Introduction
Glioblastoma (GBM) is considered to be the most
aggressive and common type of primary malignant brain tumor in
adult humans (1). With the
administration of the standard treatment of maximal safe surgical
resection followed by radiotherapy combined with concomitant and
adjuvant temozolomide (TMZ) chemotherapy, newly diagnosed patients
with GBM demonstrate an average survival time of only 12–14 months
(2). Almost one-half of patients with
GBM do not survive the first year subsequent to diagnosis (3–6).
It has previously been hypothesized that GBM
initiation and recurrence is dependent upon a group of tumor stem
cells that are resistant to current standard treatments (7,8). Certain
studies have demonstrated that glioma stem cells (GSCs) are highly
resistant to radiotherapy due to the enhanced checkpoint response
to radiation (9). Additional
investigation of the pathways involved in radioresistance is
required.
In cancer cells, DNA damage may cause cell
apoptosis, which is the purpose of chemotherapy or radiotherapy.
Thus, the condition of DNA damage may affect radiosensitivity in
GSCs (10). To investigate the
alteration of the DNA damage signaling pathway in irradiated GSCs
and to profile the genes of this pathway, the present study
cultured the human glioblastoma U87 and U251 cell lines in
serum-free medium (SFM) and obtained U87 or U251-glioblastoma
stem-like cells (GSLCs). Irradiated U87 GSLCs were tested using
gene chip polymerase chain reaction (PCR) array for the human DNA
damage signaling pathway, and irradiated U251 GSLCs were used in
quantitative PCR (qPCR) confirmatory studies.
Materials and methods
Cell culture
U87 and U251 human glioblastoma cell lines were
purchased from American Type Culture Collection and cultured in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA,
USA) with 10% fetal bovine serum (FBS) supplemented with 1%
Penicillin-Streptomycin-Glutamine (100X; Gibco Life Technologies,
Carlsbad, CA, USA) at 37°C in a 5% CO2 atmosphere. U87
and U251 GSLCs were induced from U87 and U251 cells, respectively,
in SFM composed of DMEM/F12 (Gibco Life Technologies), 20 ng/ml
basic fibroblast growth factor (FGF), 20 ng/ml epidermal growth
factor (EGF; Gibco Life Technologies), 2 µg/ml heparin
(Sigma-Aldrich, St. Louis, MO, USA) and B27 supplement (50X;
Invitrogen, Carlsbad, CA, USA). The study was approved by the
ethics committee of the Second Affiliated Hospital of Soochow
University (Suzhou, China).
Cell growth
Cell growth analysis was performed using cell
counting kit-8 (CCK-8) from Dojindo Laboratories (Kumamoto,
Kumamoto, Japan), according to the manufacturer's instructions.
Briefly, the cells were cultured at a density of 5,000 cells/well
in 200 µl of culture medium and treated with 10 Gy X-ray radiation
or left untreated. The cells were then cultured in a humidified
incubator with a 5% CO2 atmosphere. After 72 h, 20 µl
CCK-8 solution was added to each well and the cells were incubated
for 1 h in the incubator. The optical density value was determined
by measuring the absorbance at 450 nm using an ELISA plate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell sorting analysis by flow
cytometry
Cells were cultured, collected and incubated with a
phycoerythrin-labeled mouse anti-human CD133 IgG1
monoclonal antibody (1:1,000; catalog no. 130-080-801; Miltenyi
Biotec GmbH, Bergisch Gladbach, Germany) or fluorescein
isothiocyanate-labeled mouse anti-human Nestin IgG1
monoclonal antibody (1:1,000; catalog no. MAB1259; R&D Systems,
Inc., Minneapolis, MN, USA) for flow cytometry for 1 h at 37°C.
Subsequent to washing, the cells were analyzed by
fluorescence-activated cell sorting (FACS) using a BD FACSAria Cell
Sorter (BD Biosciences, San Jose, CA, USA).
X-ray irradiation
The cells were cultured in SFM and grown as GSLCs.
The GSLCs were then exposed to 6 MV X-ray irradiation, produced by
a Siemens-Primus linear accelerator (Siemens, Munich, Bavaria,
Germany), at a dose rate of 2 Gy/min. The total dose administered
was 10 Gy. The GSLCs were then harvested 72 h later for PCR array
analysis or qPCR.
