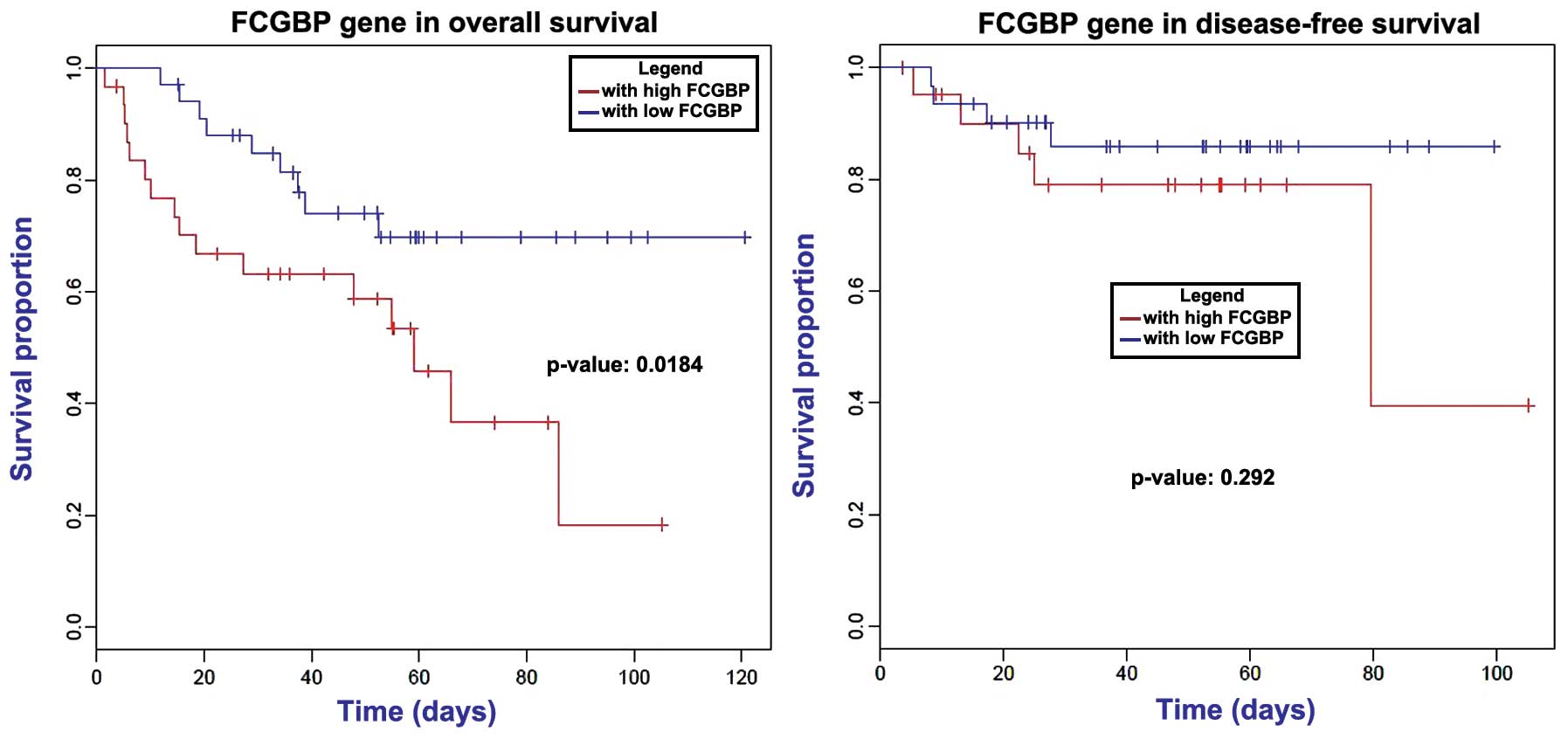
|
1
|
National Cancer Institute. SEER Cancer
Statistics Factsheets: Colon and Rectum Cancer, National Cancer
Institute (Bethesda, MD, USA). simpleseer.cancer.gov/statfacts/html/colorect.html
|
|
2
|
U.S. Cancer Statistics Working Group:
United States cancer statistics: 1999–2008 incidence and mortality
web-based report. U.S. Department of Health and Human Services.
