Introduction
Ovarian cancer is one of the leading causes of
cancer incidence and cancer-associated mortality in women. Surgery
combined with chemotherapy is the standard therapeutic strategy,
and recurrence and chemoresistance in ovarian cancer patients are
the main factors affecting the prognosis (1). Therefore, anti-resistance has become a
prominent target for research into ovarian cancer (2,3).
The combination data mining method increases the
probability of identifying the biological processes and functional
candidate genes that could represent the high-throughput data and
have a great effect on the studied disease. A number of studies
have revealed that applying in silico bioinformatic
approaches in mining data from high-throughput microarray profiles
is reliable and effective in predicting disease-causing biomarkers
and has high accuracy (4,5). Qu et al (6) used microarray technology to profile
microRNA (miRNA/miR) expression between CNE-2R and its parental
cell line, CNE-2, and miR-205 was found to contribute to the
radioresistance of nasopharyngeal carcinoma by directly targeting
PTEN. In order to find feasible approaches for solving the
chemoresistance in ovarian carcinoma, more and more studies have
been performed in the last decade. Using microarray profiles,
multiple potential biomarkers have been reported to be involved in
chemoresistant ovarian carcinoma, including miR-106a, miR-591
(7), miR-23b, miR-27a (8), ARID1A (9)
and Notch3 (10). However, the
biological mechanisms of the biomarkers in chemoresistant ovarian
carcinoma remain unclear.
The present study aimed to extract
differentially-expressed miRNAs from microarray datasets from the
Gene Expression Omnibus (GEO) database to probe their biological
function in the development and progression of chemoresistant
ovarian carcinoma. Information retrieved via miRNAs expression
data, PPI interaction network construction and pathway enrichment
analysis was combined to screen out potential biomarkers for
chemoresistant ovarian carcinoma. This research will assist in
disclosing the biomarkers of chemoresistance in ovarian
carcinoma.
Materials and methods
miRNA expression profiles
The miRNA expression profile of the GSE43867 dataset
was obtained from the GEO database (http:www.ncbi.nlm.nih.gov/geo/), which is based on the
GPL16566 Applied Biosystems TaqMan Array Human miRNA A/B Cards v2.0
platform (Applied Biosystems Life Technologies, Foster City, CA,
USA). This dataset included the miRNA profile expression
microarrays from formalin-fixed and paraffin-embedded blocks of 86
chemotherapy-treated cases with serous epithelial ovarian
carcinomas, which were submitted by Vecchione et al
(11).
Screening of differentially-expressed
miRNAs
GEO2R (http:www.ncbi.nlm.nih.gov/geo/geo2r/) is an
interactive web tool that performs comparisons on original
submitter-supplied processed data tables using the GEO query and
limma R packages from the Bioconductor project (12). GEO2R was used to analyze the published
microarray data of the GSE43867 dataset from the GEO database. In
total, 86 chemotherapy-treated patients with serous epithelial
ovarian carcinomas were divided into two groups: The response group
consisted of 36 complete response cases and 12 partial response
cases, while the non-response group consisted of 10 stable cases
and 28 progressive disease cases. The results were downloaded in
text format, and the miRNAs that met the cut-off criteria of
P<0.05 and a |log fold-change| of >1.0 were screened out as
differentially-expressed miRNAs.
Prediction of target genes of
differentially-expressed miRNAs
Targets of miRNAs are predicted by an online target
prediction tool, TargetScan 6.2 (http:www.targetscan.org/) (13,14), which
predicts the biological targets of miRNAs by searching for the
presence of conserved 8mer and 7mer sites that match the seed
region of each miRNA. A prediction score of >0.5 is selected as
a criterion for target genes with each miRNA.
Construction of a protein-protein
interaction (PPI) network
STRING is a database of known and predicted PPIs
based on the sources derived from the genomic context,
high-throughput experiments, coexpression and previous knowledge
(15). STRING quantitatively
integrates interaction data from these sources for a large number
of organisms, and transfers information between these organisms
where applicable (16). The latest
version, STRING9.1 (http:string-db.org/), covers 5,214,234 proteins from
1,133 organisms (17). In the present
study, a PPI network of the miRNA target genes was constructed by
STRING9.1, and highly-correlated genes/proteins (confidence score,
>0.7) were selected as inclusion criteria for PPI network
analysis.