RNA isolation
The cells were cultured and harvested, and then
added to 1 ml TRIzol reagent (Invitrogen) prior to being
homogenized for 1 min at room temperature. Chloroform was added,
and the sample was mixed with thermal agitation for 15 sec,
followed by incubation at room temperature for 3 min and
centrifugation at 14,000 × g for 15 min at 4°C. The upper aqueous
phase was collected, and the RNeasy Micro kit (Qiagen, Valencia,
CA, USA) was used for the purification of total RNA from cell
samples, according to the manufacturer's instructions.
Human DNA damage signaling pathway PCR
array
The RNA samples were tested for the relative
expression of genes involved in DNA damage signaling by the Human
DNA Damage Signaling RT2 Profiler™ PCR Array
(SABiosciences, Frederick, MD, USA) according to the manufacturer's
instructions. Briefly, the RNA was reverse transcribed to form
cDNA. The samples were diluted in qPCR master mix (RT2 SYBR Green;
SABiosciences), according to the supplier's instructions, and
pipetted into 96-well PCR array plates. qPCR was performed in
technical duplicates (ABI Prism 7900HT Sequence Detection System;
Applied Biosystems, Foster City, CA, USA). Raw data from the
real-time PCR was uploaded using the RT2 Profiler PCR
Array Data Analysis template (SABiosciences). A large number of
gene expression analyses have been performed and the results
published using the arrays and web-based automated RT2
Profiler PCR Array Data Analysis method. The integrated web-based
software package for the PCR array system automatically performed
all comparative threshold cycle-based fold-change calculations from
the uploaded data. For these calculations, the average expression
of the reference gene β-actin was used for normalization of the
data. Subsequent to normalization, the relative expression of each
gene was averaged for the three samples in each cell group. Fold
changes in average gene expression were expressed as the difference
in expression of untreated U87 GSLCs compared with the difference
in expression of radiation treated cells.
RT-qPCR
The miScript Reverse Transcription kit (Qiagen) was
used to generate cDNA from RNA for qPCR. The cDNA template (100 ng)
was used to perform RT-qPCR analyses for XPA, RAD50, PPP1R15A and
α-tubulin using their specific forward primers and reverse primers,
according to the manufacturer's instructions (Qiagen). The 5′-3′
primer sequences were as follows: XPA forward,
ATGTAAAAGCAGCCCCAAAGA, and reverse, TGGCAAATCAAAGTGGTTCATA (191 bp
amplicon); RAD50 forward, GCGGAGTTTTGGAATAGAGGAC, and reverse,
GAGCAACCTTGGGATCGTGT (185 bp amplicon); PPP1R15A forward,
GATGGCATGTATGGTGAGCG, and reverse, GGTGTGATGGTGGATAAGAGAACT (208 bp
amplicon); and α-tubulin forward, AGATCATTGACCTCGTGTTGGA, and
reverse, ACCAGTTCCCCCACCAAAG (101 bp amplicon) (National Institutes
of Health, Bethesda, MD, USA). α-tubulin was used as a control to
normalize the levels of the genes. The StepOnePlus RT-PCR system
(Applied Biosystems) was used to perform qPCR, using a hot start at
95°C for 15 min, then denaturation at 95°C for 15 sec, with
annealing for 30 sec at 60°C and extension for 30 sec at 72°C for
40 cycles, followed by a melt curve analysis. Data analysis for the
differences in gene expression between the control and
radiation-treated cells was performed using Microsoft Excel
(Microsoft, Redmond, WA, USA). Gene expression was quantified using
the following equation: ΔΔCt = (Ctradiation-treated
sample - Ctinternal control) - (Ctcontrol
sample - Ctinternal control), where Ct is the mean
threshold cycle from each group. The fold change was calculated as
2−ΔΔCt.
Statistical analysis
All data were expressed as the mean ± standard
deviation and analyzed using one-way analysis of variance, followed
by the Student-Newman-Keuls post-hoc test. P<0.05 was considered
to indicate a statistically significant difference. SPSS 15.0
(SPSS, Inc., Chicago, IL, USA) was used for all statistical
analyses.
