Introduction
Breast cancer has a major impact on the health of
women worldwide. It is the most frequently diagnosed cancer and a
leading cause of cancer-related death, ranking second in Caucasian
(1) and Saudi female patients
(Cancer Incidence Report, NCR, 2004). The incidence and mortality
rates vary between various ethnically and geographically distinct
populations, with the lowest incidence reported among Asians and
the highest among North Americans (2). Multiple causes characterize breast
carcinomas, which may be either familial or sporadic. Genetic
predisposition accounts for only about 5–10% of breast cancer,
whereas 90% of breast cancer cases are sporadic and their origin
remains to be determined (3). The
Saudi population comprises more than 50% of females younger than 20
years old. In this population, the majority of breast cancer cases
diagnosed are at advanced stages and at an early age (4). Similar characteristics have been found
in African-American female individuals (5,6).
Breast carcinogenesis is associated with various
types of somatic genetic alterations, such as mutations in
oncogenes and tumor suppressor genes (7). The most frequently mutated gene in
human malignancies, including breast cancer, is the TP53
gene (8). This important tumor
suppressor gene is a multifunctional transcription factor involved
in the control of cell cycle progression, DNA repair, apoptosis and
angiogenesis (9). The proportion of
TP53 mutations in various cancer tissues ranges from 10 to
80% (10), while that of
TP53 mutations reported in breast tumors ranges from 15 to
71%, with significant differences among populations. Over 1,400
TP53 mutations have been identified in breast cancer
(11). Of these mutations, 80% are
clustered within exons 5–8 (12).
Notably, the proportion of TP53 mutations is higher in
younger patients and those with advanced breast cancer (13); these patients comprise the prevalent
breast cancer patient group among the Saudis. Furthermore,
variations in patterns and distribution of p53 mutations in breast
cancer occur according to ethnicity and geographical location,
indicating the effect of genetic and environmental factors
(14).
Cells lacking normal p53 function have a selective
growth advantage and are more resistant to ionizing radiation and
frequently used anticancer drugs compared to cells with wild-type
p53 protein (15). TP53 gene
mutations predict the response of breast cancer patients to
treatment with various chemotherapeutic agents (16,17).
Furthermore, it has been shown that the TP53 mutation status
is a crucial survival marker of breast cancer that may provide
prognostic data which complements clinical variables (18).
In the present study, the prevalence of TP53
mutations in Arab breast cancer patients was among the highest in
the world (40%), and occurred more frequently in young patients.
Notably, 7 novel mutations, including a 15-bp deletion, were
identified in these sporadic breast cancer patients.
Materials and methods
Sample collection
A total of 119 archived breast tumor samples were
collected from Arab patients living in Saudi Arabia and suffering
invasive ductal carcinoma. All of these patients were diagnosed at
King Faisal Specialist Hospital and Research Center in Riyadh. The
experimental protocol was approved by the institutional Basic
research and Ethics Protocol Committees (RAC proposal no. 2040037).
The age of the patients at the time of diagnosis ranged from 22 to
80 years (median 51). A total of 108 fresh blood samples (5 ml)
were collected from volunteer healthy Arab female individuals, and
used as controls. The age of the healthy Saudi female individuals
(controls) ranged from 17 to 76 years (median 47).
DNA purification
Genomic DNA was purified using the Gentra Puregen
kit according to the manufacturer’s instructions (Gentra Puregene
blood kit; Qiagen, Valencia, CA, USA; cat. no D-50K1–4).
