Introduction
Numerous studies have reported that the repopulation
of malignant cells, such as through radiation-induced cancer
progression, limits the effectiveness of radiation therapy
(1–5).
Malaise et al (6) first
revealed that the regrowth of irradiated transplantable mouse
fibrosarcoma cells was faster compared with the regrowth of
non-irradiated cells. Withers et al (4) performed a retrospective analysis and
concluded that the repopulation of squamous cell carcinoma of the
head and neck accelerated following a lag period of ~4 weeks
subsequent to the initiation of radiotherapy. One possible
interpretation of this study is that the protracted treatment
schedule may permit a constant rate of repopulation throughout the
administration of conventional fractionated radiotherapy. However,
the molecular basis of the repopulation phenomenon remains
unclear.
Over the previous decade, accumulated data have
demonstrated that the 5′ and 3′-untranslated regions (UTRs) play
important roles in the expression of several genes, particularly in
the post-transcriptional state (7–9). A
previous study has demonstrated the reciprocal regulation of
hypoxia-inducible factor (HIF)-1α expression in a 5′- and
3′-UTR-dependent fashion (10). This
previous observation also suggested a possible correlation between
UTR-dependent regulation of HIF-1α and cancer malignancy.
The present study aimed to demonstrate the enhanced
expression of several transcription factors involved in cancer stem
cell maintenance through a 5′-UTR-dependent mechanism in irradiated
mouse fibrosarcoma cells.
Materials and methods
Plasmids and reagents
Luciferase reporter plasmids (PathDetect pAP-1and
PathDetect p53) were purchased from Agilent Technologies (Santa
Clara, CA, USA). Hypoxia responsible elements, Ets binding
sequences and 536 bp (1457–1992 bp) of the human matrix
metalloproteinase 1 (MMP1) enhancer sequence (GenBank ID,
AY769434.1) were cloned into a pGL3 basic vector (Promega, Madison,
WI, USA). Full length HIF-1α, which consisted of 294 bp of the
5′-UTR, 2,481 bp of the coding region and 1,195 bp of the 3′-UTR of
HIF-1α, was cloned into the pcDNA3 vector (Invitrogen, Carlsbad,
CA, USA) and designated as pcDNA HIF-5C3. pcDNA HIF-5C lacked the
3′-UTR of HIF-1α and pcDNA HIF-C3 lacked the 5′-UTR of HIF-1α.
pcDNA HIF-C contained only the coding region for HIF-1α (10). The full-length expression plasmids for
mouse c-fos and Ets2 were purchased from Origene (Rockville, MD,
USA). Four 3′-UTR sequences, consisting of the 3′-UTR HIF-1α, cMyc,
Ets2 and c-fos sequences, were cloned downstream of the luciferase
gene of the pmir GLO vector (Promega). Cobalt(II) chloride
hexahydrate and G418 disulfate salt were purchased from
Sigma-Aldrich (St. Louis, MO, USA).
Cell culture and cell cloning
The transplantable fibrosarcoma QRsP cell line,
which was established by the present authors and is described
elsewhere (11,12) and NIH3T3 cells (RCB0150: Riken,
Tsukuba, Japan), was cultured with Dulbecco's modified Eagle's
medium (DMEM) containing 8% fetal bovine serum (FBS). In total,
1×106 QRsP cells were irradiated at a dose of 10 Gy
using a LINAC system (Toshiba, Tokyo, Japan). Trypsinized QRsP
cells were seeded onto 10 cm dishes 1 h subsequent to irradiation
and cultured for 14 days. Well-demarcated colonies were trypsinized
using a cloning cylinder and grown in DMEM for 14 days.
Subsequently, 6 independent cell lines were established and
classified as QRsPIR-1 to QRsPIR-6. The parental QRsP cells and the
QRsPIR-1 and QRsPIR-2 cell lines were implanted subcutaneously into
the dorsal area of 6-week-old female c57bl/6 mice (CLEA Japan,
Inc., Tokyo, Japan). The cells were recovered from the tumor mass
of each animal 28 days subsequent to implantation and designated as
QRsPV, QRsPV-IR1 and QRsPV-IR2. All cells were stored at −80°C for
additional analysis. In order to mimic hypoxic conditions, the
cells were exposed to 200 µM CoCl2 diluted in DMEM, for
16 h. The animal experiments were strictly compliant with the
animal care guidelines of Hokkaido University (Sapporo, Hokkaido,
Japan).