Centers for Disease Control and Prevention and National Cancer
Institute (Atlanta, GA). 2012.
|
|
3
|
Lansdorp-Vogelaar I, van Ballegooijen M,
Zauber AG, Habbema JDF and Kuipers EJ: Effect of rising
chemotherapy costs on the cost savings of colorectal cancer
screening. J Natl Cancer Inst. 101:1412–1422. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pickhardt PJ, Hassan C, Halligan S and
Marmo R: Colorectal cancer: CT colonography and colonoscopy for
detection - systematic review and meta-analysis. Radiology.
259:393–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Nicolantonio F, Martini M, Molinari F,
Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L,
Frattini M, Siena S, et al: Wild-type BRAF is required for response
to panitumumab or cetuximab in metastatic colorectal cancer. J Clin
Oncol. 26:5705–5712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoff PM, Ansari R, Batist G, Cox J, Kocha
W, Kuperminc M, Maroun J, Walde D, Weaver C, Harrison E, et al:
Comparison of oral capecitabine versus intravenous fluorouracil
plus leucovorin as first-line treatment in 605 patients with
metastatic colorectal cancer: Results of a randomized phase III
study. J Clin Oncol. 19:2282–2292. 2001.PubMed/NCBI
|
|
7
|
Zhou M, Wang X, Hu J, et al: Geographical
distribution of cancer mortality in China, 2004–2005. Zhonghua yu
fang yi xue za zhi. Chin J Prev Med. 44:303–308. 2010.(In
Chinese).
|
|
8
|
Zhang W, Winder T, Ning Y, Pohl A, Yang D,
Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, et al: A let-7
microRNA-binding site polymorphism in 3-untranslated region of KRAS
gene predicts response in wild-type KRAS patients with metastatic
colorectal cancer treated with cetuximab monotherapy. Ann Oncol.
22:104–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami T, Kawada K, Iwamoto M, Akagami
M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM, et
al: The role of CXCR3 and CXCR4 in colorectal cancer metastasis.
Int J Cancer. 132:276–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dahlmann M, Okhrimenko A, Marcinkowski P,
Osterland M, Herrmann P, Smith J, Heizmann CW, Schlag PM and Stein
U: RAGE mediates S100A4-induced cell motility via MAPK/ERK and
hypoxia signaling and is a prognostic biomarker for human
colorectal cancer metastasis. Oncotarget. 5:3220–3233. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stange DE, Engel F, Longerich T, Koo BK,
Koch M, Delhomme N, Aigner M, Toedt G, Schirmacher P, Lichter P, et
al: Expression of an ASCL2 related stem cell signature and IGF2 in
colorectal cancer liver metastases with 11p15.5 gain. Gut.
59:1236–1244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Del Rio M, Mollevi C, Vezzio-Vie N, Bibeau
F, Ychou M and Martineau P: Specific extracellular matrix
remodeling signature of colon hepatic metastases. PLoS One.
8:e745992013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen DT, Hernandez JM, Shibata D, McCarthy
S, Humphries LA, Clark W, Elahi A, Gruidl M, Coppola D and Yeatman
T: Complementary strand microRNAs mediate acquisition of metastatic
potential in colonic adenocarcinoma. J Gastrointest Surg.
6:905–912. 2012. View Article : Google Scholar
|
|
15
|
Del Rio M, Molina F, Bascoul-Mollevi C,
Copois V, Bibeau F, Chalbos P, Bareil C, Kramar A, Salvetat N,
Fraslon C, et al: Gene expression signature in advanced colorectal
cancer patients select drugs and response for the use of
leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 25:773–780.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kroon J, in't Veld LS, Buijs JT, Cheung H,
van der Horst G and van der Pluijm G: Glycogen synthase kinase-3β
inhibition depletes the population of prostate cancer
stem/progenitor-like cells and attenuates metastatic growth.
Oncotarget. 5:8986–8994. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Rogoff HA, Keates S, Gao Y,
Murikipudi S, Mikule K, Leggett D, Li W, Pardee AB and Li CJ:
Suppression of cancer relapse and metastasis by inhibiting cancer
stemness. Proc Nat Acad Sci USA. 112:1839–1844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McDonald PC, Winum J-Y, Supuran CT and
Dedhar S: Recent developments in targeting carbonic anhydrase IX
for cancer therapeutics. Oncotarget. 3:84–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lou Y, McDonald PC, Oloumi A, Chia S,
Ostlund C, Ahmadi A, Kyle A, dem Keller Auf U, Leung S, Huntsman D,
et al: Targeting tumor hypoxia: Suppression of breast tumor growth
and metastasis by novel carbonic anhydrase IX inhibitors. Cancer
Res. 71:3364–3376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tafreshi NK, Lloyd MC, Bui MM, Gillies RJ
and Morse DL: Carbonic anhydrase IX as an imaging and therapeutic
target for tumors and metastases. Carbonic Anhydrase. Mechanism.