Results
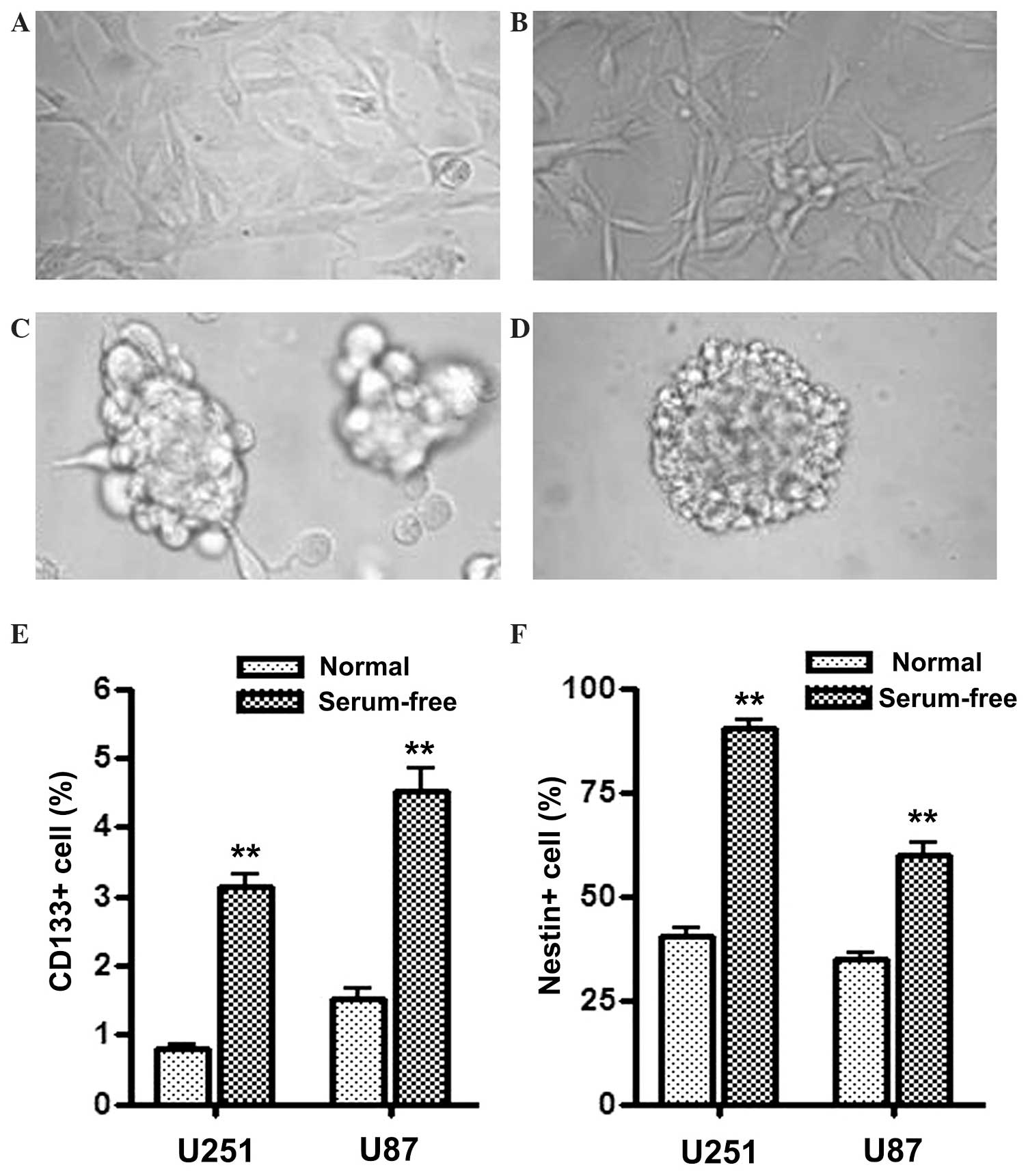
Serum-free culture induces U251 and
U87 cells with GSC properties
U251 and U87 human glioblastoma cells were cultured
in serum-free medium supplemented with EGF and FGF2, which
comprises tumor sphere medium, and demonstrated sphere formation
(Fig. 1C and D). Flow cytometry was
used to score the proportion of CD133+ and
Nestin+ cells in normal medium or serum-free medium
cultured cells. As the results in Fig. 1E
and F demonstrate, the proportion of CD133+ and
Nestin+ cells were all significantly increased in U251
and U87 cells cultured in serum-free medium compared with the cells
cultured in normal medium. Since CD133 and Nestin are the most
widely used markers to identify GSCs, the present results indicate
that U251 and U87 cells cultured in tumor sphere medium became
enriched in properties of GSCs. In the current study, these sphere
formatted cells cultured in serum-free medium are referred to as
GSLCs.