Colony assay
QRsPV cells were transfected with pcDNA HIF-5C, and
24 h later, 1×106 cells were irradiated at a dose of 4,
8 or 10 Gy using the LINAC system (Toshiba, Tokyo, Japan). In
total, 1×106 trypsinized cells were seeded onto 60-mm
culture dishes 1 h subsequent to irradiation, and 1×104
non-irradiated cells that had been transfected with the pcDNA3
vector were also seeded onto 60-mm culture dishes to obtain the
plating efficiency. The plating efficiency was calculated as the
percentage of cells seeded that grow into colonies under G418
selection conditions. The cells were cultured for 2 weeks with DMEM
containing G418 at a final concentration of 1 mg/ml, followed by
methanol fixation and Giemsa staining (2% Giemsa's solution:
Merck-Millipore, Darmstadt, Germany).
Histopathological examination
Semi-confluent QRsPV or QRsPV-IR2 cells were
trypsinized and resuspended with phosphate-buffered saline (PBS).
In total, 1×104 PBS-suspended cells were injected
subcutaneously into the dorsal region of 6-week-old female c57bl/6
mice, with 3 mice per group. On day 28, all animals were sacrificed
and the tumor masses were dissected. The tumor tissues were fixed
in 10% formaldehyde, with occasional de-calcification, and then
embedded in paraffin, according to routine pathological procedure.
The specimens were sliced into 5-µm thick sections and stained with
hematoxylin and eosin.
Western blotting
The cells were lysed in a buffer containing 250 mM
NaCl, 50 mM HEPES (pH 7.0) and 0.1% Nonidet P-40 with a protease
inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). The samples
were then subjected to western blotting using anti-human HIF-1α
mouse monoclonal antibody was purchased from BD Biosciences (Cat
no. 610959, Sparks, MD, USA) and anti-Von Hippel-Lindau (VHL)
monoclonal antibody was purchased from Santa Cruz Biotechnology
(Cat no. sc-17780, Dallas, TX, USA). Both primary antibodies were
used at dilutions of 1:1,000. Horseradish Peroxidase conjugated
donkey anti-mouse secondary antibody was purchased from Jackson
Immunoresearch (West Grove, PA, USA) and was used at dilutions of
1:2,000. Amersham ECL reagents were purchased from GE Healthcare
Bio-Sciences (Pittsburgh, PA, USA).
Reporter assay
The QRsPV cells were plated at a density of
1×105 cells per well in 24-well plates on the day prior
to transfection. The cells were transiently transfected with
Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA)
containing 75 ng of luciferase reporters and 7.5 ng of pGL4.72
luciferase reporter vector (Promega) and 300 ng of a series of
HIF-1α expression plasmids (Fig. 2A).
The cell lysates were subjected to luciferase assays using a
dual-luciferase reporter assay system (Promega) 24 h subsequent to
transfection, according to the manufacturer's instructions.
Statistical analysis
Statistical differences were analyzed using
Student's t-test. P≤0.05 was considered to indicate a
statistically significant difference.
Results
Cloning of repopulated cancer
cells
Accumulated data have suggested that accelerated
repopulation of irradiated cancer cells occurs 3 weeks subsequent
to irradiation (4,5). To analyze this phenomenon, irradiated or
non-irradiated QRsP cells were injected into the dorsal region of
c57bl/6 mice. After 28 days, the tumor masses were collected for
tissue culture and designated as QRsPV, QRsPV-IR1 and QRsPV-IR2
tumors. The cells were re-inoculated into mice, with 3 mice each in
the QRsPV, QRsPV-IR1 and QRsPV-IR2 groups, for 28 days. During the
incubation period, 1 out of 3 animals inoculated with QRsPV-IR2
cells demonstrated severe leg paralysis. As indicated in Fig. 1A, the QRsPV-IR2 cells demonstrated an
infiltrative morphology, whereas QRsPV cells revealed
well-demarcated expansive growth, without evident infiltration.
