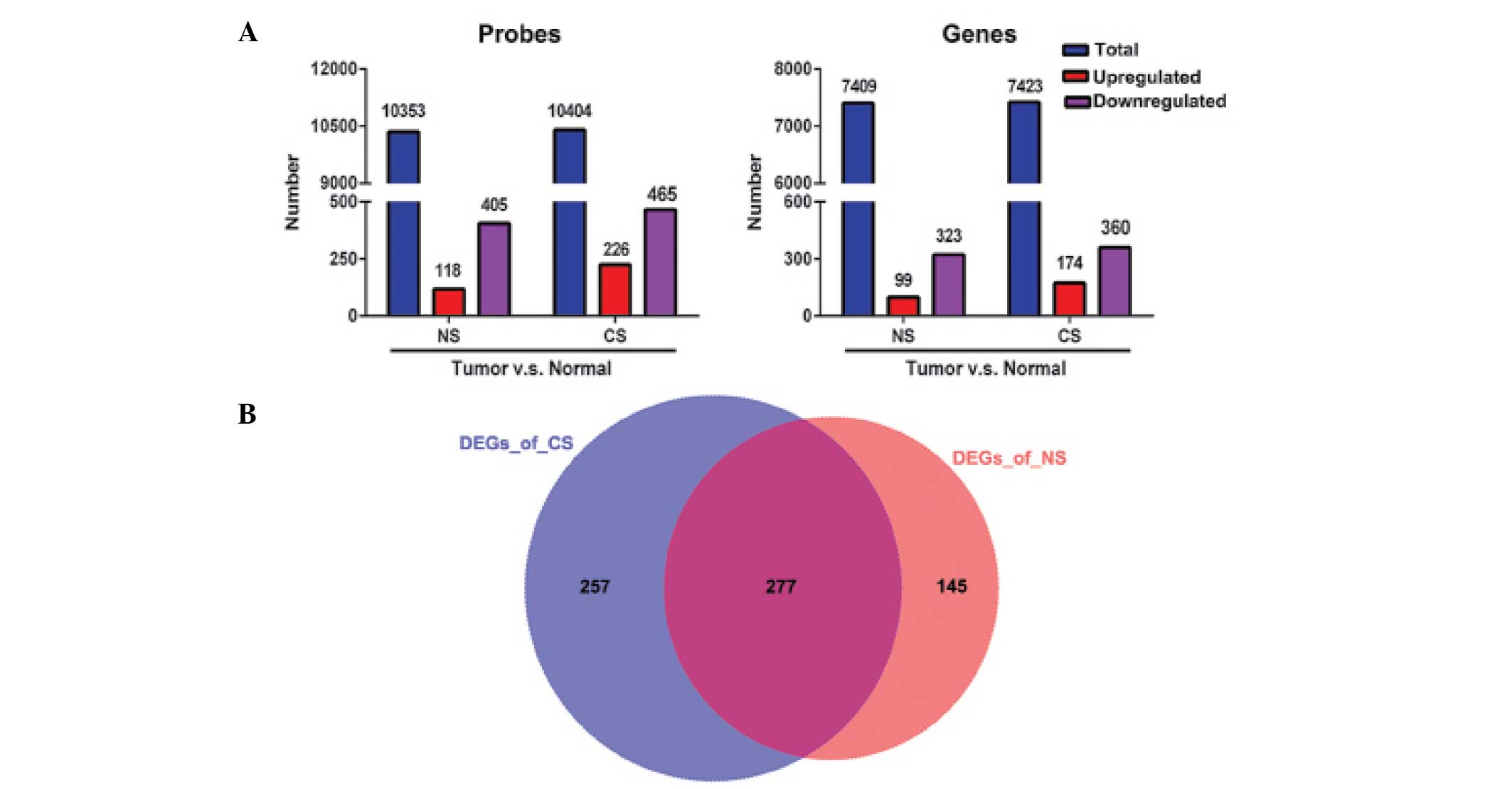
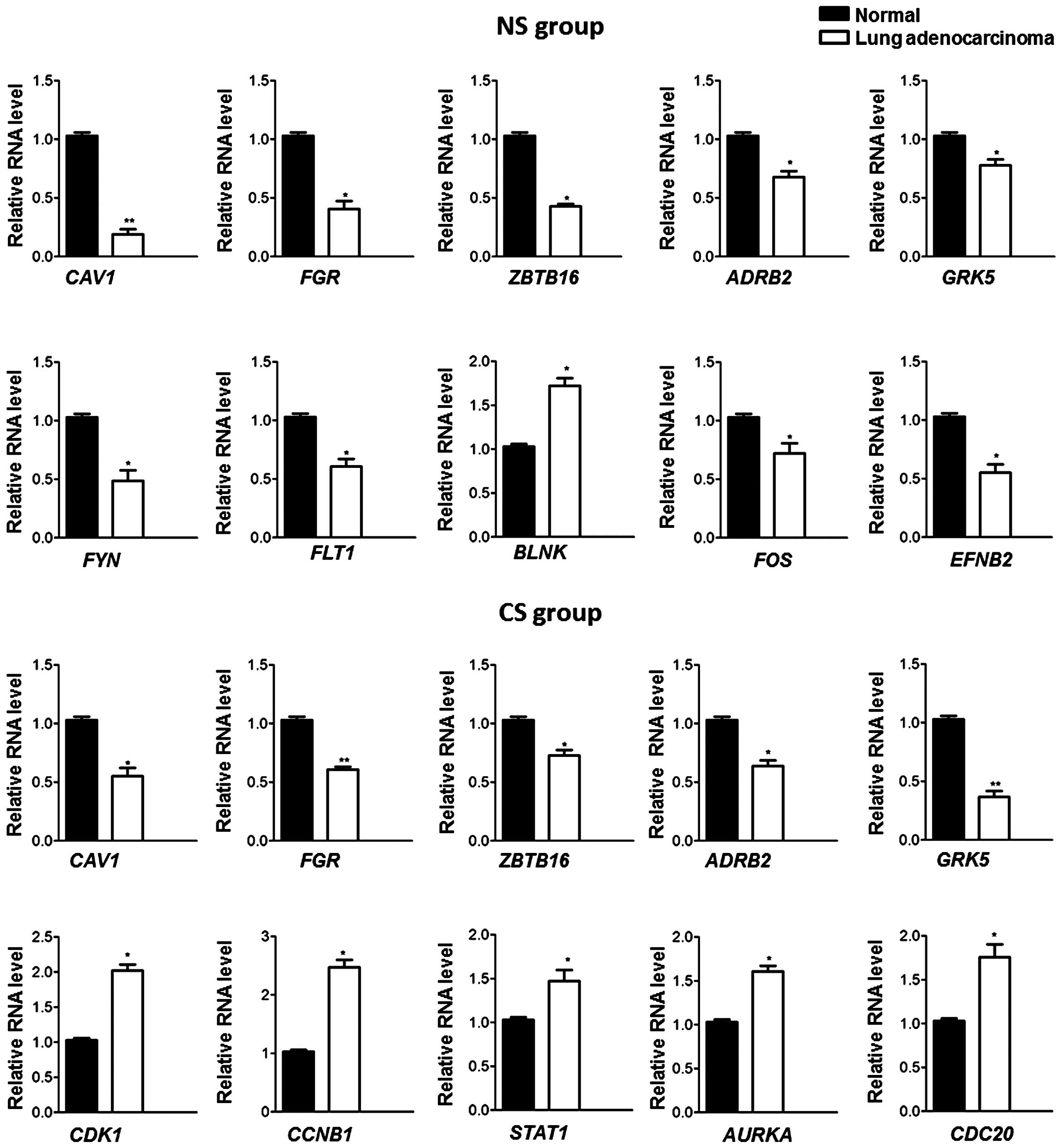