Radiation treatment alters the
expression profile of DNA damage-associated genes in GSLCs
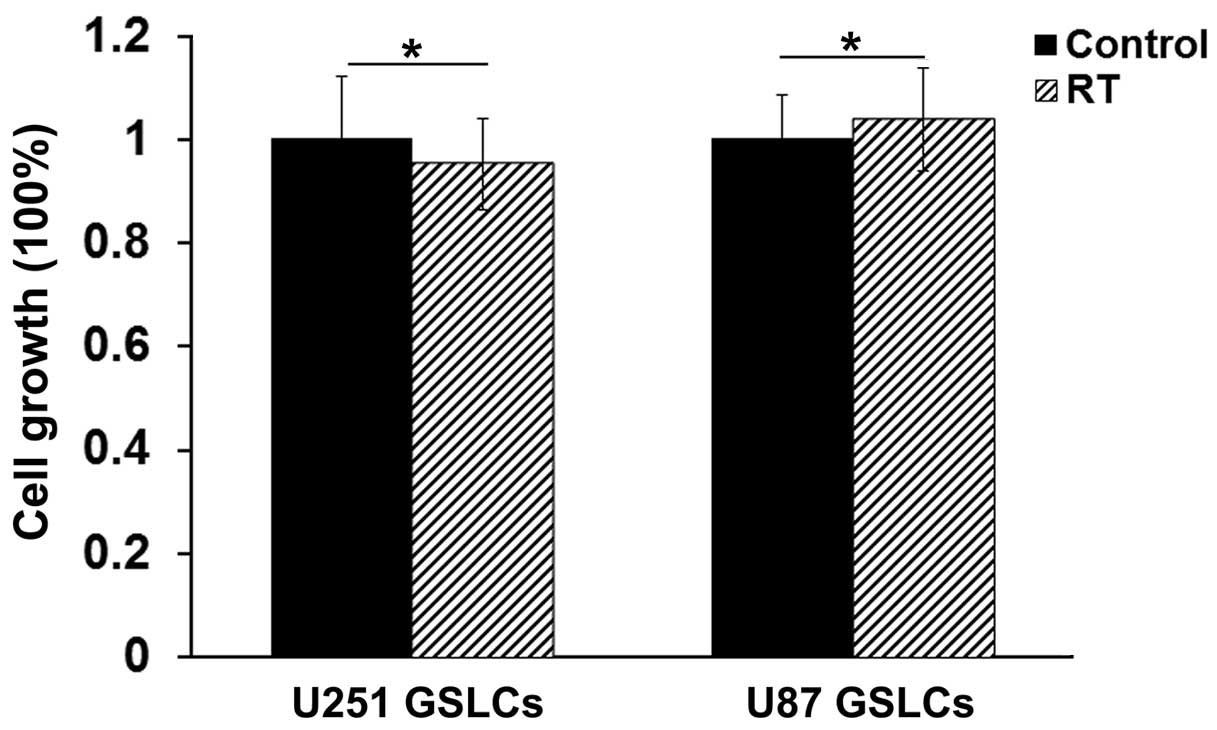
The U87 GSLCs were cultured to a population of
2–5×106 cells and treated with 10 Gy radiation. After 72
h, the cells were harvested. Cell growth was detected using a CCK-8
kit, which revealed that 10 Gy radiation induced no cell growth
suppression, indicating the radioresistance of U87 GSLCs (Fig. 2). The detection of the expression
profiles of DNA damage-associated genes in cells were performed
using DNA damage signaling pathway chip PCR array, which profiled
the histone modification status, also termed histone code, of 90
genes involved in DNA damage signaling pathways. The detected genes
are listed in Table I, with H7-9
acting as the reverse transcription control and H10-12 acting as
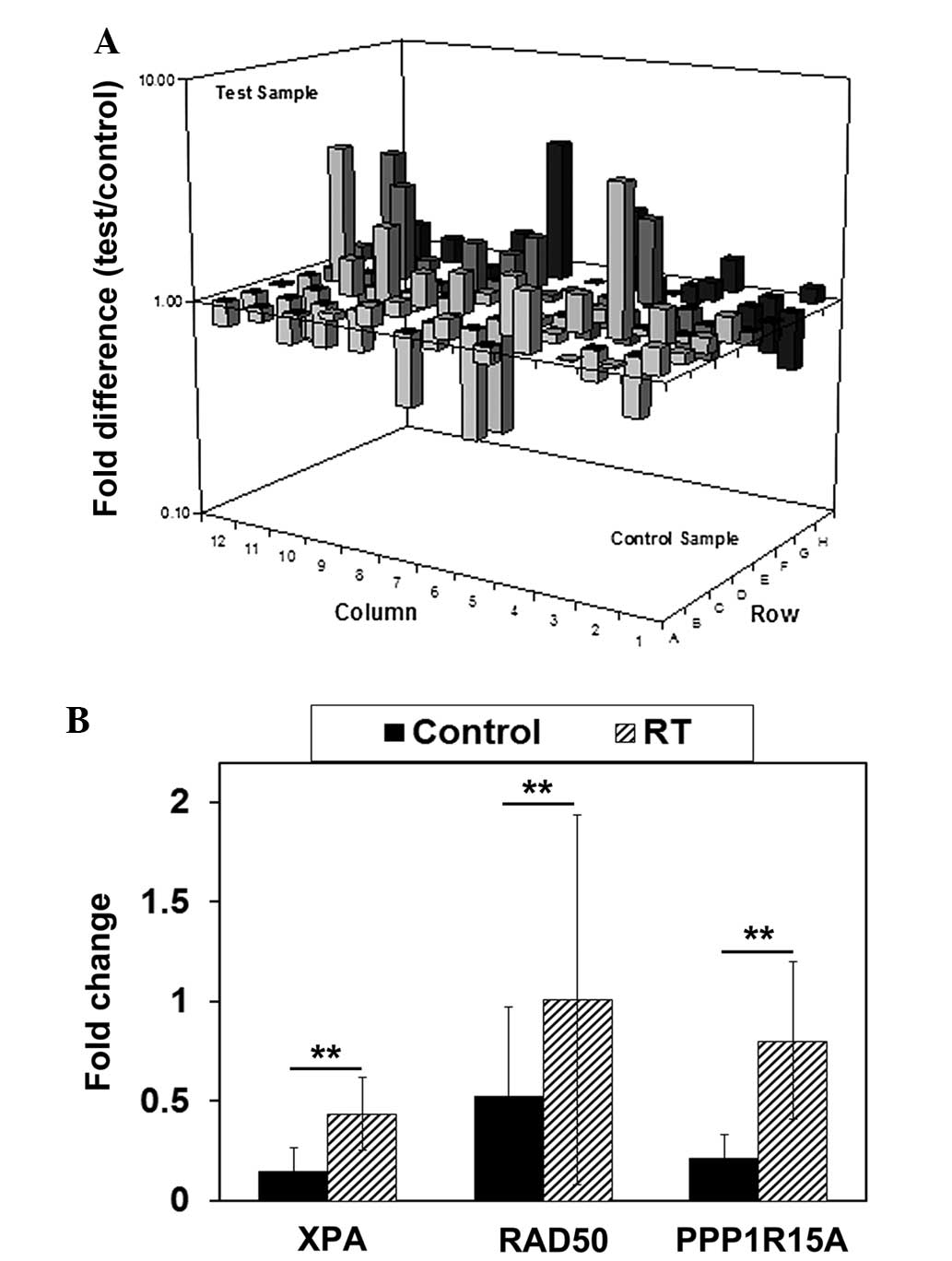
the PCR positive control. Fig. 3A
reports the alteration of the expression profile of DNA
damage-associated genes. Gene expression levels that were increased
≥2-fold in the irradiated U87 GSLCs compared with the untreated U87
GSLCs are listed in Table II.
 | Table I.Sets of genes analyzed in the Human
