
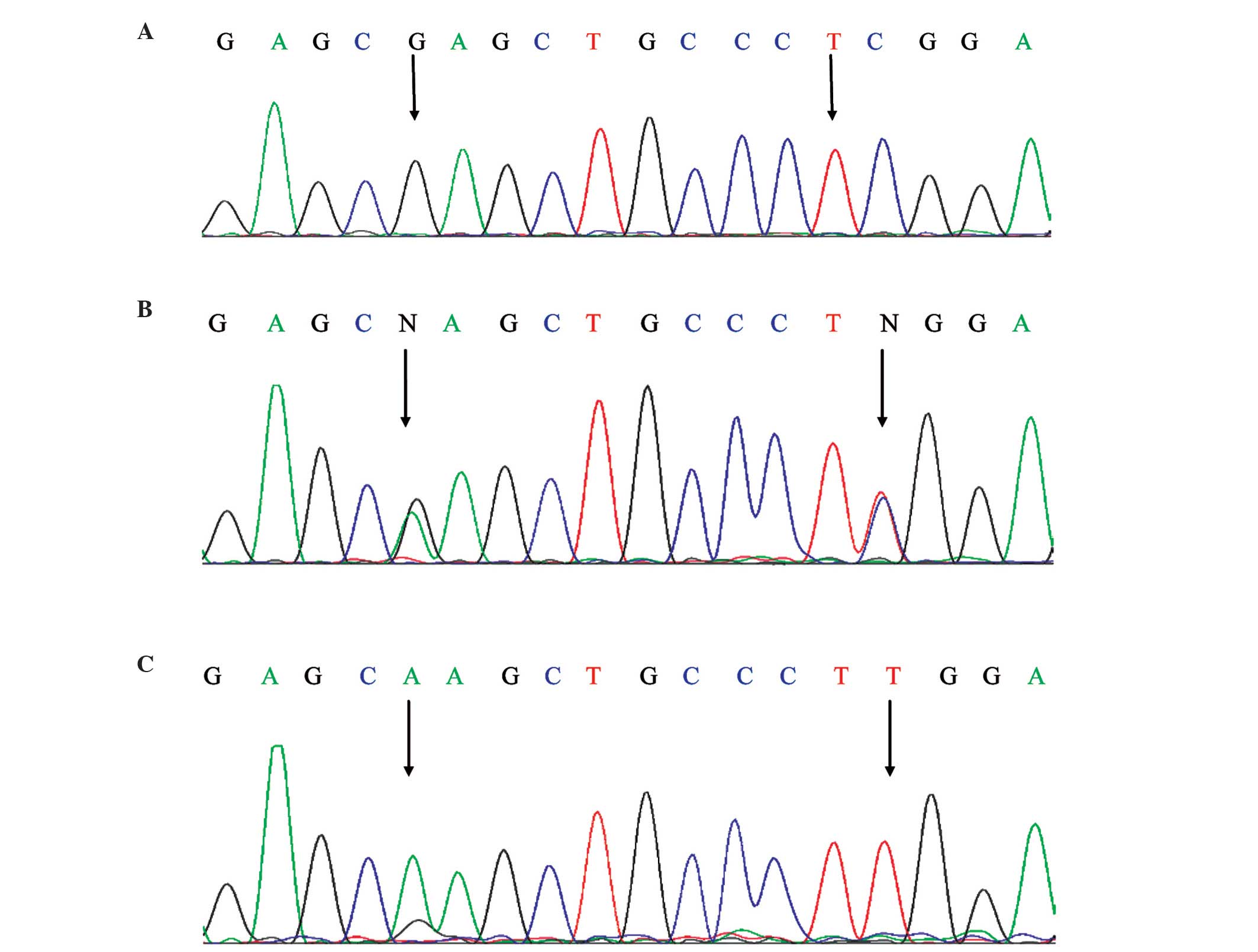
three bands of 428, 270 and 193 bp. In order to ensure the accuracy
of the genotyping data, the data was confirmed by direct sequencing
analysis by Sangon Biotech Co., Ltd.) (Fig. 2). SPSS 17.0 software was used to
analyze the p73 genotypes and the results are shown in Table II.
 | Table II.Association between p73 polymorphisms
and risk of non-small cell lung cancer. |
Table II.