Functional enrichment and pathway
enrichment analysis
Functional enrichment (GO biological process terms)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis (KEGG and Panther pathways) were performed for
the genes in the PPI network using the GeneCodis3 web tool
(http:genecodis.dacya.ucm.es/) (18,19). and
the statistical test used for the enrichment was based on the
hypergeometric distribution to compute P-values, which were
corrected by the Benjamini and Hochberg false discovery rate method
for multiple hypothesis testing (α=0.05). Only those terms with a
value of P<0.05 and a count number of ≥5 genes were selected for
analysis.
Results
Differentially-expressed miRNAs
A total of 6 differentially-expressed miRNAs were
identified in the chemotherapy response cases compared with the
non-response cases (control) in the serous epithelial ovarian
carcinomas according to the criteria. The 6 miRNAs were
hsa-miR-760, hsa-miR-483-5p, hsa-miR-766, hsa-miR-198,
hsa-miR-129-3p and hsa-miR-642. The information for these 6
differentially-expressed miRNAs is presented in Table I.
 | Table I.Differentially-expressed miRNAs
obtained from the GSE43867 dataset. |
Table I.
Differentially-expressed miRNAs
obtained from the GSE43867 dataset.
| miRNA | |logFC| | P-value |
|---|
| hsa-miR-642 | 1.01956 | 0.00848 |
| hsa-miR-198 | 1.23491 | 0.01075 |
| hsa-miR-483-5p | 1.35478 | 0.01347 |
| hsa-miR-129-3p | 1.11770 | 0.02684 |
| hsa-miR-760 | 1.59264 | 0.03878 |
| hsa-miR-766 | 1.24657 | 0.03971 |
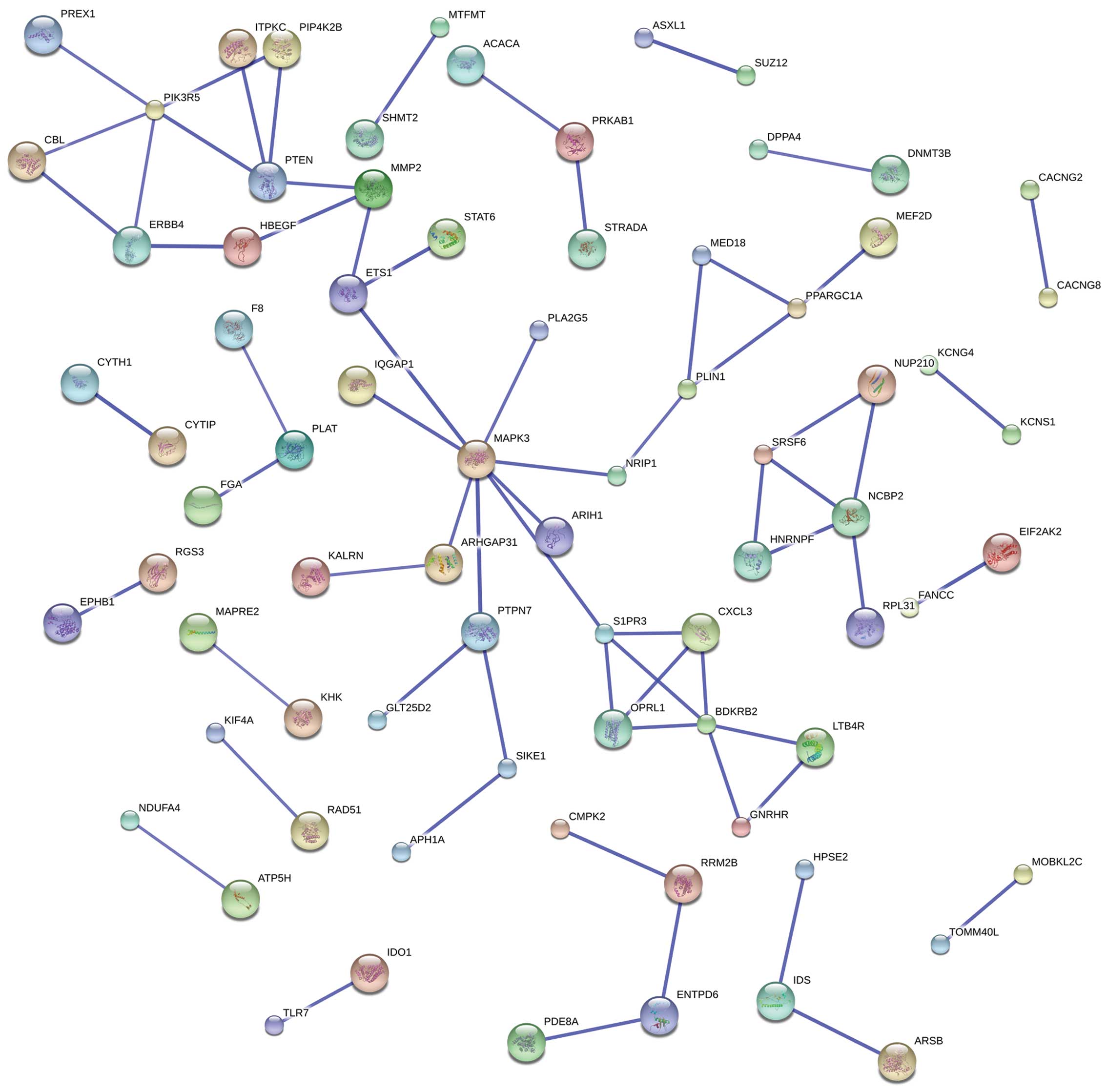
Target genes and PPI network
construction
A total of 317 target genes met the criteria of the
6 differentially-expressed miRNAs. The target genes were uploaded
to the STRING online tool: A total of 67 interactions were found to
exist among 76 proteins (genes) through the PPI network analysis,
and 6 of them were potential key genes (PIK3R5, PTEN, MAPK3, S1PR3,
BDKRB2 and NCBP2). Additionally, 37 proteins/genes were involved in
the construction of the MAPK3 and NCBP2 module PPI network, as
shown in Fig. 1.
GO term enrichment and KEGG
analysis
The GeneCodis 3 online tool was used to characterize
the biological functions of the genes that were included in the PPI
network of the aforementioned potential key genes. Functional
analysis demonstrated that the genes in this module could be