DNA Damage Signaling RT2 Profiler™ polymerase chain
reaction array. |
Table I.
Sets of genes analyzed in the Human
DNA Damage Signaling RT2 Profiler™ polymerase chain
reaction array.
| Set | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|
| A | ABL1 | ANKRD17 | APEX1 | ATM | ATR | ATRX | BRCA1 | BTG2 | CCNH | CDK7 | CHEK1 | CHEK2 |
| B | CIB1 | CIDEA | CRY1 | DDB1 | DDIT3 | DMC1 | ERCC1 | ERCC2 | EXO1 | FANCG | FEN1 | XRCC6 |
| C | GADD45A | GADD45G | GML | GTF2H1 | GTF2H2 | GTSE1 | HUS1 | IGHMBP2 | IP6K3 | XRCC6BP1 | LIG1 | MAP2K6 |
| D | MAPK12 | MBD4 | MLH1 | MLH3 | MNAT1 | MPG | MRE11A | MSH2 | MSH3 | MUTYH | N4BP2 | NBN |
| E | NTHL | OGG1 | PCBP4 | PCNA | AIFM1 | PMS1 | PMS2 | PMS2L3 | PNKP | PPP1R15A | PRKDC | RAD1 |
| F | RAD17 | RAD18 | RAD21 | RAD50 | RAD51 | RAD51L1 | RAD9A | RBBP8 | REV1 | RPA1 | SEMA4A | SESN1 |
| G | SMC1A | SUMO1 | TP53 | TP73 | TREX1 | UNG | XPA | XPC | XRCC1 | XRCC2 | XRCC3 | ZAK |
| H | B2M | HPRT1 | RPL13A | GAPDH | ACTB | HGDC | RTC | RTC | RTC | PPC | PPC | PPC |
 | Table II.Genes that demonstrated a >2-fold
increase in expression in irradiated cells compared with the
untreated U87 glioblastoma stem-like cells. |
Table II.
