Introduction
Nasopharyngeal carcinoma (NPC) is a rare condition
worldwide, but it is endemic in certain populations (1). The development of NPC may be attributed
to the dynamic interplay of environmental factors and genetic
susceptibility. Host factors, including tobacco smoking,
consumption of salt-preserved fish, history of chronic respiratory
tract diseases and Epstein-Barr virus (EBV) infection, are
well-established risk factors for the development of NPC (2,3). However,
not all individuals exposed to EBV infection and environmental
carcinogenic factors develop NPC, which indicates that genetic
susceptibility may also contribute. Glutathione S-transferases
(GSTs) are a large family of phase II detoxification enzymes that
regulate the conversion of toxic compounds to hydrophilic
metabolites (4,5). GSTM1 is one of the main subtypes of
GSTs, and is involved in protecting hosts against cancer (4). The GSTM1 gene is located on the short
arm of chromosome 1 (1p13.3), and displays several polymorphisms
(6). The most common polymorphism in
the GSTM1 gene is a null variant, which has been widely
investigated as a risk biomarker for various types of cancer
(7,8).
The GSTM1 null variant may lead to the absence of enzymatic
activity, and individuals who carry this variant are thought to be
at increased risk of developing cancer (6). Numerous studies have previously
evaluated the association of the GSTM1 polymorphism with the
susceptibility to NPC, but the results are inconsistent, possibly
due to the limited sample size and low power of these studies,
which are insufficient to detect the precise effect in a single
study (9–23). Thus, in the present study, a
meta-analysis was performed in order to quantitatively evaluate the
association between the GSTM1 polymorphism and the susceptibility
to NPC.
Materials and methods
Search strategy
A comprehensive search of PubMed, Embase, Web of
Science and China National Knowledge Infrastructure (CNKI)
databases was performed up to May 13th, 2014, in order
to identify potentially relevant studies on the association between
the GSTM1 polymorphism and the risk of developing NPC. The
literature search was conducted independently by 2 investigators,
and the disagreements were resolved by consensus. The search terms
were as follows: Glutathione S-transferase, GST, GSTs, glutathione
S-transferase M1, GSTM, GSTM1, polymorphism, mutation or variation;
with nasopharyngeal cancer, nasopharyngeal carcinoma or NPC. A
manual search of the references of all the retrieved publications
was conducted to identify additional studies. There were no
language or sample size limitations.
Inclusion and exclusion criteria
Studies were included in the present meta-analysis
according to the following inclusion criteria: i) Studies that
evaluated the association between the GSTM1 polymorphism and the
risk of developing NPC; ii) those that were case-control studies;
iii) those that described the diagnoses of NPC and the sources of
cases and controls; iv) those that contained available data for
acquiring numbers of the null and present genotypes of GSTM1; and
iv) those that presented sufficient information for the calculation
of the odds ratio (OR) with its corresponding 95% confidence
interval (CI). Single cases and family-based studies were excluded
from the meta-analysis. In those cases of overlapping studies, the
latest or the most complete one was selected for inclusion in the
analysis.
Data extraction
The following data were extracted from the relevant
publications: First authors name; year of publication; ethnicity
and country of origin of cohort; numbers of cases and controls;
genotyping method; source of controls; and genotype distributions
in cases and controls. Disagreements were settled through
discussion among the 2 investigators mentioned above.
Statistical analysis
The strength of the associations between the GSTM1
polymorphism and the risk of developing NPC was estimated by OR and
the corresponding 95% CI, based on the frequencies of the null and
present genotypes in cases and controls. The pooled ORs for the
null vs. present genotype were calculated using the fixed- or
random-effects model (known as the Mantel-Haenszel and DerSimonian
and Laird method, respectively), and determined by the Z test
(24,25). Q and I2 tests were adopted
to assess the heterogeneity among the studies included in the
meta-analysis. P<0.01 and I2>50% were considered
to indicate a statistically significant difference in heterogeneity
(26,27). Subgroup analyses were also conducted
according to ethnicity: Of the 12 included studies, 10 were
conducted in Asians (11–16,18,20–22),
1 in North Africans (17) and 1 in a
mixed population (9). Publication
bias was evaluated by funnel plot (28) and Eggers linear regression test
(29). Sensitivity analysis by
omission of each individual study was performed to identify the
source of between-study heterogeneity and to confirm the stability
and reliability of the pooled results (30). The statistical analyses were performed
using STATA12.0 software (StataCorp LP, College Station, TX,
USA).