Therefore, the QRsPV-IR2 cells were utilized as a model for an
irradiated progressed cell line in subsequent experiments. In the
following experiment, several reporter constructs, consisting of
HIF-1α, p53, AP-1 and Ets consensus sequence-containing luciferase
reporters, were introduced into QRsPV and QRsPV-IR2 cells. In the
QRsPV-IR2 cells, HIF-1α, AP-1 and Ets-dependent endogenous
transcriptional reactions were significantly increased (P=0.00036,
P=0.027 and P=0.017, respectively) compared with the QRsPV cells,
whereas p53-dependent transcription was severely decreased
(P=0.017) (Fig. 1B).
HIF-1α expression was activated in
irradiated cells
To confirm the 5′- and 3′-UTR dependent expression
regulation in the QRsPV cells, HIF-1α plasmids (Fig. 2A; HIF5C3, 5C, C and C3) were
transfected and cell lysates underwent western blot analysis. As
indicated in Fig. 2B, cells
transfected with HIF-5C3 and HIF-5C expressed an increased amount
of HIF-1α protein compared with HIF-C. Low expression of HIF-1α was
demonstrated in QRsPV cells (Fig.
2B). In the subsequent experiment, QRsPV and QRsPV-IR2 cells
were transfected with several HIF-1α plasmids combined with an HRE
luciferase reporter and a Renilla luciferase plasmid.
Accelerated 5′-UTR-dependent translation of HIF-1α is evident in
irradiated QRsPV-IR2 cells (Fig. 2C).
Increased HIF-1α expression was observed in QRsPV-IR2 cells
transfected with HIF-5C compared with the cells transfected with
HIF-C (P=0.025). The difference in HIF-1α expression was not
significant between the HIF-5C-transfected and HIF-C-transfected
QRsPV cells (P=0.065). These observations encouraged the
investigation of the accumulation of HIF-1α under hypoxia-mimicking
conditions in QRsPV and QRsP-IR2 cells. As demonstrated in Fig. 2D, >2 fold HIF-1α-dependent
luciferase activity was detected in QRsPV-IR2 cells under
hypoxia-mimicking conditions, which was a significant difference
(P=0.025).
Enhanced AP-1 and Ets-associated gene
expression in irradiated cells
Since activated AP-1 and Ets-dependent transcription
was observed in QRsPV-IR2 cells, the UTR-dependent expressional
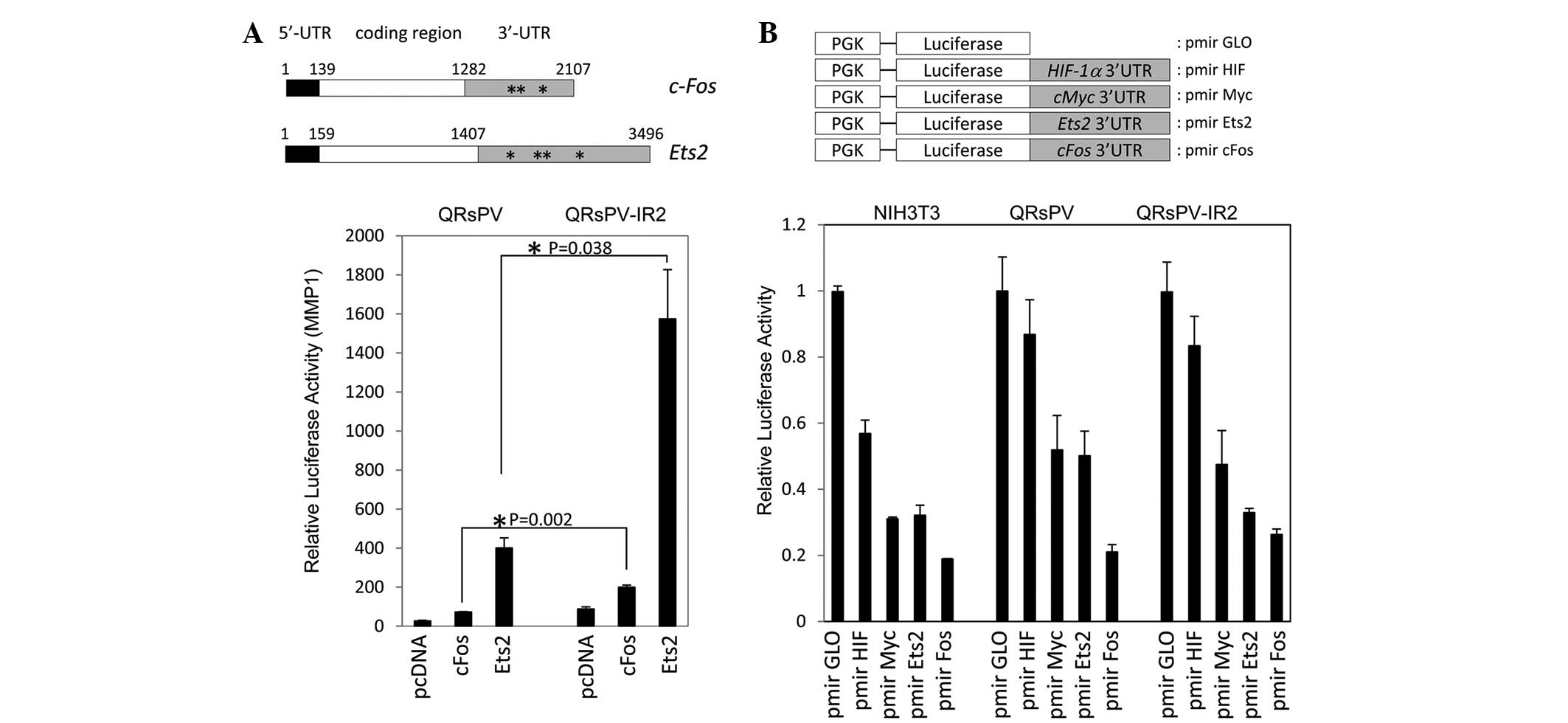