DNA amplification and sequencing of the
TP53 gene
Standard PCR was performed to amplify exons 4–9 and
their intron/exon borders of the TP53 gene, using the
HotStar Taq polymerase kit (Qiagen, Chatsworth, CA, USA). The
primers used for this amplification are listed in Table I. Each PCR reaction was performed in
a total volume of 25 μl containing 4 ng of genomic DNA, 0.5 mM
dNTPs, 1 mM primers, 0.04 units Taq DNA polymerase and
MgCl2 (1.5–3 mM). MgCl2 concentrations were
optimized according to the different primers (Table I). Following a denaturation step of
10 min at 94°C, the PCR amplification consisted of 35 cycles of 45
sec at 94°C, 45 sec at 62°C, 45 sec at 72°C, followed by a final
extension step of 10 min at 72°C. The PCR products were then
directly sequenced using the ABI Prism BigDye Terminator v3.1 cycle
sequencing kit (Applied Biosystems, Foster City, CA, USA). The
unincorporated dye labeled terminators were removed using the DyeEx
96 kit (Qiagen). The reaction product was resuspended in a
formamide loading buffer, and then separated and detected in the
ABI 3730x1 DNA analyzer (Applied Biosystems). The analysis of the
obtained sequence was carried out using the GeneBank database,
NT_010718. TP53 somatic mutations were confirmed by two
independent experiments.
 | Table ITP53 primers used in the PCR
reactions. |
Table I
TP53 primers used in the PCR
reactions.
| Primer | Length (bp) | Sequence (5′ to
3′) | Size | Annealing
temperature (°C) | MgCl2
(mM) |
|---|
| Exon 4 | | | 370 | 62 | 1.5 |
| Forward | 20 | TGA GGA CCT GGT CCT
CTG AC | | | |
| Reverse | 20 | CGG CCA GGC ATT GAA
GTC TC | | | |
| Exon 5 | | | 330 | 62 | 3.0 |
| Forward | 20 | TGT TCC AGT TGC TTT
ATC TG | | | |
| Reverse | 20 | AGA GCA ATC AGT GAG
GAA TC | | | |
| Exon 6 | | | 180 | 56–62 | 2.0 |
| Forward | 20 | GGC CTC TGA TTC CTC
ACT GA | | | |
| Reverse | 20 | GGT CCC CTA AGC AGC
AGG AG | | | |
| Exon 7 | | | 257 | 62 | 2.5 |
| Forward | 20 | CAG GTC TCC CCA AGG
CGC AC | | | |
| Reverse | 20 | TGG AAG AAA TCG GTA
AGA GG | | | |
| Exon 8,9 | | | 391 | 56–62 | 2.5 |
| Forward | 20 | CCT TAC TGC CTC TTG
CTT CT | | | |
| Reverse | 20 | TGT TAG ACT GGA AAC
TTT CC | | | |
Statistical analysis
Statistical analysis was carried out using the SPSS
program version 17. The Chi-square test (χ2) was used to
test for an association between categorical data. P≤0.05 was
considered to be statistically significant.
Results
Prevalence of TP53 mutations is high
among Arab breast cancer patients
Screening for TP53 mutations was carried out
on exons 4–9.
DNA from 119 breast carcinoma tumor samples was
amplified and sequenced. A total of 40 of 119 (33.61%) patients
harbored mutations in the TP53 gene; with 6 patients
harboring more than one mutation. Subsequently, 47 substitutions
were identified in the samples obtained from these 40 patients.