Genes that demonstrated a >2-fold
increase in expression in irradiated cells compared with the
untreated U87 glioblastoma stem-like cells.
| Gene | Full name | Location | Fold change |
|---|
| BRCA1 | Breast cancer 1,
early onset | 17q21-q24 | −2.13 |
| DMC1 | DMC1 dosage
suppressor of mck1 homolog, meiosis-specific homologous
recombination (yeast) | 22q13.1 | −3.11 |
| GML |
Glycosylphosphatidylinositol anchored
molecule like | 8q24.3 |
4.71 |
| GTSE1 | G-2 and S-phase
expressed 1 | 22q13.2-q13.3 | −3.24 |
| IP6K3 | Inositol
hexakisphosphate kinase 3 | 6p21.31 |
2.19 |
| N4BP2 | NEDD4 binding
protein 2 | 4p14 |
4.24 |
| NBN | Nibrin | 8q21-q24 | −2.05 |
| PPP1R15A | Protein phosphatase
1, regulatory subunit 15A | 19q13.2 |
2.74 |
| RAD50 | RAD50 homolog
(S. cerevisiae) | 5q23-q31 |
2.42 |
| SEMA4A | Sema domain,
immunoglobulin domain (Ig), transmembrane domain (TM) and short
cytoplasmic domain, (semaphorin) 4A | 1q22 |
3.46 |
| TREX1 | Three prime repair
exonuclease 1 | 3p21.31 |
2.34 |
| XPA | Xeroderma
pigmentosum, complementation group A | 9q22.3 |
4.31 |
Confirmatory studies by TaqMan
qPCR
To confirm the results of the PCR array, the RT-qPCR
assay was performed using U251 GSLCs to assess the expression of
the XPA, RAD50 and PPP1R15A genes. The
expression of these genes increased >2-fold and the genes were
associated with radiation-induced DNA damage, as determined by gene
functional searching from Entrez Gene and the published literature
in PubMed Central (National Center for Biotechnology Information,
Bethesda, MD, USA). To detect the genes, the U251 GSLCs were
cultured to a population of 2–5×106 cells and treated
with 10 Gy radiation. After 72 h, the cells were respectively
harvested and the qPCR was performed, as well as the cell growth
detection (Fig. 2). As the results in
Fig. 3B demonstrate, after 72 h the
expression of all three genes had significantly increased
(P<0.01).
Discussion
Cancer stem cells are considered to be a
subpopulation of cancer cells that is highly enriched with
tumorigenic potential (9). GSCs have
been found to demonstrate improved survival subsequent to
irradiation and chemotherapy and contribute to the recurrence of
GBM (11). In addition, GSCs
frequently reside in perivascular and hypoxic regions, actively
promoting angiogenesis and facilitating the survival of cancer
cells in harsh environments (12).
The presence of GSCs was considered to be the origin of the
difficulty in treating GBM. Previously, GSCs were reported to
isolate, in the form of spheres, from glioma tissues cultured in
SFM containing the stem cell mitogens EGF and FGF, which is the
same method used to isolate neural stem cells from brain tissue
(13). In the present study, GSLCs
were isolated from the human GBM U87 and U251 cell lines using the
previously described GSCs-isolation SFM method, and the GSLCs were
characterized by sphere formation ability and positivity for CD133
or other putative stem markers such as Sox2 and Nestin (13). Flow cytometry was used to score the
proportion of CD133+ and Nestin+ cells, and
it was found that each of these proportions was significantly
increased in GSLCs cultured in serum-free medium compared with
those cultured in normal medium. Since CD133 and Nestin are
considered to be valuable stem cell-specific markers for
determining the clinical outcome of glioma patients, these markers
were used in the present study for cell identification (14).
The identified GSLCs isolated from the U87 and U251
cells were subsequently used in additional investigations. The
cells were irradiated with 10 Gy X-rays after 72 h, and the U87 and
U251 GSLCs demonstrated radioresistance with no cell growth
suppression. The DNA damage signaling pathway chip PCR array, which
profiled the histone modification status, also termed the histone
code, of 90 genes involved in DNA damage signaling pathways, was
performed on the irradiated U87 GSLCs. According to the PCR array
results, the expression of three genes, consisting of XPA,
RAD50 and PPP1R15A, was increased >2-fold and
these genes were involved in radiation-associated DNA damage. These
genes were selected to undergo qPCR confirmatory investigation in
U251 GSLCs.