regulation of c-fos and Ets2 was investigated. As
described in Fig. 3A, the two mRNA
sequences contained relatively long 5′- and 3′-UTRs. Typical
AU-rich (ARE) elements were identified in the two mRNA sequences,
with 3 AREs in c-fos and 4 AREs in Ets2 (Fig. 3A). The two plasmids were transfected
together with a MMP1-luciferase reporter, as AP-1 and Ets binding
sites are available on this construct, and a Renilla plasmid
into the QRsPV and QRsPV-IR2 cells. Fig.
3A reveals the significantly enhanced luciferase activity
associated with c-fos (P=0.002) and Ets2 (P=0.038) expression in
QRsPV-IR2 cells compared with QRsPV cells. These observations
prompted the confirmation of whether the 3′-UTR-dependent
repressive system is activated in irradiated cells. Several 3′-UTR
sequences were cloned downstream of luciferase sequences in pmir
GLO vectors (Fig. 3B). All constructs
were introduced into NIH3T3 negative control, QRsPV and QRsPV-IR2
cells. As indicated in Fig. 3B, there
was a significant repressive effect via HIF1α 3′UTR in NIH3T3
(P=0.03), but not in QRsPV (P=0.14) and QRsP-IR2 cells (P=0.10).
These results indicate that 3′-UTR-dependent regulatory machinery
may not be involved in the enhanced gene expression in irradiated
cells.
Selective effect of X-ray
irradiation
Since cancer tissues consist of a heterogeneous cell
population (13,14), the majority of which are proliferating
cancer cells and CSCs, it is possible that the high-dose X-ray
irradiation affects only the proliferating cancer cells under the
present experimental conditions. Therefore, certain components of
the QRsP parent cell population, in particular the CSC-like
population of QRsP cells, should exhibit enhanced 5′-UTR dependent
HIF-1α expression, as observed in QRsPV-IR2 cells. To address this
issue, 24 independent single-cell-derived QRsPV cells, designated A
through X, were cloned and reporter assays were performed. As
indicated in Fig. 4A, 2 out of the 24
clones (A and M1)exhibited similar luciferase activity to that
exhibited by the QRsPV-IR2 cells. It should be noted that there was
no evident difference between the endogenous expression levels of
VHL in these cell lines (Fig.