enriched into 5 functional GO terms, as shown in Table II. The target genes were identified
to be associated with GO categories such as regulation of
transcription, apoptotic process and signal transduction, as shown
in Table II. Based on KEGG pathway
analysis, the target genes were found to be involved in the
gonadotropin-releasing hormone signaling pathway, the ErbB
signaling pathway and in other cancer pathways, as well as in the
regulation of the actin cytoskeleton, as shown in Table III.
 | Table II.Significant GO terms obtained from the
microRNA targets in the protein-protein interaction network. |
Table II.
Significant GO terms obtained from the
microRNA targets in the protein-protein interaction network.
| GO term | Genes | P-value |
|---|
| GO:0048011-nerve
growth factor receptor signaling pathway | MAPK3, APH1A, KALRN,
PTEN, PREX1 | 0.000876 |
| GO:0045944-positive
regulation of transcription from RNA polymerase II promoter | NRIP1, MAPK3,
PPARGC1A, MEF2D, STAT6, ETS1 | 0.004176 |
| GO:0045893-positive
regulation of transcription, DNA-dependent | NRIP1, MAPK3,
PPARGC1A, ERBB4, ETS1 | 0.005182 |
| GO:0006915-apoptotic
process | APH1A, KALRN, PREX1,
MEF2D, ERBB4 | 0.008394 |
| GO:0007165-signal
transduction | IQGAP1, KALRN,
ARHGAP31, ERBB4, HBEGF, STAT6 | 0.013630 |
 | Table III.KEGG pathways obtained from the miRNA
targets. |
Table III.
KEGG pathways obtained from the miRNA
targets.
| Pathway | Genes | P-value |
|---|
| Kegg:04912 GnRH
signaling pathway | MAPK3, MMP2, HBEGF,
PLA2G5, GNRHR |
2.78×10−12 |
| Kegg:04012 ErbB
signaling pathway | MAPK3, PIK3R5, ERBB4,
HBEGF, CBL |
3.05×10−12 |
| Kegg:05200 Pathways
in cancer | MAPK3, PIK3R5,
MMP2, PTEN, ETS1, CBL |
3.21×10−10 |
| Kegg:04810
Regulation of actin cytoskeleton | MAPK3, PIK3R5,
BDKRB2, PIP4K2B, IQGAP1 |
5.97×10−10 |
Discussion
The present study identified 6
differentially-expressed miRNAs from the GEO GSE43867 dataset,
which included a miRNA profile expression microarray for
chemoresistant serous epithelial ovarian carcinomas, by
bioinformatics analysis. In order to confirm the functions of these
miRNAs, the target genes were predicted and the PPI network was
constructed. Finally, functional annotations and pathway analysis
were conducted to determine the biological mechanisms contributing
to chemoresistant ovarian carcinoma.