XPA encodes a zinc finger protein involved in
DNA excision repair. The XPA protein is a component of the
nucleotide excision repair (NER) complex, which is responsible for
the repair of ultraviolet (UV) radiation-induced photoproducts and
DNA adducts induced by chemical carcinogens (15). The XPA protein is one of eight factors
that were found to be deficient in XP disorders, which are
characterized by an increased sensitivity to UV radiation and a
predisposition to development of skin cancers (16). Functionally, XPA is considered to play
roles in verifying DNA damage, stabilizing repair intermediates,
and recruiting other NER factors to the damaged DNA (17).
The protein encoded by RAD50 is involved in
DNA double-strand break repair. The RAD50 protein forms a complex
with Mre11/NBS1 to form the Mre11/RAD50/NBS1 (MRN) complex
(18). The protein complex binds to
DNA and exhibits numerous enzymatic activities that are required
for the non-homologous joining of DNA ends. The MRN complex is a
conserved protein complex that plays a key role in the response to
DNA damage (19). Kuroda1 et
al (20) demonstrated that
degradation of MRN complex results in the inability to repair DNA
double-strand breaks (DSB) and leads to radiosensitization, which
indicates that RAD50 plays a key role in radioresistance.
PPP1R15A, also termed GADD34, is a
member of a group of genes that demonstrate increased transcript
levels following stressful growth arrest conditions and treatment
with DNA-damaging agents (21). The
expression of the PPP1R15A protein increases in response to medium
depletion or DNA damage, heat shock and several other treatments
that induce apoptosis (22).
Overexpression of PPP1R15A enhances the apoptotic response
to DNA damage following ionizing radiation (23). The transcriptional induction of
PPP1R15A is strongly associated with apoptosis (24).
The present data obtained from RT-qPCR confirmatory
studies in U251 GSLCs revealed that 72 h after the administration
of 10 Gy radiation, the expression of all three genes was
significantly increased, with a 1.98-fold increase in XPA
expression, 1.67-fold increase in RAD50 expression and
2.78-fold increase in PPP1R15A expression. The upregulation
of the three genes in irradiated GSLCs after 72 h may indicate the
alteration of DNA damage repair responses. The induction of
XPA, RAD50 and PPP1R15A by X-ray treatment may
confer radioresistance in GSLCs. In conclusion, the present study
revealed the alteration of the DNA damage signaling pathway profile
in radiation treated GSLCs. Out of the genes that demonstrated a
significant increase in expression, the XPA, RAD50
and PPP1R15A radiation-associated genes were selected for
additional study. The expression of these three genes was
significantly increased in U87 and U251 radiation-resistant GSLCs,
indicating three potential targets for overcoming GSLCs
radioresistance. It is necessary to investigate these genes
separately in radiation treated GSCs.
Acknowledgements
The authors would like to thank the Department of
Radiotherapy at the Second Affiliated Hospital of Soochow
University for providing the radiotherapy equipment and techniques.
This study was supported by the Research Fund of the National
Natural Science Foundation of China (grant nos. 81472739, 81272799
and 81172400), the Scientific and Technological Development Program
of Suzhou, China (grant nos. SYS201477 and SYSD2012090), and the
Jiangsu Provincial Special Program of Clinical Medical Science
(grant no. BL2014040).
References
|
1
|
DeAngelis LM: Brain tumors. N Engl J Med.
344:114–123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Mason WP, van den Bent MJ, et al:
European Organisation for Research and Treatment of Cancer Brain
Tumor and Radiotherapy Groups; National Cancer Institute of Canada
Clinical Trials Group: Radiotherapy plus concomitant and adjuvant
temozolomide for glioblastoma. N Engl J Med. 352:987–996. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ravizza R, Cereda E, Monti E and Gariboldi
MB: The piperidine nitroxide Tempol potentiates the cytotoxic
effects of temozolomide in human glioblastoma cells. Int J Oncol.
25:1817–1822. 2004.PubMed/NCBI
|
|
4
|
Bruyere C, Mijatovic T, Lonez C, et al:
Temozolomide-induced modification of the CXC chemokine network in
experimental gliomas. Int J Oncol. 38:1453–1464. 2011.PubMed/NCBI
|
|
5
|
Ashizawa T, Akiyama Y, Miyata H, et al:
Effect of the STAT3 inhibitor STX-0119 on the proliferation of a
temozolomide-resistant glioblastoma cell line. Int J Oncol.