4B).
These results encouraged the investigation of
whether the cells expressing high HIF-1α levels were capable of
surviving the lethal damage of ionizing irradiation. Colony assays
were performed using QRsPV cells transfected with HIF-1α or a
control vector. The transfected cells were irradiated at a dose of
4, 8 or 10 Gy, followed by selection for 10 days in G418 and Giemsa
staining. As indicated in Fig. 4C,
HIF-1α-transfected cells revealed a radio-resistant phenotype
compared with the control at radiation doses of 4, 8 and 10 Gy
(P=0.046, P=0.025 and P=0.044, respectively). The mean size of
colonies consisting of HIF-1α-transfected cells was increased
compared with control transfectants (data not shown). It is likely
that the total number of cells surviving irradiation, rather than
the colony number, was increased by HIF-1α overexpression.
Discussion
CSCs are considered to be responsible for the onset,
self-renewal, mutation accumulation and metastasis of tumors. CSCs
may exist as dormant cells within the primary tumor mass (13,14), but
they may transition between a dormant state and an actively
proliferating state due to genotoxic damage caused by ionizing
irradiation or chemotherapeutic treatment. If the genotoxic damage
kills the majority of proliferating cancer cells, but not the CSCs,
over the prolonged course of therapy, the CSCs may be activated and
the percentage of CSCs in the residual tumor mass may be increased.
The existence of a large number of CSCs in the primary lesion may
explain the high frequency of relapse, distant metastasis and
resistance to cancer therapy. These concepts demonstrate an
extremely good fit for the conventional repopulation theory.
Previously, the reciprocal regulation of HIF-1α by
5′- and 3′-UTR-dependent mechanisms and the possible correlation
between 5′ UTR-dependent translational regulation of HIF-1α and
tumor malignancies have been reported (10). In the present study, the cells that
survived radiation, the QRsPV-IR2 cells, demonstrated a more
aggressive phenotype compared with the non-irradiated QRsPV cells.
The 5′-UTR-dependent translational activity of HIF-1α was markedly
increased in the QRsPVIR-2 cells, even under a normoxic
environment. Increased endogenous HIF-1α expression in the
QRsPV-IR2 cells was also identified under hypoxia-mimicking
conditions. Overall, these results suggest that the QRsPV-IR2 cells
that survived radiation may express larger levels of HIF-1α, Ets2
and c-fos proteins and may exhibit a more aggressive phenotype
compared with the parental cell line. It was also notable that a
repression of p53 dependent transcriptional activity was observed
in QRsPV-IR2 cells. Previous studies have reported the involvement
of cMyc, HIF-1α and Ets2 transcription factors in CSC maintenance
(15–17). By contrast, p53 has been regarded as
the barrier to CSC formation (15,18,19). It is
likely that the expression pattern found in QRsPV-IR2 cells is
suitable for maintaining CSC-like phenotypes.
It was also found in the present study that parental
QRsPV cells contained a small population that exhibited the same
phenotype as radiation-resistant CSC-like QRsPV-IR2 cells. Notably,
exogenous expression of HIF-1α was sufficient to activate the
resistant phenotype of QRsPV cells against high-dose X-ray
irradiation. These observations prompted the hypothesis that 10-Gy
irradiation is lethal for the majority of proliferating QRsPV
cells, but not for CSC-like QRsPV cells expressing an increased
level of HIF-1α. It appears likely that the QRsPV-IR2 tumors
contained a larger number of CSCs. Whole genomic sequencing using
next generation sequencing techniques is required to eliminate the
possibility that X-irradiation induced the genomic mutations or
genomic modification of cancer cells under the present experimental
conditions.
A large number of transcription factors may
contribute to the maintenance of the stemness of CSCs (15–17). It
appears possible that several transcription factors that are
involved in CSC maintenance, are regulated by the same mechanism in
an UTR-dependent manner. If these CSC maintenance genes are
regulated simultaneously by the same molecular mechanism, such a
novel pathway may be a promising future target for the suppression
of CSCs.