Results
Study characteristics
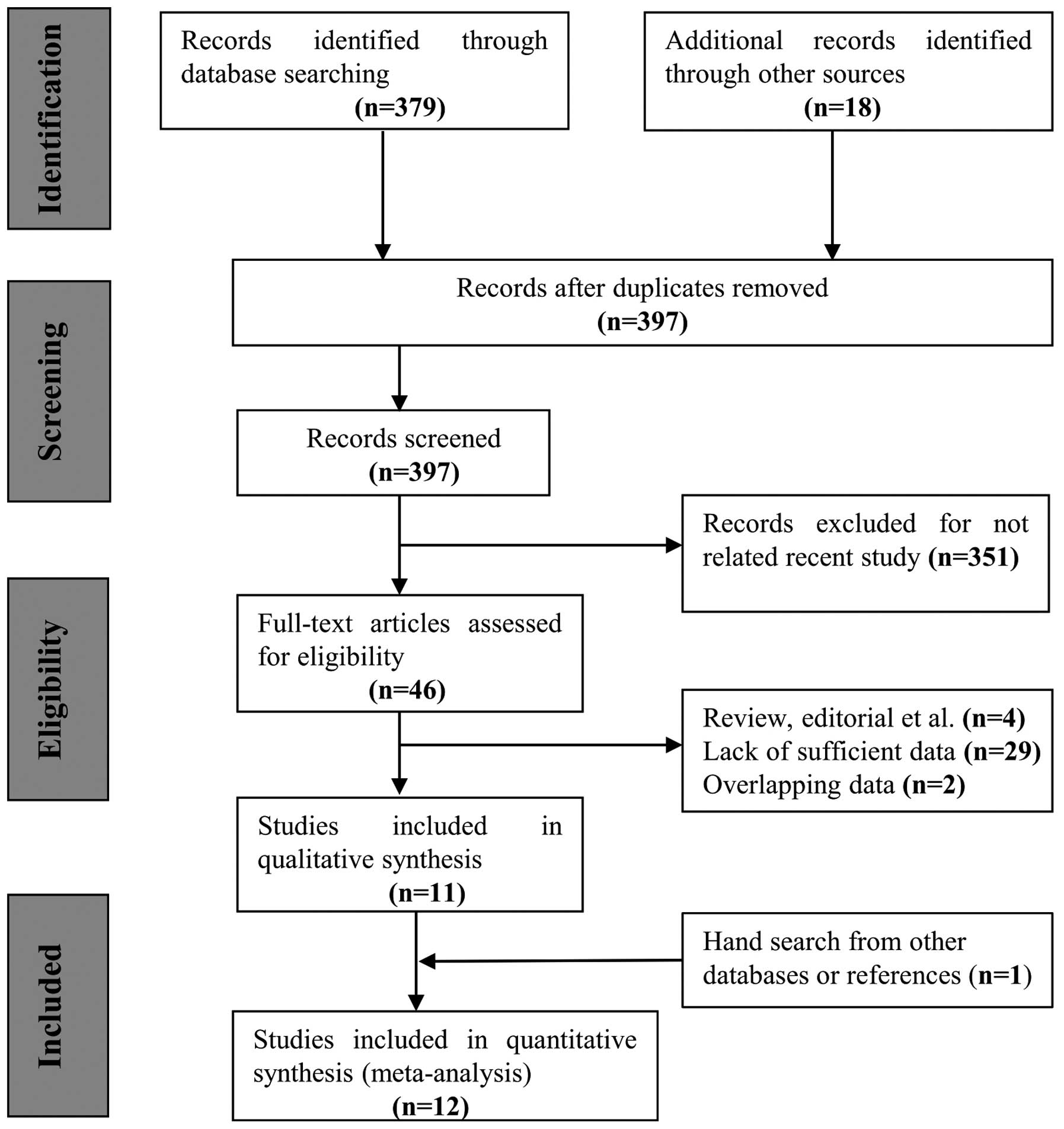
Relevant studies were retrieved following a
systematic literature search of PubMed, Embase, Web of Science and
CNKI databases. The selection process for the articles included in
the present meta-analysis is depicted in Fig. 1. The initial search yielded 394 study
titles and abstracts, which were then subjected to an independent
review. Based on the inclusion criteria, 12 individual publications
studying the association of the GSTM1 polymorphism and the risk of
developing NPC in a total of 1,593 cases and 2,868 individuals were
available for analysis (9,11–17,20–22).
The clinical characteristics of these studies are listed in
Table I. Among the 12 included
studies, 10 were conducted in Asians (11–16,18,20–22),
1 in North African (17) and 1 in a
mixed population (9).
 | Table I.Summary of the main characteristics of
the studies included in the present meta-analysis. |
Table I.