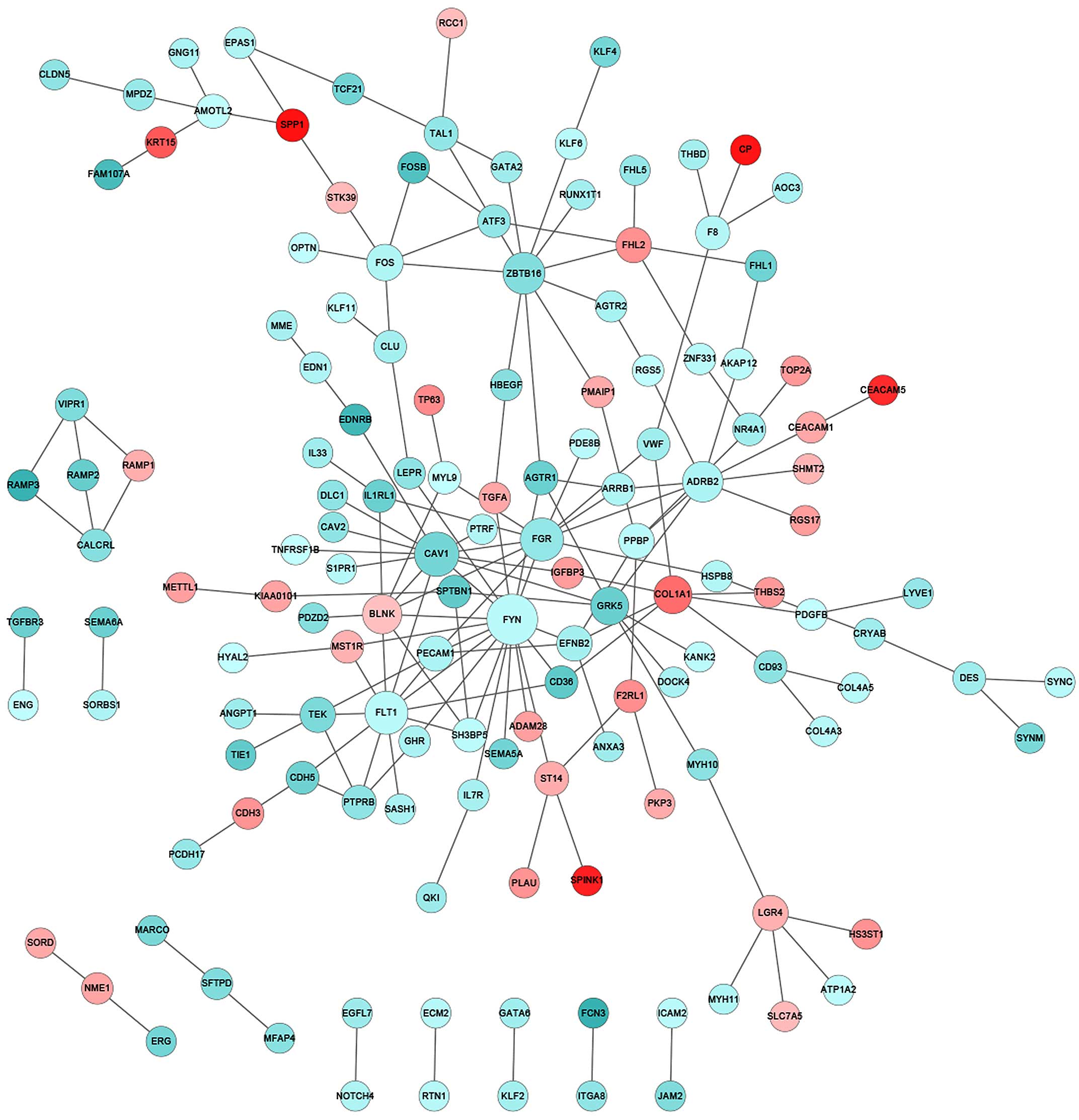
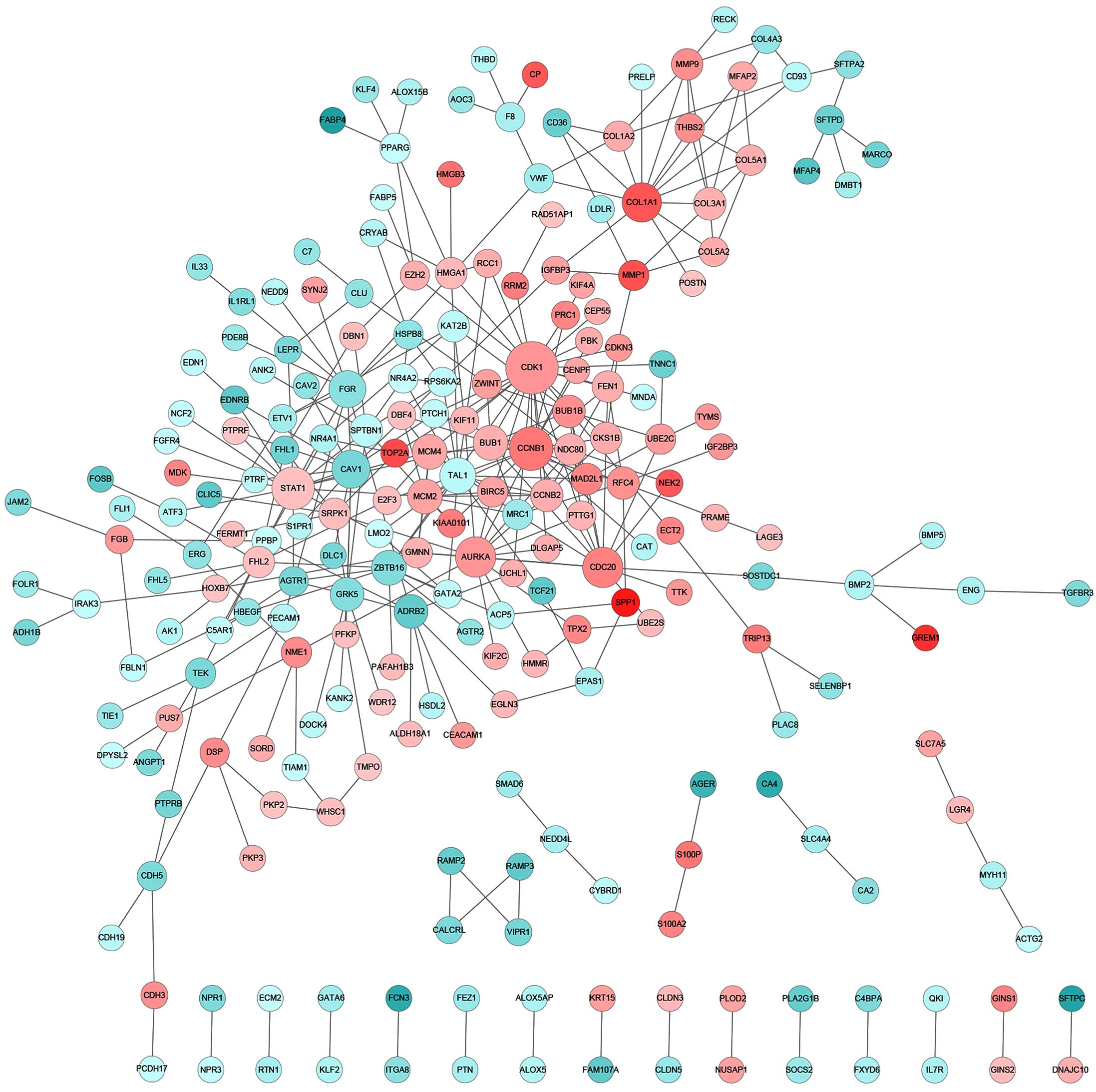