The 6 differentially-expressed miRNAs were
hsa-miR-760, hsa-miR-483-5p, hsa-miR-766, hsa-miR-198,
hsa-miR-129-3p and hsa-miR-642. Iwaya et al (20) reported that miR-760 was downregulated
in the bone marrow and primary tumor of advanced gastric cancer,
while Wang et al (21) found
that plasma miR-601 and miR-760 were of high value for
discriminating advanced adenomas from normal controls. Zheng et
al (22) found high expression
levels of miR-483-5p in tumor tissues, plasma and cell lines of
nasopharyngeal carcinoma patients. The results of a study by Wang
et al (23) confirmed that
miR-483-5p suppresses the proliferation of glioma cells via
directly targeting extracellular signal-regulated kinase 1 (ERK1).
The microarray results of a study by Yu et al (24) indicated that miR-483-5p was
upregulated in ovarian serous carcinoma at stage III compared with
stage I. miR-766 has been shown to be associated with esophageal
cancer, cutaneous squamous cell carcinoma and lung adenocarcinoma
(25–27), while miRNA-198 has been reported to be
of high value for predicting the clinical outcomes of cancer
(28,29). Several studies have demonstrated the
use of miR-129 as a diagnostic and prognostic biomarker for various
tumors, such as renal cell carcinoma and gastric cancer (30,31). It
has also been reported that increasing miR-642 improves cisplatin
sensitivity in advanced bladder cancer and cell lines (32).
In order to ascertain the function of the
differentially-expressed miRNAs in the present study, the target
genes of these miRNAs were retrieved by TargetScan 6.2, and 317
target genes were obtained. Through the construction of a PPI
network of the target genes of 6 differentially expressed miRNAs,
76 genes/proteins were shown to be involved in the PPI network, and
6 of these were screened out as potential key genes for
chemoresistant ovarian carcinoma. PIK3R5, PTEN, MAPK3, S1PR3,
BDKRB2 and NCBP2 formed a module in the PPI network construction,
indicating that the two genes, PIK3R5 and MAPK3, may play important
roles in the development of chemoresistance in ovarian carcinoma.
Results obtained from GO biological process and pathway enrichment
analyses also lead to the identification of MAPK3 and PIK3R5 as key
genes, which were involved in the majority of GO terms and KEGG
pathways associated with chemoresistance in ovarian carcinoma.
MAPK3, also known as ERK1, is a member of the
mitogen-activated protein kinase family. Amsterdam et al
(33) reported that intense
phosphorylated ERK1 and ERK2 was detected in the peripheral areas
of stage II ovarian carcinoma. Jeong et al (34) found that ERK1/2 are important in
docetaxel resistance in MCF-7 spheroids. In recent years, MAPK/ERK1
has been reported to be associated with various cancer types, and
it is believed that this gene participates in the process of cancer
metastasis (35–37). PIK3R5 is a protein-coding gene. PIK3R5
has been reported to be associated with ataxia-oculomotor apraxia
(38). Shull et al (39) reported that PIK3R5 mutations
co-occurred with BRAF mutations, indicating that the gene may be a
potential chemotherapeutic target for melanoma patients resistant
to BRAF inhibitors. Through functional enrichment analysis, the
present study identified the aforementioned pathways associated
with the hub genes and their regulators. Therefore, MAPK3 and
PIK3R5 may be selected as therapeutic targets for chemoresistant
ovarian carcinoma. We speculate that MAPK3 and PIK3R5 may be
associated with the occurrence and development of chemoresistant
ovarian carcinoma.
In conclusion, a bioinformatic approach was applied
to identify the differentially-expressed miRNAs in serous
epithelial ovarian carcinomas samples of chemotherapy-treated
responsive cases compared to those of non-responsive cases. The
results suggested that MAPK3 and PIK3R5 may be associated with
chemoresistant ovarian carcinoma. However, as of yet, there are no
other studies to prove that MAPK3 and PIK3R5 are associated with
chemoresistant ovarian carcinoma. The present study may provide
novel insights into the molecular mechanism of chemoresistant
ovarian carcinoma, thus, further studies regarding the association
of the two genes and chemoresistant ovarian carcinoma are
required.