Summary of the main characteristics of
the studies included in the present meta-analysis.
|
|
|
|
|
|
| GSTM1
polymorphism |
|---|
|
|
|
|
|
|
|
|
|---|
|
|
| Cohort | Sample size | Null | Present |
|---|
|
|
|
|
|
|
|
|---|
| First author | Ref. | Country | Ethnicity | Case | Control | Case | Control | Case | Control |
|---|
| Nazar-Stewart V | 9 | USA | Mixed | 83 | 142 | 45 | 63 | 38 | 79 |
| Da SJ | 11 | China | Asian | 80 | 80 | 48 | 36 | 32 | 44 |
| Cheng YJ | 12 | Taiwan | Asian | 314 | 337 | 173 | 169 | 141 | 168 |
| Liu ZG | 13 | China | Asian | 46 | 53 | 28 | 18 | 18 | 35 |
| Deng ZL | 14 | China | Asian | 91 | 135 | 56 | 64 | 35 | 71 |
| Deng ZL | 15 | China | Asian | 127 | 207 | 78 | 95 | 49 | 112 |
| Tiwawech D | 16 | Thailand | Asian | 78 | 145 | 50 | 74 | 28 | 71 |
| Bendjemana K | 17 | Tunisia | African | 45 | 100 | 23 | 33 | 22 | 67 |
| Guo X | 18 | China | Asian | 341 | 590 | 204 | 328 | 137 | 262 |
| Jiang Y | 20 | China | Asian | 182 | 366 | 97 | 157 | 85 | 215 |
| Wei YP | 21 | China | Asian | 126 | 641 | 78 | 305 | 48 | 336 |
| Liao ZL | 22 | China | Asian | 80 | 72 | 50 | 32 | 30 | 40 |
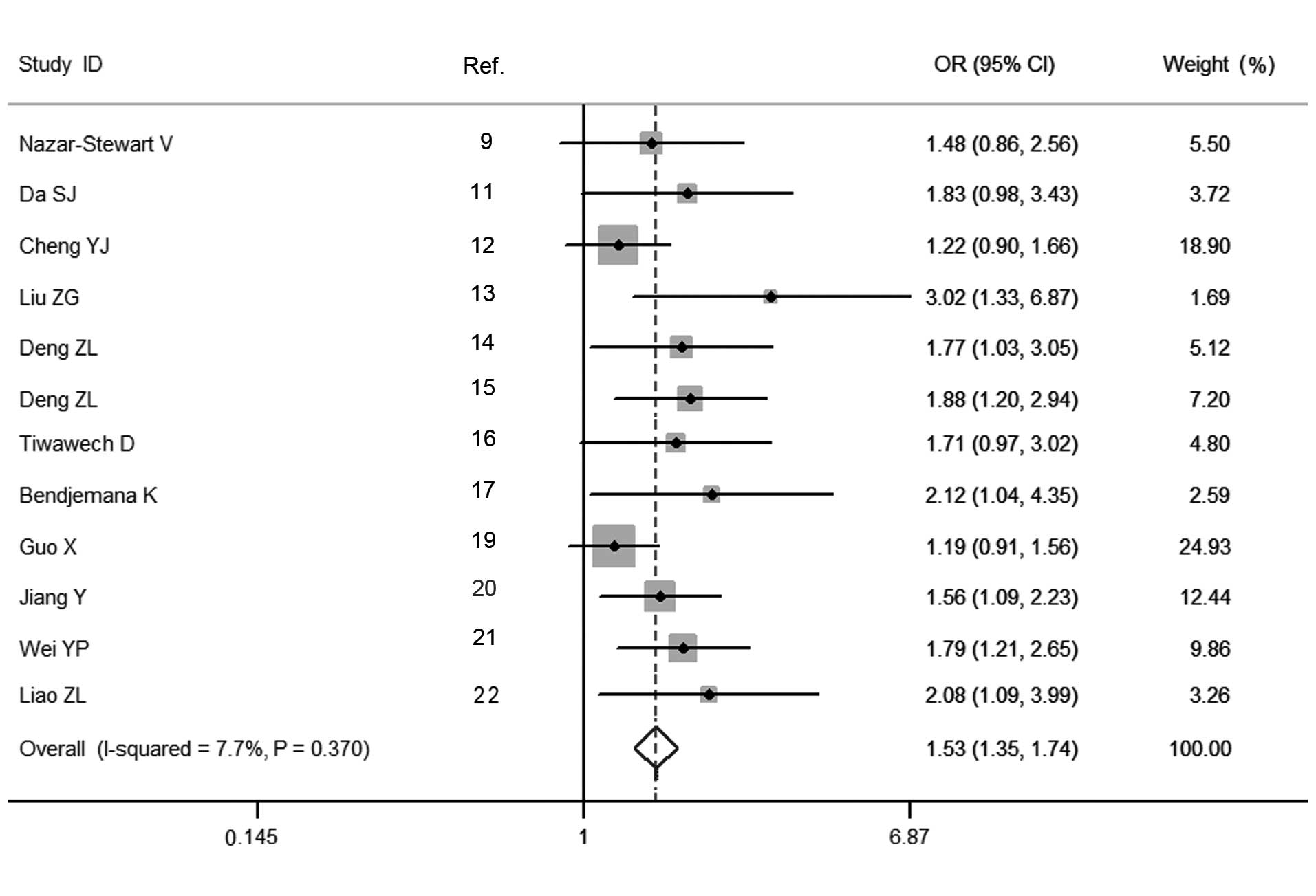
Pooled analysis results
Estimation of the association between the GSTM1
polymorphism and the susceptibility to NPC was performed, and the
pooled OR for the GSTM1 polymorphism suggested a significantly
increased risk of developing NPC for carriers of the null genotype,
compared with the present genotype (OR=1.530, 95% CI=1.348–1.737,
Pheterogeneity=0.370; Fig.