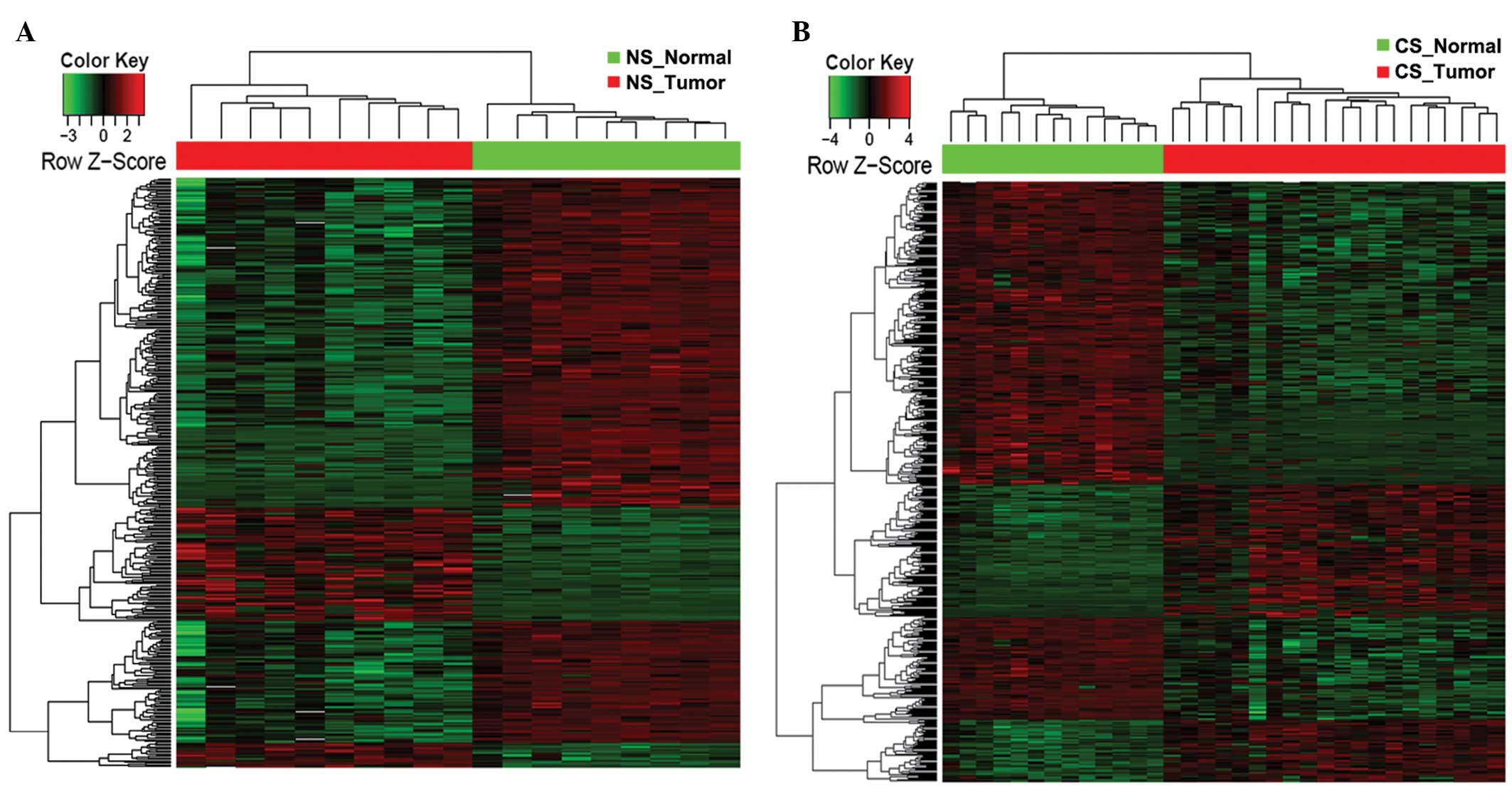
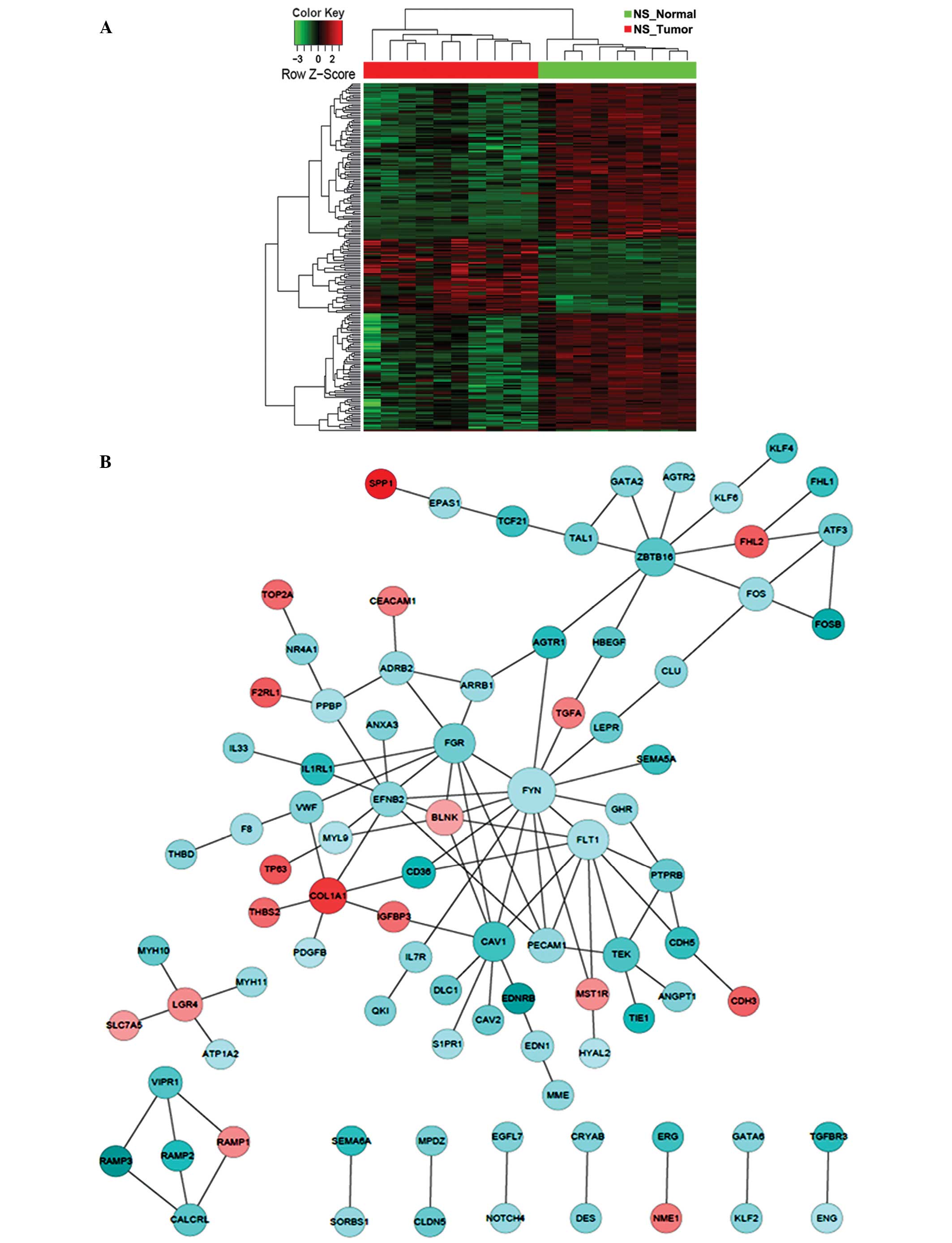