Association between p73 polymorphisms
and risk of non-small cell lung cancer.
|
| Cases (%) | Controls (%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Genotype | n=186 | n=198 | P-value | OR (95%CI) |
|---|
| GC/GC | 108 (58.1) | 104 (52.5) | – | 1 |
| GC/AT | 68 (36.6) | 68 (34.3) | 0.864 | 0.963
(0.626–1.481) |
| AT/AT | 10 (5.4) | 26 (13.1) | 0.010 | 0.370
(0.170–0.806) |
| GC allele | 284 (76.3) | 276 (69.7) | – | 1 |
| AT allele | 88 (23.7) | 120 (30.3) | 0.038 | 0.713
(0.517–0.983) |
Association between p73 polymorphisms
and NSCLC risk
The present study aimed to determine whether the p73
G4C14-to-A4T14 polymorphism was associated with susceptibility to
NSCLC. The genotypes and allele distributions of the p73
polymorphism in patients and controls are shown in Table II; genotype frequencies in patients
and controls were in accordance with the Hardy-Weinberg
equilibrium. The results demonstrated a significant association
between AT/AT genotype and a reduced risk of NSCLC (P=0.037;
OR=0.481; 95% CI=0.240–0.965) compared with the GC/GC genotype. In
addition, patients carrying the AT allele (GC/AT or AT/AT) had a
significantly decreased risk of NSCLC (P=0.038; OR=0.713; 95%
CI=0.517–0.983) compared with that of the p73 GC allele group,
which indicated that the AT allele may be a protective factor
against NSCLC.
Association between p73 polymorphisms
and clinicopathological parameters
The association between p73 SNPs and the
clinicopathological features of NSCLC is depicted in Table III. The results revealed that there
was no significant association between the p73 genotypes and tumor
histological type (P=0.410), tumor stage (P=0.192) or lymph node
metastasis (P=0.298).
 | Table III.Association between p73 polymorphisms
and clinicopathological parameters of non-small cell lung
cancer. |
Table III.
Association between p73 polymorphisms
and clinicopathological parameters of non-small cell lung
cancer.
| Variable | n (%) | GC/GC (%) | GC/AT (%) | AT/AT (%) | P-value |
|---|
| Histological
(n=174) |
|
|
|
| 0.410 |
| Squamous
cell | 116 (66.7) | 70 (60.3) | 38 (32.8) | 8 (6.9) |
|
|
Adenocarcinoma | 58
(33.3) | 32 (55.2) | 24 (41.4) | 2 (3.4) |
|
| Tumor stage
(n=186) |
|
|
|
| 0.192 |
| I+II | 150 (80.6) | 90 (60) | 54 (36) | 6 (4) |
|
|
III+IV | 36
(19.4) | 18 (50) | 14 (38.9) | 4 (11.1) |
|
| Lymph node
metastasis (n=186) |
|
|
|
| 0.298 |
|
Yes | 48
(25.8) | 24 (50) | 22 (45.8) | 2
(4.2) |
|
| No | 138 (74.2) | 84 (60.9) | 46 (33.3) | 8 (5.8) |
|
Discussion
The p73 gene is known to activate the promoters of
certain genes that are responsive to p53, including those involved
in cell cycle control, DNA repair and apoptosis; therefore, p73
attenuates cell growth through the induction of apoptosis or G1
cell cycle arrest in a manner comparable to that of the p53 gene
(2). Although the accurate functional
association between the risk of cancer and p73 G4C14-to-A4T14
genetic variants remains to be elucidated, it was proposed that the
change of GC to AT may result in the formation of a stem-loop
structure and may therefore influence p73 translation efficiency
and gene expression (2).