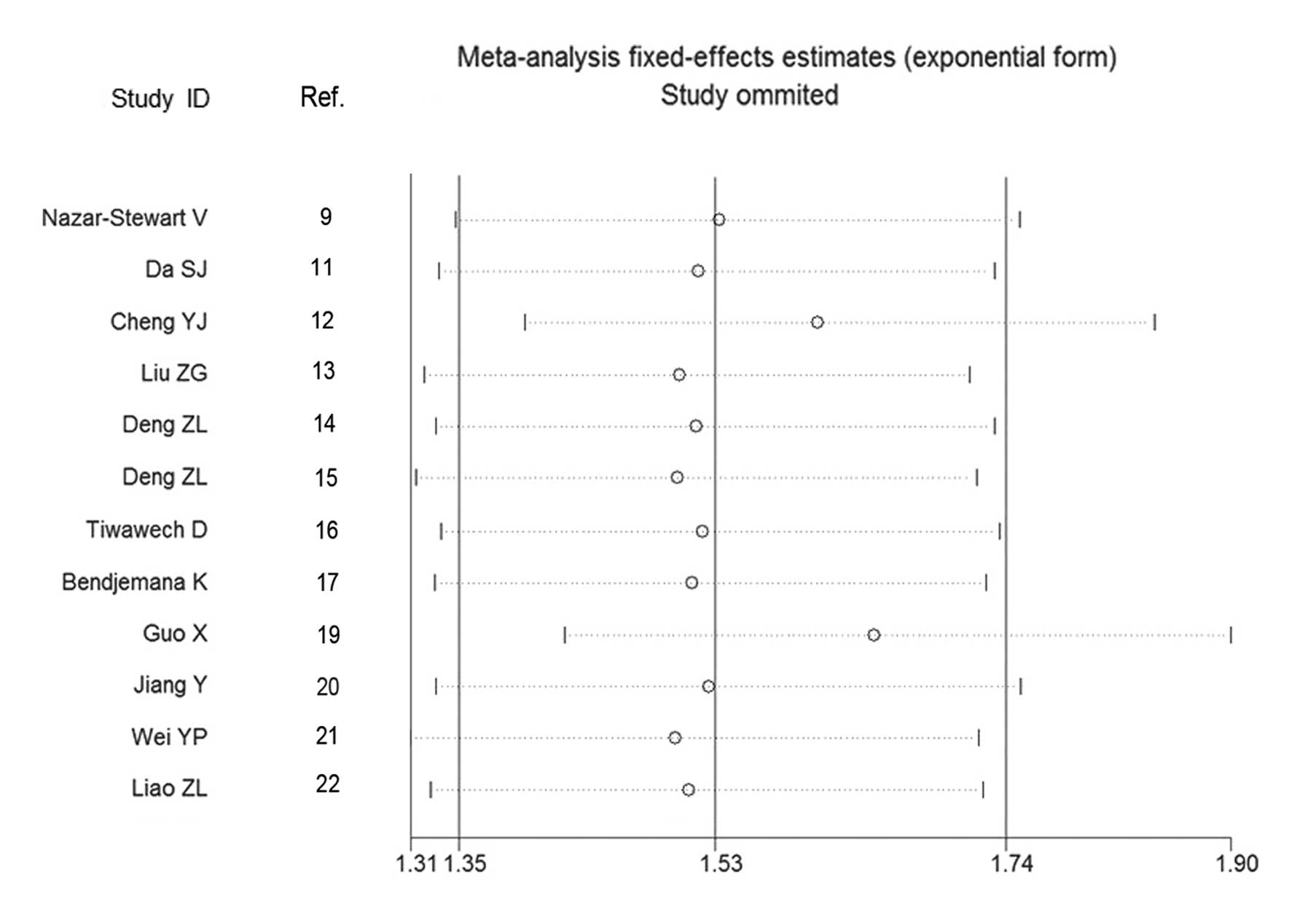
2). Sensitivity analysis by omission of each individual study
further confirmed this significant association (Fig. 3). In the subgroup analyses by
ethnicity, a significantly increased risk of developing NPC was
observed for the polymorphism of GSTM1 in the Asian population
(OR=1.516, 95% CI=1.328–1.731, Pheterogeneity=0.270).
Subgroup analyses in the North African and mixed populations were
not performed due to insufficient data availability.