Notably, only 19 exonic mutations of these substitutions were
previously identified in breast cancer patients (Table II). Different types of mutations
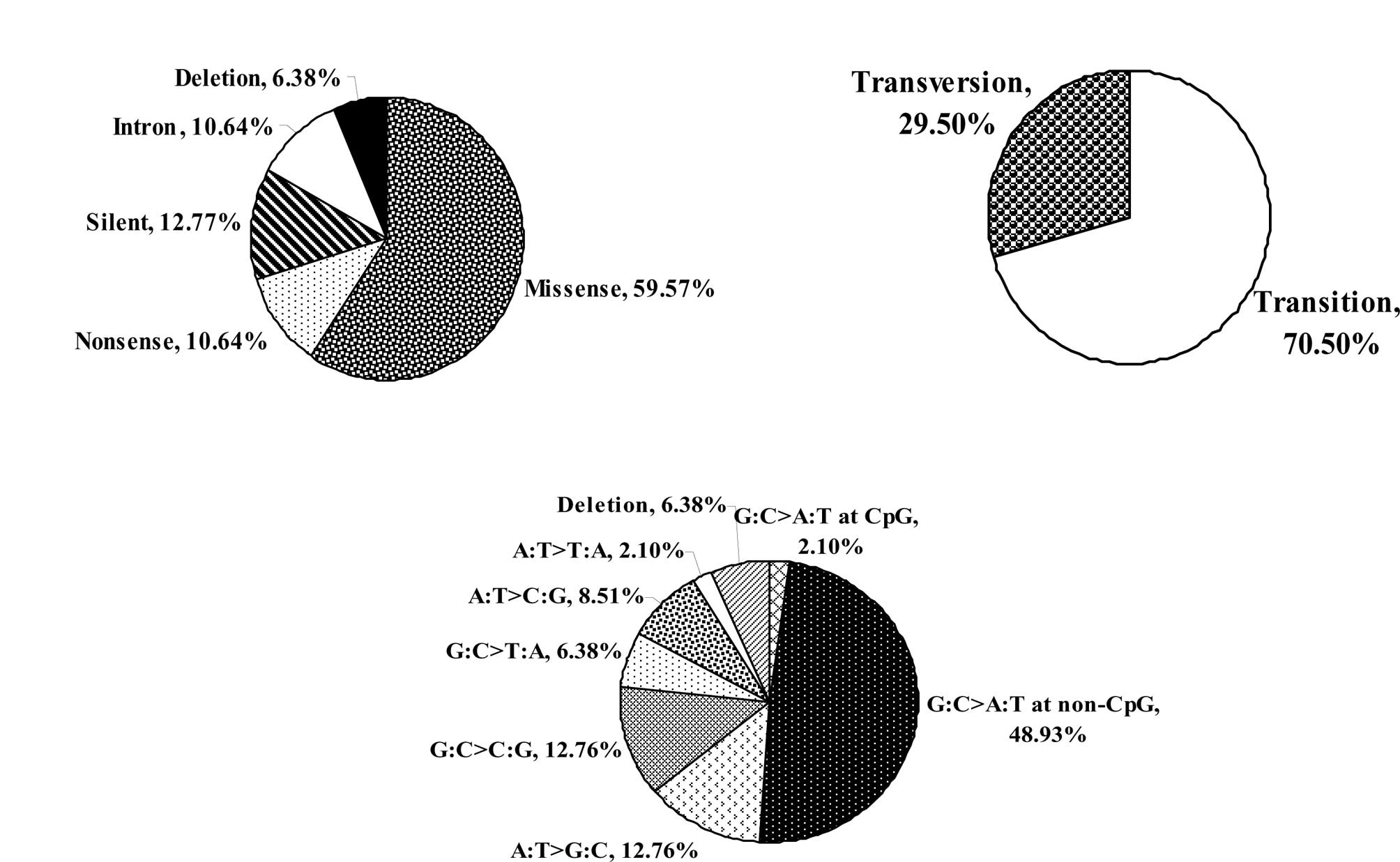
were detected: 28 (59.57%) were missense mutations, 6 (12.77%) were
silent, 5 (10.64%) were nonsense (stop) mutations, and 3 (6.38%)
deletions and 5 mutations (10.64%) were found in the intron-exon
intersections (Fig. 1A). Two of the
3 deletions led to a premature stop codon (frame shift) (Table II).
 | Table IISummary of TP53 mutations and
their nature/location found in breast cancer tissues. |
Table II
Summary of TP53 mutations and
their nature/location found in breast cancer tissues.