Previous studies have provided evidence to suggest
that the p73 gene has a crucial role in the pathogenesis of various
types of cancer, such as lung cancer (6–9). It is
therefore plausible that an association exists between the p73
G4C14-to-A4T14 polymorphism and the incidence of lung cancer. p73
is a member of the p53 family of tumor suppressor genes, which has
two distinct promoters that may be expressed in different
N-terminal isoforms (20). The
transactivation proficient isoform (TAp73) has pro-apoptotic
functions, whereas the N-terminally truncated isoform-ΔN p73 exerts
an anti-apoptotic effect. It has therefore been proposed that the
altered expression ratio of ΔN:TA may have a role in the
pathogenesis of certain types of cancers (21–24), such
as lung cancer (8). As indicated by
the role of TAp73 in cancer development, it was hypothesized that
individuals with the AT allele may have decreased TAp73 levels and
a reduced ratio of TAp73:ΔTAp73, which may result in an increased
susceptibility to lung cancer.
The p73 gene was suggested to be a susceptibility
candidate gene for lung cancer. However, the results of previous
studies regarding the association between p73 G4C14-to-A4T14
polymorphism and susceptibility to lung cancer are inconsistent
(10,15–18). Two
previous studies observed that the AT allele was associated with a
significantly increased incidence of lung cancer compared with the
GC/GC allele in a Northern Chinese population (10) and a non-Hispanic Caucasian population
(16). By contrast, one study
reported a marked association between the AT allele and the
decreased risk of developing lung cancer in a Chinese population
(15), which is consistent with the
results of the present study. However, a further two studies failed
to identify any associations between the p73 G4C14-to-A4T14
polymorphism and the risk of lung cancer in Korean and Japanese
populations (17,18). These findings suggested that the role
of the p73 G4C14-to-A4T14 polymorphism may vary between different
ethnic populations.
In the present case-control study, it was
investigated whether the p73 G4C14-to-A4T14 polymorphism was
associated with the risk of NSCLC in a Chinese population. The
results revealed that the genotype distribution was markedly
different between the 186 NSCLC patients and the 198 healthy
controls; this therefore indicated that the p73 AT/AT genotype was
associated with a decreased risk of NSCLC. In addition, no
significant association was detected between p73 gene SNPs and
tumor histological type, tumor stage or lymph node metastasis of
NSCLC. Thus, whether the p73 gene polymorphism affected the
occurrence of lung cancer requires further verification in terms of
clinical development and survival.
In conclusion, the results of the present study
indicated that the p73 AT allele may be a reduced genetic risk
factor for the tumorigenesis and development of NSCLC in Chinese
patients. Further studies are required in order to confirm the
association between p73 polymorphism and the risk of lung cancer.
However, if confirmed by subsequent studies, this genetic risk
factor may be valuable for explaining the pathogenesis of lung
cancer, which may contribute to future cancer diagnosis techniques
and therapies.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 61171061), the
Natural Science Foundation of Hunan Province of China (nos.
12JJ4082 and 14JJ2049), the Outstanding Youth Project Supported by
Scientific Research Fund of Hunan Provincial Education Department
(no. 14B050) and the Natural Science Foundation of Hunan University
of Technology (no. 2013HZX04).
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaghad M, Bonnet H, Yang A, et al:
Monoallelically expressed gene related to p53 at lp36, a region
frequently deleted in neuroblastoma and other human cancers. Cell.
90:809–819. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bénard J, Douc-Rasy S and Ahomadegbe JC:
TP53 family members and human cancers. Hum Mutat. 21:182–191. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moll UM, Erster S and Zaika A: p53, p63
and p73 - solos, alliances and feuds among family members. Biochim
Biophys Acta. 1552:47–59. 2001.PubMed/NCBI
|
|
5
|
Shibukawa K, Miyokawa N, Tokusashi Y, et
al: High incidence of chromosomal abnormalities at lp36 and 9p21 in
early-stage central type squamous cell carcinoma and squamous
dysplasia of bronchus detected by autofluorescence bronchoscopy.