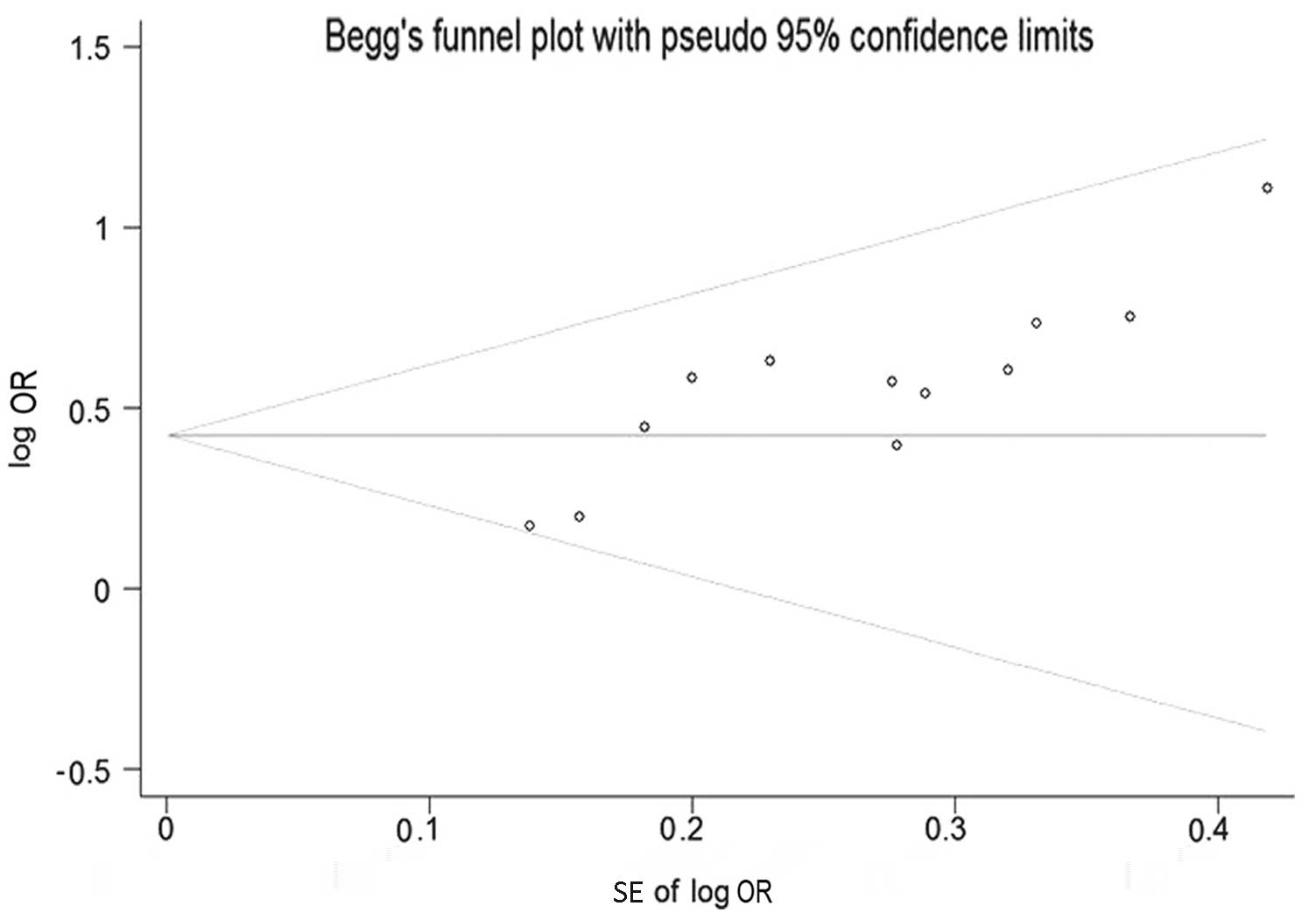
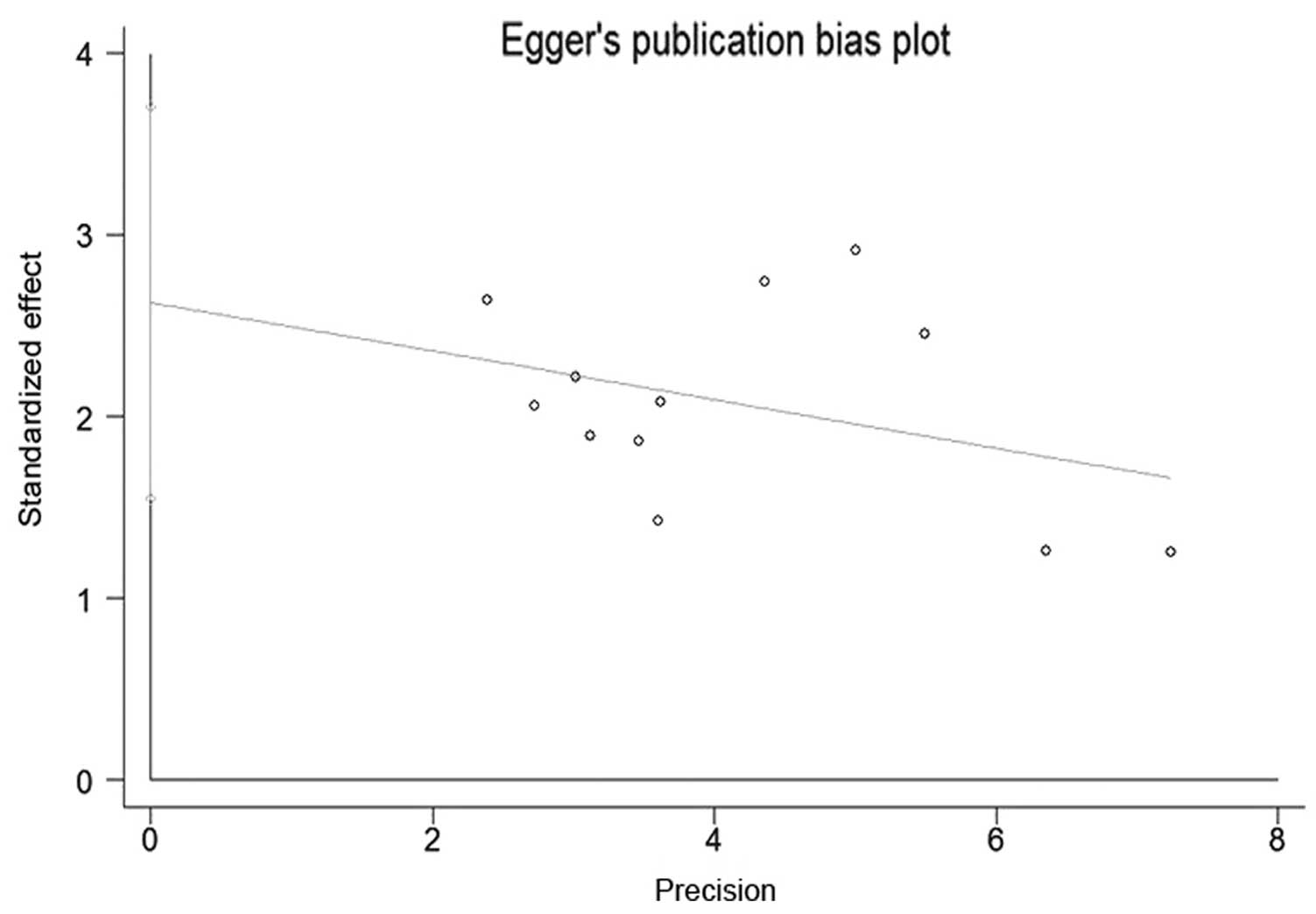
Publication bias
Beggs funnel plot and Eggers test were used to
estimate the publication bias of all the studies on the
associations of the GSTM1 polymorphism and the risk of developing
NPC that were included in the present meta-analysis. The funnel
plot shapes of the Beggs test did not reveal any evidence of
asymmetry (Fig. 4). Furthermore, the
P-value of the Eggers test was <0.05, indicating absence of
publication bias among the included studies (Fig. 5).
Discussion
GSTs belong to the biotransformation family of
enzymes. GSTs are phase II enzymes with catalytic and noncatalytic
activities in vivo, which are involved in the detoxification
of electrophilic compounds by glutathione conjugation, including
carcinogens and cytotoxic drugs (31,32). Since
the presumed function of GSTs is to protect tissues against toxic
and carcinogenic compounds, they are considered to be important
determinants in the development of prostate cancer. GSTM1, one of
the main subtypes of GSTs, participates in the protection of the
host against cancer (4). The GSTM1
gene displays several polymorphisms, among which, the null variant
is the most common one, and it has been widely investigated as a
risk biomarker for various types of cancer (7,8). The GSTM1
null variant may result in the absence of enzymatic activity, and
individuals who carry the null variant are thought to be at
increased risk of developing cancer, since the polymorphic
deletions of GSTs may affect their ability to detoxify
electrophilic carcinogens, which may lead to an increase in the
hosts susceptibility to environmental toxins and carcinogens
(6,33). Mutations in GSTM1 are the most
commonly studied polymorphisms of GSTs, regarding genetic
susceptibility to cancer (34–36).
Numerous individual case-control studies on the GSTM1 polymorphism
in association with the risk of developing NPC have been conducted
in the past 2 decades (9–23). Certain studies have previously
suggested that the GSTM1 null genotype was associated with
increased risk of developing NPC (9,20), while
other studies did not observe any significant associations between
various genetic polymorphisms of GSTM1 and the risk of developing
NPC (12).
These discrepancies may arise from the small size of
the sample in individual case-control studies, which may result in
insufficient statistical power, particularly for weak correlations.