References
|
1
|
Pignata S, Cannella L, Leopardo D, Pisano
C, Bruni GS and Facchini G: Chemotherapy in epithelial ovarian
cancer. Cancer Lett. 303:73–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khaider NG, Lane D, Matte I, Rancourt C
and Piché A: Targeted ovarian cancer treatment: The TRAILs of
resistance. Am J Cancer Res. 2:75–92. 2012.PubMed/NCBI
|
|
3
|
Kigawa J: New strategy for overcoming
resistance to chemotherapy of ovarian cancer. Yonago Acta Med.
56:43–50. 2013.PubMed/NCBI
|
|
4
|
Saei AA and Omidi Y: A glance at DNA
microarray technology and applications. Bioimpacts. 1:75–86.
2011.PubMed/NCBI
|
|
5
|
Cao WJ, Wu HL, He BS, Zhang YS and Zhang
ZY: Analysis of long non-coding RNA expression profiles in gastric
cancer. World J Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qu C, Liang Z, Huang J, Zhao R, Su C, Wang
S, Wang X, Zhang R, Lee MH and Yang H: MiR-205 determines the
radioresistance of human nasopharyngeal carcinoma by directly
targeting PTEN. Cell Cycle. 11:785–796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huh JH, Kim TH, Kim K, Song JA, Jung YJ,
Jeong JY, Lee MJ, Kim YK, Lee DH and An HJ: Dysregulation of
miR-106a and miR-591 confers paclitaxel resistance to ovarian
cancer. Br J Cancer. 109:452–461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH,
Kwon YD, Lee C, Kim OJ and An HJ: MicroRNAs overexpressed in
ovarian ALDH1-positive cells are associated with chemoresistance. J
Ovarian Res. 6:182013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Katagiri A, Nakayama K, Rahman MT, Rahman
M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K,
Kobayashi H, et al: Loss of ARID1A expression is related to shorter
progression-free survival and chemoresistance in ovarian clear cell
carcinoma. Mod Pathol. 25:282–288. 2012.PubMed/NCBI
|
|
10
|
Rahman MT, Nakayama K, Rahman M, Katagiri
H, Katagiri A, Ishibashi T, Ishikawa M, Iida K, Nakayama S, Otsuki
Y and Miyazaki K: Notch3 overexpression as potential therapeutic
target in advanced stage chemoresistant ovarian cancer. Am J Clin
Pathol. 138:535–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vecchione A, Belletti B, Lovat F, Volinia
S, Chiappetta G, Giglio S, Sonego M, Cirombella R, Onesti EC,
Pellegrini P, et al: A microRNA signature defines chemoresistance
in ovarian cancer through modulation of angiogenesis. Proc Natl
Acad Sci USA. 110:9845–9850. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grimson A, Farh KK, Johnston WK,
GarrettEngele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
TabasMadrid D, NogalesCadenas R and
Pascual-Montano A: GeneCodis3: A non-redundant and modular
enrichment analysis tool for functional genomics. Nucleic Acids
Res. 40:W478–W483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
NogalesCadenas R, CarmonaSaez P, Vazquez
M, Vicente C, Yang X, Tirado F, Carazo JM and Pascual-Montano A:
GeneCodis: Interpreting gene lists through enrichment analysis and
integration of diverse biological information. Nucleic Acids Res.