| Base change | Structural
change | Mutation type | Exon/Intron | Coding
Description | Mut _ ID |
|---|
| GAC>GGCa | D48G | Transition | E4 | c.143A>G | 449 |
| TGG>TAGa | W53X | Transition | E4 | c.158G>A | 502 |
| ACT>CCTc | T55P | Transversion | E4 | c.163A>C | – |
| CCA>CTAc | P58S | Transition | E4 | c.173C>T | – |
| CCC>CCTc | P64L | Transition | E4 | c.192C>T | – |
| GCA>GGAa | A76G | Transversion | E4 | c.227C>G | 753 |
| GCA>GCGc | A78A | Transition | E4 | c.234A>G | – |
| GCC>GCTa | A84A | Transition | E4 | c.252C>T | 843 |
| Del of Ca | A88TdelfsX33 | Deletion | E4 | c.263del1 | 887 |
| CCC>CCTa | P89P | Transition | E4 | c.267C>T | 899 |
| TAC>TCCc | Y107S | Transversion | E4 | c.320A>C | – |
| GGG>AGGa | G117R | Transition | E4 | c.349G>A | 1209 |
| C>T | No change | Transition | IVS 4-3 | c.376-3C>T | 5820 |
| C>T | - | Transition | IVS 4–14 | c.376-14C>T | – |
| TCC>TTCb | S127F | Transition | E5 | c.380C>T | 1341 |
| Del CAAc |
L130-N131delLfsX15 | Deletion | E5 | c.390–392del3 | – |
| GTG>GCGb | V143A | Transition | E5 | c.428T>C | 1590 |
| CCC>CCTb | P153P | Transition | E5 | c.459C>T | 1761 |
| ACC>ATCb | T155I | Transition | E5 | c.464C>T | 1794 |
| ACC>ACTb | T155T | Transition | E5 | c.465C>T | 1799 |
| ACC>AACa | T155N | Transversion | E5 | c.464C>A | 1792 |
| Del 15 bpc | V157-A161del | Deletion | E5 | c.469–483del15 | – |
| ATC>ATTb | I162I | Transition | E5 | c.486C>T | 1932 |
| CAG>TAGb,d | Q165X | Transition | E5 | c.493C>T | 1972 |
| TGC>TACb | C176Y | Transition | E5 | c.527G>A | 2166 |
| CAT>CGTb | H193R | Transition | E6 | c.578A>G | 2410 |
| CAT>TATb | H193Y | Transition | E6 | c.577C>T | 2408 |
| TAT>GATa | Y220D | Transversion | E6 | c.658T>G | 2819 |
| TAT>TGTb | T220C | Transition | E6 | c.659A>G | 2821 |
| GAG>GCGa,d | E221D | Transversion | E6 | c.662A>C | 2833 |
| TAC>TAGb,d | Y234X | Transversion | E7 | c.702C>G | 3029 |
| TAC>AACb | Y234N | Transversion | E7 | c.700T>A | 3020 |
| TGT>TTTb | C238F | Transversion | E7 | c.713G>T | 3108 |
| C>T | - | Transition | IVS 7–15 | c.783-15C>T | – |
| GTG>TTGb | V272L | Transversion | E8,9 | c.814G>T | 3713 |
| AGA>GGAb | R280G | Transition | E8,9 | c.838A>G | 3844 |
| AGA>ACAa | R280T | Transversion | E8,9 | c.839G>C | 3850 |
| CCT>CTTb | P295L | Transition | E8,9 | c.884C>T | 4084 |
| CAC>TACa | H297Y | Transition | E8,9 | c.889C>T | 4106 |
| CCC>CTCa | P316L | Transition | E8,9 | c.947C>T | 4348 |
| CAG>CGGa | Q317R | Transition | E8,9 | c.950A>G | 4363 |
| ACC>ATCa | T329I | Transition | E8,9 | c.986C>T | 4501 |
| G>A | - | Transition | IVS 8+18 | c.919+18G>A | – |
| G>A | - | Transition | IVS 9+28 | c.993+28G>A | – |
The majority of the identified mutations were
transitions (Fig. 1B). Only one
transition mutation of proline-153 occurred at a CpG site.