In addition, differences in genetic background, study design and
source of cases and controls may also contribute to these
controversial and inconclusive findings. Therefore, in the present
study, a meta-analysis was conducted by pooling the ORs with 95%
CIs of all the currently available case-control studies on GSTM1
and NPC, in order to clarify these apparently contradictory
findings. A total of 12 eligible studies with 1,593 cases and 2,868
controls were analyzed to quantitatively evaluate the association
between the GSTM1 polymorphism and the susceptibility to NPC. The
results suggested that the GSTM1 null genotype was significantly
associated with the risk of developing NPC.
However, there were several limitations in the
present meta-analysis: i) The adjusted estimation, which is a more
precise way of estimating the association, was not pooled; thus,
further studies are required to calculate the pooled OR, adjusting
for other confounding factors; ii) a potential source of bias in
studies of genotypes may be due to the inclusion of individuals
from different ethnic backgrounds; and iii) gene-gene and
gene-environment interactions were not analyzed in the present
meta-analysis due to the unavailability of relevant studies.
Therefore, additional case-control studies are required to further
analyze these potential interactions and their role in the
association between GSTM1 and NCP.
In summary, the results from the present
meta-analysis indicate that the GSTM1 polymorphism participates in
the development of NPC. However, the significant associations of
the GSTM1 polymorphism with the risk of developing NPC must be
validated in future studies, which must also account for the
influence of gene-gene and gene-environment interactions in this
potential association.
Acknowledgements
The authors would like to thank Dr Qiliang Cai
(Department of Urology, The Second Hospital of Tianjin Medical
University, Tianjin Institute of Urology, Tianjin, China) for the
invaluable discussions regarding study design and statistical
analyses.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
ONeil JD, Owen TJ, Wood VH, Date KL,
Valentine R, Chukwuma MB, Arrand JR, Dawson CW and Young LS:
Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription
factor pathway in nasopharyngeal carcinoma cells and enhances
angiogenesis in vitro. J Gen Virol. 89:2833–2842. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strange RC, Spiteri MA, Ramachandran S and
Fryer AA: Glutathione-S-transferase family of enzymes. Mutat Res.
482:21–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oakley A: Glutathione transferases: A
structural perspective. Drug Metab Rev. 43:138–151. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hayes JD and Strange RC: Glutathione
S-transferase polymorphisms and their biological consequences.
Pharmacology. 61:154–166. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang ZJ, Hao K, Shi R, Zhao G, Jiang GX,
Song Y, Xu X and Ma J: Glutathione S-transferase M1 (GSTM1) and
glutathione S-transferase T1 (GSTT1) null polymorphisms, smoking,
and their interaction in oral cancer: A HuGE review and
meta-analysis. Am J Epidemiol. 173:847–857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taioli E, Flores-Obando RE, Agalliu I,
Blanchet P, Bunker CH, Ferrell RE, Jackson M, Kidd LC, Kolb S,
Lavender NA, et al: Multi-institutional prostate cancer study of
genetic susceptibility in populations of African descent.
Carcinogenesis. 32:1361–1365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nazar-Stewart V, Vaughan TL, Burt RD, Chen
C, Berwick M and Swanson GM: Glutathione S-transferase M1 and
susceptibility to nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 8:547–551. 1999.PubMed/NCBI
|
|
10
|
Guo XL: Genetic polymorphism of cytochrome
P450 1A1 and glutathione S-transferase Ml related with
susceptibility of nasopharyngeal carcinoma. Hebei Med Univ.
2001.
|
|
11
|
Da SJ, Liang B, Wu HL and Guan LL:
Relationship between GSTM1 gene polymorphism and genetic
susceptibility in nasopharyngeal carcinoma. Prac J of Cancer.
17:617–619. 2002.(In Chinese).
|
|
12
|
Cheng YJ, Chien YC, Hildesheim A, Hsu MM,
Chen IH, Chuang J, Chang J, Ma YD, Luo CT, Hsu WL, et al: No
association between genetic polymorphisms of CYP1A1, GSTM1, GSTT1,
GSTP1, NAT2, and nasopharyngeal carcinoma in Taiwan. Cancer
Epidemiol Biomarkers Prev. 12:179–180. 2003.PubMed/NCBI
|
|
13
|
Liu ZG, Wei YP and Ma Y: Population with
GSTT1 gene deletion and the relationship to hepatocellular
carcinoma from Guangxi. J Guangxi Med Univ. 20:161–163. 2003.(In
Chinese).
|
|
14
|
Deng ZL, Wei YP and Ma Y: Frequent genetic
deletion of detoxifying enzyme GSTM1 and GSTT1 genes in
nasopharyngeal carcinoma patients in Guangxi Province, China.
Zhonghua Zhong Liu Za Zhi. 26:598–600. 2004.(In Chinese).
PubMed/NCBI
|
|
15
|
Deng ZL, Wei YP, Luo W, Liao ZL and Ma Y:
Glutathione S-transferase M1 and T1 gene deletion associated with
increased susceptibility to nasopharyngeal carcinoma.