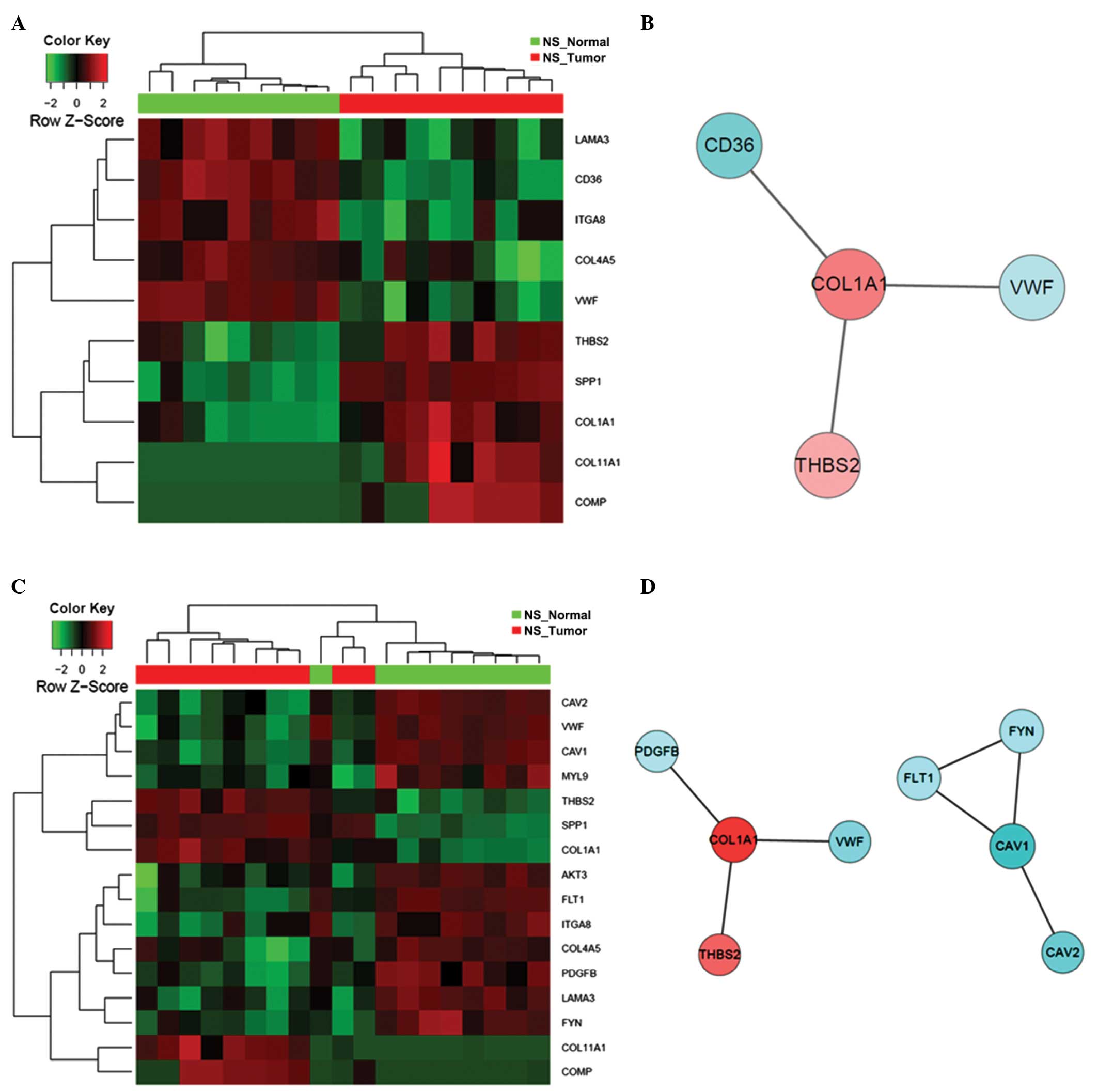
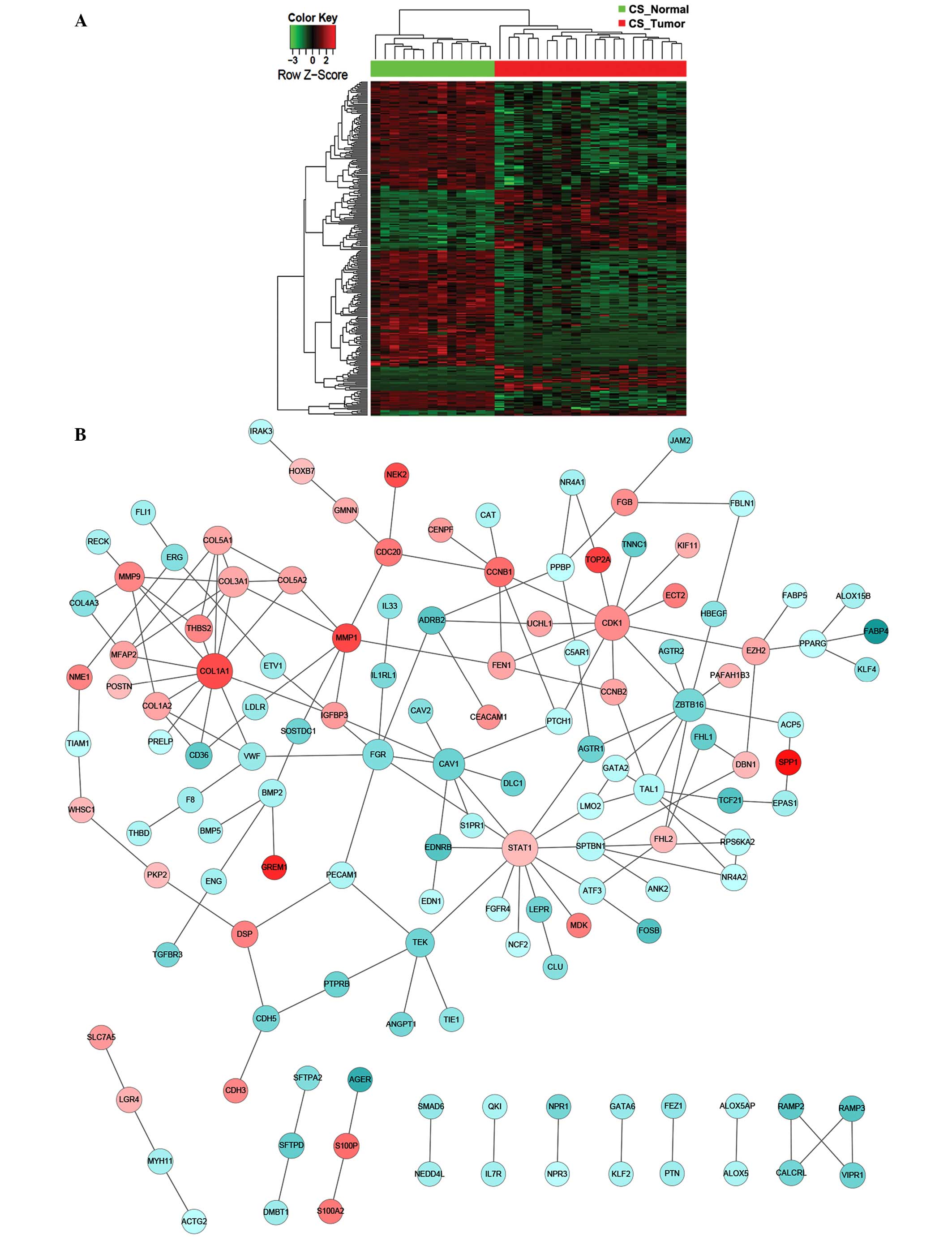
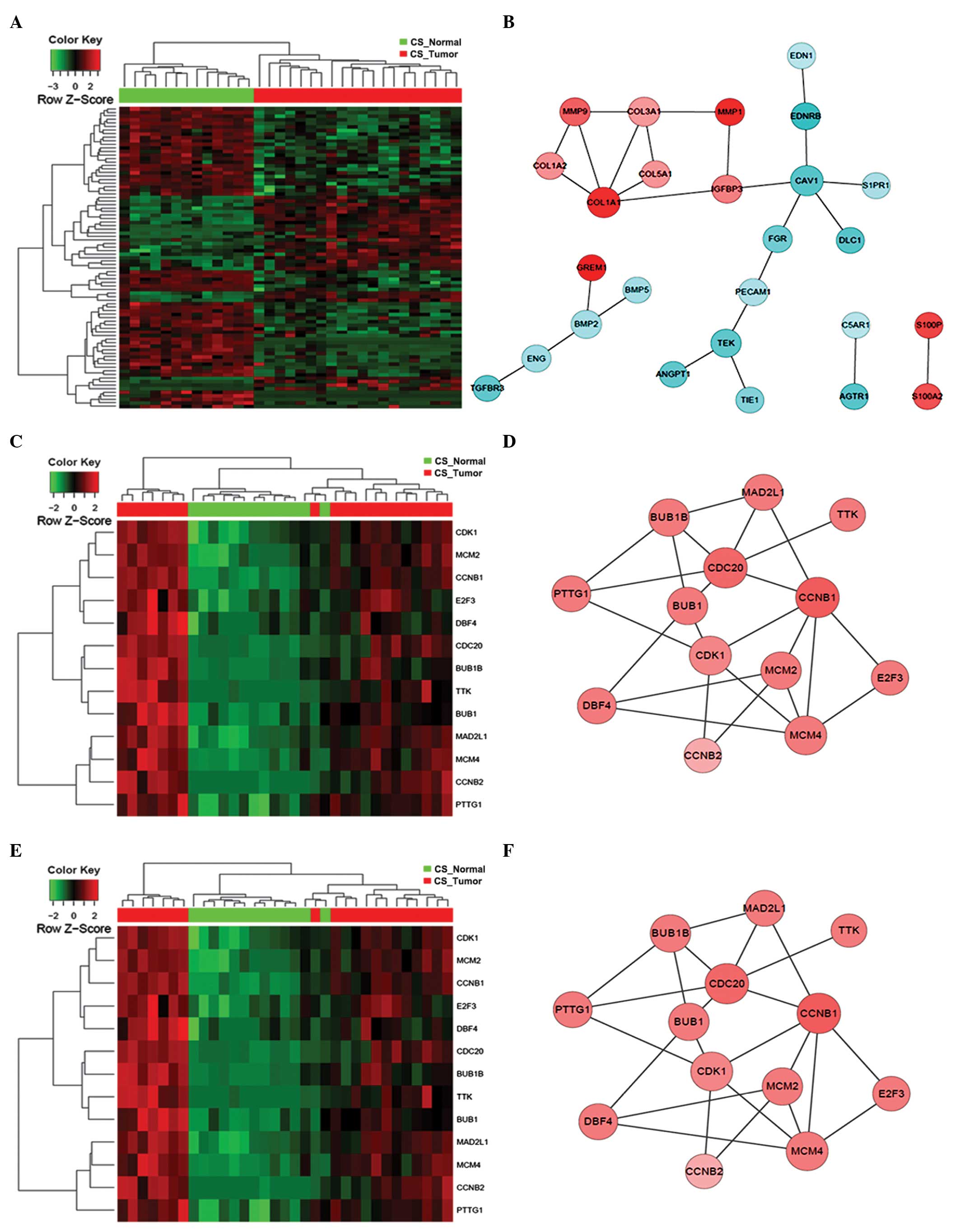
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Surveillance, Epidemiology, and End
Results Program, . SEER Stat Fact Sheets. Lung and Bronchus Cancer.