Acknowledgements
This study was supported by a Grant-in-Aid for
Scientific Research (grant no. B24390285) provided by the Ministry
of Education, Science, and Culture of Japan.
References
|
1
|
Allam A, Perez LA, Huang P, Taghian A,
Azinovic I, Freeman J, Duffy M, Efird J and Suit HD: The effect of
the overall treatment time of fractionated irradiation on the tumor
control probability of a human soft tissue sarcoma xenograft in
nude mice. Int J Radiat Oncol Biol Phys. 32:105–111. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beck-Bornholdt HP, Omniczynski M, Theis E,
Vogler H and Würschmidt F: Influence of treatment time on the
response of rat rhabdomyosarcoma R1H to fractionated irradiation.
Acta Oncol. 30:57–63. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Begg AC, Hofland I and Kummermehr J:
Tumour cell repopulation during fractionated radiotherapy:
Correlation between flow cytometric and radiobiological data in
three murine tumours. Eur J Cancer. 27:537–543. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Withers HR, Maciejewski B, Taylor JM and
Hliniak A: Accelerated repopulation in head and neck cancer. Front
Radiat Ther Oncol. 22:105–110. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Withers HR, Taylor JM and Maciejewski B:
The hazard of accelerated tumor clonogen repopulation during
radiotherapy. Acta Oncol. 27:131–146. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Malaise E and Tubiana M: Growth of the
cells of an experimental irradiated fibrosarcoma in the C3H mouse.
C R Acad Sci Hebd Seances Acad Sci D. 263:292–295. 1966.(In
French). PubMed/NCBI
|
|
7
|
Barreau C, Paillard L and Osborne HB:
AU-rich elements and associated factors: Are there unifying
principles? Nucleic Acids Res. 33:7138–7150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsieh AC, Liu Y, Edlind MP, Ingolia NT,
Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et
al: The translational landscape of mTOR signalling steers cancer
initiation and metastasis. Nature. 485:55–61. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yasuda M, Hatanaka T, Shirato H and
Nishioka T: Cell type-specific reciprocal regulation of HIF1A gene
expression is dependent on 5′- and 3′-UTRs. Biochem Biophys Res
Commun. 447:638–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Habelhah H, Okada F, Kobayashi M, Nakai K,
Choi S, Hamada J, Moriuchi T, Kaya M, Yoshida K, Fujinaga K and
Hosokawa M: Increased E1AF expression in mouse fibrosarcoma
promotes metastasis through induction of MT1-MMP expression.
Oncogene. 18:1771–1776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishioka T, Miyai Y, Haga H, Kawabata K,
Shirato H, Homma A, Shibata K and Yasuda M: Novel function of
transcription factor ATF5: blockade of p53-dependent apoptosis
induced by ionizing irradiation. Cell Struct Funct. 34:17–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sagar J, Chaib B, Sales K, Winslet M and
Seifalian A: Role of stem cells in cancer therapy and cancer stem
cells: A review. Cancer Cell Int. 7:92007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sales KM, Winslet MC and Seifalian AM:
Stem cells and cancer: an overview. Stem Cell Rev. 3:249–255. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akita H, Marquardt JU, Durkin ME, Kitade
M, Seo D, Conner EA, Andersen JB, Factor VM and Thorgeirsson SS:
MYC activates stem-like cell potential in hepatocarcinoma by a
p53-dependent mechanism. Cancer Res. 74:5903–5913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Keith B and Simon MC: Hypoxia-inducible
factors, stem cells and cancer. Cell. 129:465–472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Múnera J, Ceceña G, Jedlicka P, Wankell M
and Oshima RG: Ets2 regulates colonic stem cells and sensitivity to
tumorigenesis. Stem Cells. 29:430–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aloni-Grinstein R, Shetzer Y, Kaufman T
and Rotter V: p53: The barrier to cancer stem cell formation. FEBS
Lett. 588:2580–2589. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bonizzi G, Cicalese A, Insinga A and
Pelicci PG: The emerging role of p53 in stem cells. Trends Mol Med.
18:6–12. 2012. View Article : Google Scholar : PubMed/NCBI
|