45:411–418. 2014.PubMed/NCBI
|
|
6
|
Shi L, Zhang S, Feng K, et al:
MicroRNA-125b-2 confers human glioblastoma stem cells resistance to
temozolomide through the mitochondrial pathway of apoptosis. Int J
Oncol. 40:119–129. 2012.PubMed/NCBI
|
|
7
|
Fouse SD, Nakamura JL, James CD, Chang S
and Costello JF: Response of primary glioblastoma cells to therapy
is patient specific and independent of cancer stem cell phenotype.
Neuro Oncol. 16:361–371. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Estrada-Bernal A, Palanichamy K, Ray
Chaudhury A and Van Brocklyn JR: Induction of brain tumor stem cell
apoptosis by FTY720: a potential therapeutic agent for
glioblastoma. Neuro Oncol. 14:405–415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rich JN: Cancer stem cells in radiation
resistance. Cancer Res. 67:8980–8984. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bao S, Wu Q, McLendon RE, et al: Glioma
stem cells promote radioresistance by preferential activation of
the DNA damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Lai G, Wan F, Xiao Z, Zeng L, et al:
Knockdown of checkpoint kinase 1 is associated with the increased
radiosensitivity of glioblastoma stem-like cells. Tohoku J Exp Med.
226:267–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Soeda A, Park M, Lee D, et al: Hypoxia
promotes expansion of the CD133-positive glioma stem cells through
activation of HIF-1alpha. Oncogene. 28:3949–3959. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Christensen K, Aaberg-Jessen C, Andersen
C, Goplen D, Bjerkvig R and Kristensen BW: Immunohistochemical
expression of stem cell, endothelial cell and chemosensitivity
markers in primary glioma spheroids cultured in serum-containing
and serum-free medium. Neurosurgery. 66:933–947. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Song T, Yang L, et al: Nestin and
CD133: Valuable stem cell-specific markers for determining clinical
outcome of glioma patients. J Exp Clin Cancer Res. 27(85)2008.
|
|
15
|
Li Z, Musich PR, Cartwright BM, Wang H and
Zou Y: UV-induced nuclear import of XPA is mediated by
importin-alpha4 in an ATR-dependent manner. PloS One. 8:e682972013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Musich PR and Zou Y: Differential
DNA damage responses in p53 proficient and deficient cells:
Cisplatin-induced nuclear import of XPA is independent of ATR
checkpoint in p53-deficient lung cancer cells. Int J Biochem Mol
Biol. 2:138–145. 2011.PubMed/NCBI
|
|
17
|
Li Z, Musich PR, Serrano MA, Dong Z and
Zou Y: XPA-mediated regulation of global nucleotide excision repair
by ATR is p53-dependent and occurs primarily in S-phase. PloS One.
6:e283262011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gatei M, Jakob B, Chen P, et al: ATM
protein-dependent phosphorylation of Rad50 protein regulates DNA
repair and cell cycle control. J Biol Chem. 286:31542–31556. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Machida K, McNamara G, Cheng KT, et al:
Hepatitis C virus inhibits DNA damage repair through reactive
oxygen and nitrogen species and by interfering with the
ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and
hepatocytes. J Immunol. 185:6985–6998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kuroda S, Fujiwara T, Shirakawa Y, et al:
Telomerase-dependent oncolytic adenovirus sensitizes human cancer
cells to ionizing radiation via inhibition of DNA repair machinery.
Cancer Res. 70:9339–9348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grishin AV, Azhipa O, Semenov I and Corey
SJ: Interaction between growth arrest-DNA damage protein 34 and Src
kinase Lyn negatively regulates genotoxic apoptosis. Proc Natl Acad
Sci USA. 98:10172–10177. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hollander MC, Zhan Q, Bae I and Fornace AJ
Jr: Mammalian GADD34, an apoptosis- and DNA damage-inducible gene.
J Biol Chem. 272:13731–13737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hollander MC, Sheikh MS, Yu K, et al:
Activation of Gadd34 by diverse apoptotic signals and suppression
of its growth inhibitory effects by apoptotic inhibitors. Int J
Cancer. 96:22–31. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adler HT, Chinery R, Wu DY, et al:
Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and
associate with the GADD34 and hSNF5/INI1 proteins. Mol Cell Biol.
19:7050–7060. 1999.PubMed/NCBI
|