National Cancer Institute at the National Institutes of Health.
http://seer.cancer.gov/statfacts/html/lungb.htmlAccessed.
March 24–2015.
|
|
3
|
Peto R, Darby S, Deo H, Silcocks P,
Whitley E and Doll R: Smoking, smoking cessation, and lung cancer
in the UK since 1950: Combination of national statistics with two
case-control studies. BMJ. 321:323–329. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amos CI, Wu X, Broderick P, Gorlov IP, Gu
J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al:
Genome-wide association scan of tag SNPs identifies a
susceptibility locus for lung cancer at 15q25.1. Nat Genet.
40:616–622. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cornfield J, Haenszel W, Hammond EC, et
al: Smoking and lung cancer: Recent evidence and a discussion of
some questions. 1959. Int J Epidemiol. 38:1175–1191. 2009.
View Article : Google Scholar
|
|
6
|
Shigematsu H, Takahashi T, Nomura M,
Majmudar K, Suzuki M, Lee H, Wistuba II, Fong KM, Toyooka S,
Shimizu N, et al: Somatic mutations of the HER2 kinase domain in
lung adenocarcinomas. Cancer Res. 65:1642–1646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hecht SS: Tobacco smoke carcinogens and
lung cancer. J Natl Cancer Inst. 91:1194–1210. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Govindan R, Ding L, Griffith M, et al:
Genomic landscape of non-small cell lung cancer in smokers and
never-smokers. Cell. 150:1121–1134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Le Calvez F, Mukeria A, Hunt JD, Kelm O,
Hung RJ, Tanière P, Brennan P, Boffetta P, Zaridze DG and Hainaut
P: TP53 and KRAS mutation load and types in lung cancers in
relation to tobacco smoke: distinct patterns in never, former, and
current smokers. Cancer Res. 65:5076–5083. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Landi MT, Dracheva T, Rotunno M, et al:
Gene expression signature of cigarette smoking and its role in lung
adenocarcinoma development and survival. PLoS One. 3:e16512008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu J, Irizarry R, MacDonald J and Gentry
J: RIwcf JMJ. gcrma. Background Adjustment Using Sequence
Information. R package version 2.38.0.
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
January 20–2015.(Epub ahead of print). doi: 10.1093/nar/gkv007.
View Article : Google Scholar
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2013. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warnes GR, Bolker B, Bonebakker L,
Gentleman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller
S, Schwartz M and Venables B: gplots: Various R Programming Tools
for Plotting Data. R package version 2.16.0.
|
|
18
|
Carlson M: GO.db: A set of annotation maps
describing the entire Gene Ontology. R package version 3.0.0.
|
|
19
|
Carlson M: KEGG.db: A set of annotation
maps for KEGG. R package version 3.0.0.
|
|
20
|
Tenenbaum D: KEGGREST: Client-side REST
access to KEGG. R package version 1.6.4.
|
|
21
|
Mo YQ, Dai L, Zheng DH, Zhu LJ, Wei XN,
Pessler F, Shen J and Zhang BY: Synovial infiltration with
CD79a-positive B cells, but not other B cell lineage markers,
correlates with joint destruction in rheumatoid arthritis. J
Rheumatol. 38:2301–2308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han F, Gu D, Chen Q and Zhu H: Caveolin-1
acts as a tumor suppressor by down-regulating epidermal growth
factor receptor-mitogen-activated protein kinase signaling pathway
in pancreatic carcinoma cell lines. Pancreas. 38:766–774. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lobos-González L, Aguilar L, Diaz J, et
al: E-cadherin determines Caveolin-1 tumor suppression or
metastasis enhancing function in melanoma cells. Pigment Cell
Melanoma Res. 26:555–570. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rudin CM, Avila-Tang E, Harris CC, Herman
JG, Hirsch FR, Pao W, Schwartz AG, Vahakangas KH and Samet JM: Lung
cancer in never smokers: molecular profiles and therapeutic
implications. Clin Cancer Res. 15:5646–5661. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szymanowska-Narloch A, Jassem E, Skrzypski
M, Muley T, Meister M, Dienemann H, Taron M, Rosell R, Rzepko R,
Jarząb M, et al: Molecular profiles of non-small cell lung cancers
in cigarette smoking and never-smoking patients. Adv Med Sci.