Regulation, Links to Disease, and Industrial Applications. Frost SC
and McKenna R: (1st). (New York, NY). Springer. 221–254. 2014.
|
|
22
|
Hynninen P, Parkkila S, Huhtala H,
Pastorekova S, Pastorek J, Waheed A, Sly WS and Tomas E: Carbonic
anhydrase isozymes II, IX, and XII in uterine tumors. APMIS.
120:117–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou R, Huang W, Yao Y, Wang Y, Li Z, Shao
B, Zhong J, Tang M, Liang S, Zhao X, et al: CA II, a potential
biomarker by proteomic analysis, exerts significant inhibitory
effect on the growth of colorectal cancer cells. Int J Oncol.
43:611–621. 2013.PubMed/NCBI
|
|
24
|
Li XJ, Xie HL, Lei SJ, Cao HQ, Meng T-Y
and Hu YL: Reduction of CAII expression in gastric cancer:
Correlation with invasion and metastasis. Chin J Cancer Res.
24:196–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thompson J, Seitz M, Chastre E, Ditter M,
Aldrian C, Gespach C and Zimmermann W: Down-regulation of
carcinoembryonic antigen family member 2 expression is an early
event in colorectal tumorigenesis. Cancer Res. 57:1776–1784.
1997.PubMed/NCBI
|
|
26
|
Kleivi K, Lind GE, Diep CB, Meling GI,
Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE,
Skotheim RI, et al: Gene expression profiles of primary colorectal
carcinomas, liver metastases, and carcinomatoses. Mol Cancer.
6:22007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blumenthal RD, Leon E, Hansen HJ and
Goldenberg DM: Expression patterns of CEACAM5 and CEACAM6 in
primary and metastatic cancers. BMC Cancer. 7:22007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jantscheff P, Terracciano L, Lowy A,
Glatz-Krieger K, Grunert F, Micheel B, Brümmer J, Laffer U, Metzger
U, Herrmann R, et al: Expression of CEACAM6 in resectable
colorectal cancer: A factor of independent prognostic significance.
J Clin Oncol. 21:3638–3646. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schölzel S, Zimmermann W, Schwarzkopf G,
Grunert F, Rogaczewski B and Thompson J: Carcinoembryonic antigen
family members CEACAM6 and CEACAM7 are differentially expressed in
normal tissues and oppositely deregulated in hyperplastic
colorectal polyps and early adenomas. Am J Pathol. 156:595–605.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim M, Lee S, Yang S-K, Song K and Lee I:
Differential expression in histologically normal crypts of
ulcerative colitis suggests primary crypt disorder. Oncol Rep.
16:663–670. 2006.PubMed/NCBI
|
|
31
|
Risques RA, Lai LA, Himmetoglu C, Ebaee A,
Li L, Feng Z, Bronner MP, Al-Lahham B, Kowdley KV, Lindor KD, et
al: Ulcerative colitis-associated colorectal cancer arises in a
field of short telomeres, senescence, and inflammation. Cancer Res.
71:1669–1679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Selbach M and Mann M: Protein interaction
screening by quantitative immunoprecipitation combined with
knockdown (QUICK). Nat Methods. 3:981–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Spisák S, Kalmár A, Galamb O, Wichmann B,
Sipos F, Péterfia B, Csabai I, Kovalszky I, Semsey S, Tulassay Z,
et al: Genome-wide screening of genes regulated by DNA methylation
in colon cancer development. PLoS One. 7:e462152012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiong L, Wen Y, Miao X and Yang Z: NT5E
and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal
transition (EMT) are associated with tumor progression and survival
of patients with gallbladder cancer. Cell Tissue Res. 355:365–374.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Choi CH, Choi JJ, Park YA, Lee YY, Song
SY, Sung CO, Song T, Kim MK, Kim TJ, Lee JW, et al: Identification
of differentially expressed genes according to chemosensitivity in
advanced ovarian serous adenocarcinomas: Expression of GRIA2
predicts better survival. Br J Cancer. 107:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bendas G and Borsig L: Cancer cell
adhesion and metastasis: selectins, integrins, and the inhibitory
potential of heparins. Int J Cell Biol. 2012:6767312012. View Article : Google Scholar : PubMed/NCBI
|