37:W317–W322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iwaya T, Fukagawa T, Suzuki Y, Takahashi
Y, Sawada G, Ishibashi M, Kurashige J, Sudo T, Tanaka F, Shibata K,
et al: Contrasting expression patterns of histone mRNA and microRNA
760 in patients with gastric cancer. Clin Cancer Res. 19:6438–6449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang
L, Huang D, Tan C, Sheng W and Du X: Plasma miR-601 and miR-760 are
novel biomarkers for the early detection of colorectal cancer. PLoS
One. 7:e443982012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng XH, Cui C, Ruan HL, Xue WQ, Zhang
SD, Hu YZ, Zhou XX and Jia WH: Plasma microRNA profiles of
nasopharyngeal carcinoma patients reveal miR-548q and miR-483-5p as
potential biomarkers. Chin J Cancer. 33:330–338. 2014.PubMed/NCBI
|
|
23
|
Wang L, Shi M, Hou S, Ding B, Liu L, Ji X,
Zhang J and Deng Y: MiR-483-5p suppresses the proliferation of
glioma cells via directly targeting ERK1. FEBS Lett. 586:1312–1317.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu X, Zhang X, Bi T, Ding Y, Zhao J, Wang
C, Jia T, Han D, Guo G, Wang B, et al: MiRNA expression signature
for potentially predicting the prognosis of ovarian serous
carcinoma. Tumour Biol. 34:3501–3508. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M, Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
26
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sand M, Skrygan M, Georgas D, Sand D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Microarray analysis of
microRNA expression in cutaneous squamous cell carcinoma. J
Dermatol Sci. 68:119–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qi B, Yao WJ, Zhao BS, Qin XG, Wang Y,
Wang WJ, Wang TY, Liu SG and Li HC: Involvement of microRNA-198
overexpression in the poor prognosis of esophageal cancer. Asian
Pac J Cancer Prev. 14:5073–5076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han HS, Yun J, Lim SN, Han JH, Lee KH, Kim
ST, Kang MH, Son SM, Lee YM, Choi SY, et al: Downregulation of
cell-free miR-198 as a diagnostic biomarker for lung
adenocarcinoma-associated malignant pleural effusion. Int J Cancer.
133:645–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai
CH, Kao HW, Fang WL, Huang KH, Chan WC and Lin WC: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Ruan A, Wang X, Han W, Wang R, Lou
N, Ruan H, Qiu B, Yang H and Zhang X: miR-129-3p, as a diagnostic
and prognostic biomarker for renal cell carcinoma, attenuates cell
migration and invasion via downregulating multiple
metastasis-related genes. J Cancer Res Clin Oncol. 140:1295–1304.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nordentoft I, BirkenkampDemtroder K,
Agerbaek M, Theodorescu D, Ostenfeld MS, Hartmann A, Borre M,
Ørntoft TF and Dyrskjøt L: miRNAs associated with chemo-sensitivity
in cell lines and in advanced bladder cancer. BMC Med Genomics.
5:402012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Amsterdam A, Shezen E, Raanan C, Schreiber
L, Prus D, Slilat Y, BenArie A and Seger R: Nuclear localization of
phosphorylated ERK1 and ERK2 as markers for the progression of
ovarian cancer. Int J Oncol. 39:649–656. 2011.PubMed/NCBI
|
|
34
|
Jeong EK, Lee SY, Jeon HM, Ju MK, Kim CH
and Kang HS: Role of extracellular signal-regulated kinase (ERK)
1/2 in multicellular resistance to docetaxel in MCF-7 cells. Int J
Oncol. 37:655–661. 2010.PubMed/NCBI
|
|
35
|
Guégan JP, Ezan F, Théret N, Langouët S
and Baffet G: MAPK signaling in cisplatin-induced death:
Predominant role of ERK1 over ERK2 in human hepatocellular
carcinoma cells. Carcinogenesis. 34:38–47. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang D, Li X, Yao Z, Wei C, Ning N and Li
J: GABAergic signaling facilitates breast cancer metastasis by
promoting ERK1/2-dependent phosphorylation. Cancer Lett.
348:100–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu YY, Ma TL, Ge ZJ, Lin J, Ding WL, Feng
JK, Zhou SJ, Chen GC, Tan YF and Cui GX: JWA gene regulates PANC-1
pancreatic cancer cell behaviors through MEK-ERK1/2 of the MAPK
signaling pathway. Oncol Lett. 8:1859–1863. 2014.PubMed/NCBI
|
|
38
|
Al Tassan N, Khalil D, Shinwari J, Al
Sharif L, Bavi P, Abduljaleel Z, Abu Dhaim N, Magrashi A, Bobis S,
Ahmed H, et al: A missense mutation in PIK3R5 gene in a family with
ataxia and oculomotor apraxia. Hum Mutat. 33:351–354. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shull AY, LathamSchwark A, Ramasamy P,
Leskoske K, Oroian D, Birtwistle MR and Buckhaults PJ: Novel
somatic mutations to PI3 K pathway genes in metastatic melanoma.
PLoS One. 7:e433692012. View Article : Google Scholar : PubMed/NCBI
|