58:196–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woenckhaus M, Klein-Hitpass L, Grepmeier
U, Merk J, Pfeifer M, Wild P, Bettstetter M, Wuensch P, Blaszyk H,
Hartmann A, et al: Smoking and cancer-related gene expression in
bronchial epithelium and non-small-cell lung cancers. J Pathol.
210:192–204. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spira A, Beane JE, Shah V, Steiling K, Liu
G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, et al:
Airway epithelial gene expression in the diagnostic evaluation of
smokers with suspect lung cancer. Nat Med. 13:361–366. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Staaf J, Jönsson G, Jönsson M, Karlsson A,
Isaksson S, Salomonsson A, Pettersson HM, Soller M, Ewers SB,
Johansson L, et al: Relation between smoking history and gene
expression profiles in lung adenocarcinomas. BMC Med Genomics.
5:222012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du X, Qian X, Papageorge A, Schetter AJ,
Vass WC, Liu X, Braverman R, Robles AI and Lowy DR: Functional
interaction of tumor suppressor DLC1 and caveolin-1 in cancer
cells. Cancer Res. 72:4405–4416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chanvorachote P and Chunhacha P:
Caveolin-1 regulates endothelial adhesion of lung cancer cells via
reactive oxygen species-dependent mechanism. PLoS One.
8:e574662013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Z, Sokolovska A, Seymour R, Sundberg
JP and Hogenesch H: SHARPIN is essential for cytokine production,
NF-κB signaling, and induction of Th1 differentiation by dendritic
cells. PLoS One. 7:e318092012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pancotti F, Roncuzzi L, Maggiolini M and
Gasperi-Campani A: Caveolin-1 silencing arrests the proliferation
of metastatic lung cancer cells through the inhibition of STAT3
signaling. Cell Signal. 24:1390–1397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luanpitpong S, Talbott SJ, Rojanasakul Y,
Nimmannit U, Pongrakhananon V, Wang L and Chanvorachote P:
Regulation of lung cancer cell migration and invasion by reactive
oxygen species and caveolin-1. J Biol Chem. 285:38832–38840. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Z, Potter CS, Sundberg JP and
Hogenesch H: SHARPIN is a key regulator of immune and inflammatory
responses. J Cell Mol Med. 16:2271–2279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lemjabbar H, Li D, Gallup M, Sidhu S,
Drori E and Basbaum C: Tobacco smoke-induced lung cell
proliferation mediated by tumor necrosis factor alpha-converting
enzyme and amphiregulin. J Biol Chem. 278:26202–26207. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luppi F, Aarbiou J, van Wetering S, et al:
Effects of cigarette smoke condensate on proliferation and wound
closure of bronchial epithelial cells in vitro: Role of
glutathione. Respir Res. 6:1402005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schaal C and Chellappan SP:
Nicotine-mediated cell proliferation and tumor progression in
smoking-related cancers. Mol Cancer Res. 12:14–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu C, Russell RM and Wang XD: Lycopene
supplementation prevents smoke-induced changes in p53, p53
phosphorylation, cell proliferation, and apoptosis in the gastric
mucosa of ferrets. J Nutr. 136:106–111. 2006.PubMed/NCBI
|
|
41
|
Dasgupta P, Rastogi S, Pillai S, et al:
Nicotine induces cell proliferation by beta-arrestin-mediated
activation of Src and Rb-Raf-1 pathways. J Clin Invest.
116:2208–2217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Potter CS, Wang Z, Silva KA, et al:
Chronic proliferative dermatitis in Sharpin null mice: development
of an autoinflammatory disease in the absence of B and T
lymphocytes and IL4/IL13 signaling. PLoS One. 9:e856662014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Conklin BS, Zhao W, Zhong DS and Chen C:
Nicotine and cotinine up-regulate vascular endothelial growth
factor expression in endothelial cells. Am J Pathol. 160:413–418.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jull BA, Plummer HK III and Schuller HM:
Nicotinic receptor-mediated activation by the tobacco-specific
nitrosamine NNK of a Raf-1/MAP kinase pathway, resulting in
phosphorylation of c-myc in human small cell lung carcinoma cells
and pulmonary neuroendocrine cells. J Cancer Res Clin Oncol.
127:707–717. 2001.PubMed/NCBI
|
|
45
|
Furrukh M: Tobacco Smoking and Lung
Cancer: Perception-changing facts. Sultan Qaboos Univ Med J.
13:345–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pfeifer GP, Denissenko MF, Olivier M, et
al: Tobacco smoke carcinogens, DNA damage and p53 mutations in
smoking-associated cancers. Oncogene. 21:7435–7451. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Husgafvel-Pursiainen K and Kannio A:
Cigarette smoking and p53 mutations in lung cancer and bladder
cancer. Environ Health Perspect. 104 (Suppl 3):553–556. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brennan JA, Boyle JO, Koch WM, Goodman SN,
Hruban RH, Eby YJ, Couch MJ, Forastiere AA and Sidransky D:
Association between cigarette smoking and mutation of the p53 gene
in squamous-cell carcinoma of the head and neck. N Engl J Med.
332:712–717. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Muller PA and Vousden KH: p53 mutations in
cancer. Nat Cell Biol. 15:2–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Muller PA and Vousden KH: Mutant p53 in
cancer: new functions and therapeutic opportunities. Cancer Cell.
25:304–317. 2014. View Article : Google Scholar : PubMed/NCBI
|