Chinese-German J Clin Oncol. 4:276–278. 2005. View Article : Google Scholar
|
|
16
|
Tiwawech D, Srivatanakul P, Karalak A and
Ishida T: Glutathione S-transferase M1 gene polymorphism in Thai
nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 6:270–275.
2005.PubMed/NCBI
|
|
17
|
Bendjemana K, Abdennebi M, Gara S, Jmal A,
Ghanem A, Touati S, Boussen H, Ladgham A and Guemira F: Genetic
polymorphism of gluthation-S transferases and N-acetyl transferases
2 and nasopharyngeal carcinoma: The Tunisia experience. Bull
Cancer. 93:297–302. 2006.PubMed/NCBI
|
|
18
|
Guo X, OBrien SJ, Zeng Y, Nelson GW and
Winkler CA: GSTM1 and GSTT1 gene deletions and the risk for
nasopharyngeal carcinoma in Han Chinese. Cancer Epidemiol
Biomarkers Prev. 17:1760–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He Y, Zhou GQ, Li X, Dong XJ, Chai XQ and
Yao KT: Correlation of polymorphism of the coding region of
glutathione S-transferase M1 to susceptibility of nasopharyngeal
carcinoma in South China population. Ai Zheng. 28:5–7. 2009.(In
Chinese). PubMed/NCBI
|
|
20
|
Jiang Y, Li N, Dong P, Zhang N, Sun Y, Han
M, Wen J and Chen M: Polymorphisms in GSTM1, GSTTI and GSTP1 and
nasopharyngeal cancer in the East of China: a case-control study.
Asian Pacif J Cancer Prev. 12:3097–3100. 2011.
|
|
21
|
Wei YP, Long XD, Liu ZG, Ma Ya and Deng
ZL: Genetic polymorphism of glutathione-S-transferase M1 and T1 in
associated with carcinogenesis of hepatocellular carcinoma and
nasopharyngeal carcinoma. Chinese-German J Clin Oncol. 11:138–141.
2012. View Article : Google Scholar
|
|
22
|
Liao ZL, Deng ZL, Wei YP, Xie KS, Zhang B,
Dai XM and Xu CS: Relationship of GSTT1 and GSTM1 gene
polymorphisms with the development of nasopharyngeal carcinoma. J
Guangxi Med Univ. 22:372–374. 2005.
|
|
23
|
Tian SZ: The analysis of clinical factors
related to laryngeal carcinoma and research on the association
between genetic polymorphisms of xenobiotic-metabolizing enzymes.
DNA repair genes and susceptibility to laryngeal carcinoma
(Zhongshan Univ). 2001.
|
|
24
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
26
|
Cochran WG: The comparison of percentages
in matched samples. Biometrika. 37:256–266. 1950. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stuck AE, Rubenstein LZ and Wieland D:
Bias in meta-analysis detected by a simple, graphical test:
Asymmetry detected in funnel plot was probably due to true
heterogeneity. BMJ. 316:469–471. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tobias A, Saez M, Galan I and Campbell MJ:
Sensitivity analysis of common statistical models used to study the
short-term effects of air pollution on health. Int J Biometeorol.
47:227–229. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hayes JD, Flanagan JU and Jowsey IR:
Glutathione transferases. Annu Rev Pharmacol Toxicol. 45:51–88.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sharma R, Yang Y, Sharma A, Awasthi S and
Awasthi YC: Antioxidant role of glutathione S-transferases:
Protection against oxidant toxicity and regulation of
stress-mediated apoptosis. Antioxid Redox Signal. 6:289–300. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Strange RC, Lear JT and Fryer AA:
Glutathione S-transferase polymorphisms: Influence on
susceptibility to cancer. Chem Biol Interact. 111(112): 351–364.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moaven O, Raziee HR, Sima HR, Ganji A,
Malekzadeh R, A'Rabi A, Abdollahi A, Memar B, Sotoudeh M, Naseh H,
et al: Interactions between glutathione-S-transferase M1, T1 and P1
polymorphisms and smoking, and increased susceptibility to
esophageal squamous cell carcinoma. Cancer Epidemiol. 34:285–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Karami S, Boffetta P, Rothman N, Hung RJ,
Stewart T, Zaridze D, Navritalova M, Mates D, Janout V, Kollarova
H, et al: Renal cell carcinoma, occupational pesticide exposure and
modification by glutathione S-transferase polymorphisms.
Carcinogenesis. 29:1567–1571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sreeja L, Syamala V, Hariharan S, Syamala
VS, Raveendran B, Sivanandan CD, Madhavan J and Ankathil R:
Glutathione S-transferase M1, T1 and P1 polymorphisms:
Susceptibility and outcome in lung cancer patients. J Exp Ther
Oncol. 7:73–85. 2008.PubMed/NCBI
|