Oncol Rep. 22:81–87. 2009.PubMed/NCBI
|
|
6
|
Uramoto H, Sugio K, Oyama T, et al:
Expression of the p53 family in lung cancer. Anticancer Res.
26:1785–1790. 2006.PubMed/NCBI
|
|
7
|
Liu K, Zhan M and Zheng P: Loss of p73
expression in six non-small cell lung cancer cell lines is
associated with 5′CpG island methylation. Exp Mol Pathol. 84:59–63.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lo Iacono M, Monica V, Saviozzi S, et al:
p63 and p73 isoform expression in non-small cell lung cancer and
corresponding morphological normal lung tissue. J Thorac Oncol.
6:473–481. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nyman U, Sobczak-Pluta A, Vlachos P, et
al: Full-length p73alpha represses drug-induced apoptosis in small
cell lung carcinoma cells. J Biol Chem. 280:34159–34169. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang X, Li X, Wu Z, et al: The p73
G4C14-to-A4T14 polymorphism is associated with risk of lung cancer
in the Han nationality of North China. Mol Carcinog. 52:387–391.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tokuchi Y, Hashimoto T, Kobayashi Y, et
al: The expression of p73 is increased in lung cancer, independent
of p53 gene alteration. Br J Cancer. 80:1623–1629. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Q, Athan E, S, Wei M, et al: TP73
allelic expression in human brain and allele frequencies in
Alzheimer's disease. BMC Med Genet. 5:5–14. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li GJ, Sturgis EM, Wang LE, et al:
Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk
of squamous cell carcinoma of the head and neck. Carcinogenesis.
25:1911–1916. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ryan BM, McManus R, Daly JS, et al: A
common p73 polymorphism is associated with a reduced incidence of
oesophageal carcinamo. Br J Cancer. 85:1499–1503. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu Z, Miao X, Ma H, et al: Dinucleotide
polymorphism of p73 gene is associated with a reduced risk of lung
cancer in a Chinese population. Int J Cancer. 114:455–460. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li G, Wang LE, Chamberlain RM, et al: p73
G4C14-to-A4T14 polymorphism and risk of lung cancer. Cancer Res.
64:6863–6866. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi JE, Kang HG, Chae MH, et al: No
association between p73 G4C14-to-A4T14 polymorphism and the risk of
lung cancer in a Korean population. Biochem Genet. 44:533–540.
2006. View Article : Google Scholar
|
|
18
|
Hiraki A, Matsuo K, Hamajima N, et al:
Different risk relations with smoking for non-small cell lung
cancer: Comparison of TP53 and TP73 genotypes. Asian Pac J Cancer
Prev. 4:107–112. 2003.PubMed/NCBI
|
|
19
|
Lee KE, Hong YS, Kim BG, et al: p73 G4C14
to A4T14 polymorphism is associated with colorectal cancer risk and
survival. World J Gastroenterol. 16:4448–4454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Melino G, De Laurenzi V and Vousden KH:
p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2:605–615.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grob TJ, Novak U, Maisse C, et al: Human
delta Np73 regulates a dominant negative feedback loop for TAp73
and p53. Cell Death Differ. 8:1213–1223. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishimoto O, Kawahara C, Enjo K, et al:
Possible oncogenic potential of DeltaNp73: A newly identified
isoform of human p73. Cancer Res. 62:636–641. 2002.PubMed/NCBI
|
|
23
|
Tomasini R, Tsuchihara K, Wilhelm M, et
al: TAp73 knockout shows genomic instability with infertility and
tumor suppressor functions. Genes Dev. 22:2677–2691. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Concin N, Becker K, Slade N, et al:
Transdominant DeltaTAp73 isoforms are frequently up-regulated in
ovarian cancer. Evidence for their roles as epigenetic p53 inhibits
in vivo. Cancer Res. 64:2449–2460. 2004. View Article : Google Scholar : PubMed/NCBI
|