Furthermore, various base changes were identified in the 47
TP53 mutations with 24 (51.1%) C:G→T:A transitions (at the
CpG and non-CpG sites) representing the most frequent one (Fig. 1C). The frequency of this transition
reached 48.93% at the non-CpG sites (Fig. 1C).
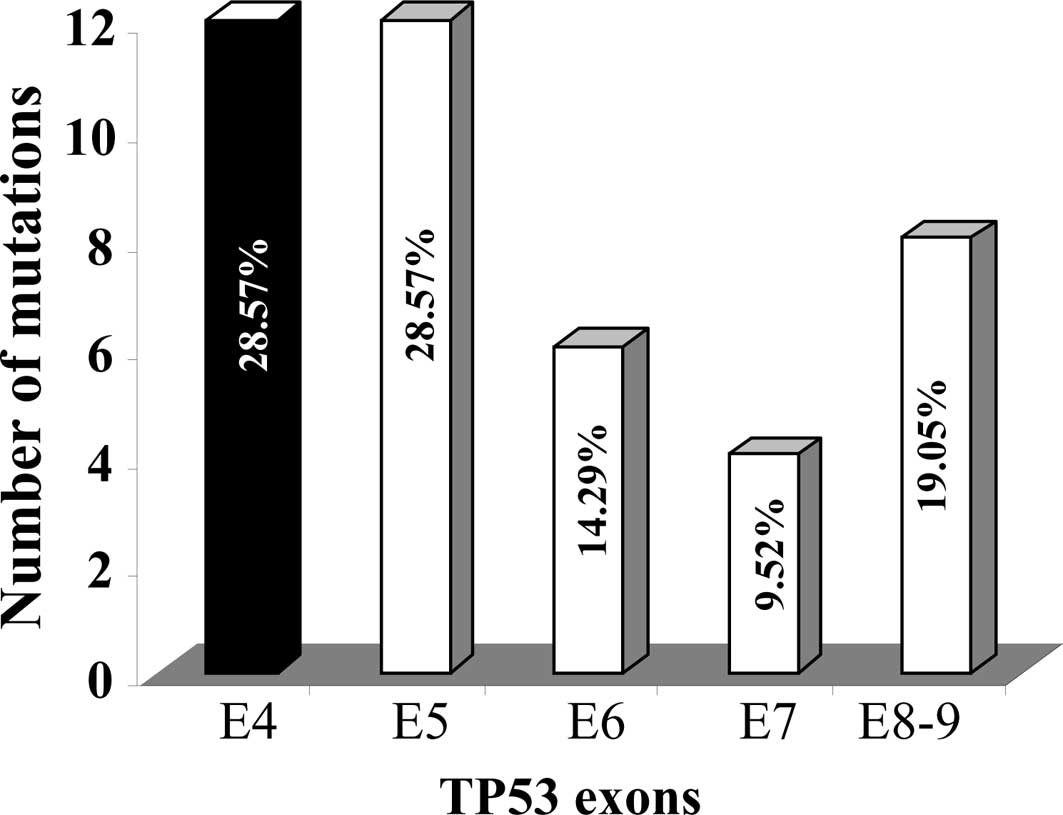
Fig. 2 shows the
distribution of the TP53 mutations within exons 4–9 of the
gene. The majority of the mutations were identified in exons 4 and
5 (12 mutations, representing 29%, in each). However, only 4
mutations were identified in exon 7, while exons 6 and 8,9 harbored
6 and 8 mutations, respectively. This finding shows that exon 4 is
a hot-spot for TP53 mutations in the Saudi Arabian
population. Furthermore, 9 of the 47 mutations were found within
the conserved regions (II, III, IV and V) of the TP53 gene.
Of the 9 mutations, 3 were identified in conserved region V at
valine 272 and argenine 280 (Table
II). Argenine 280 is a significant amino acid involved in
direct DNA binding. Another 2 mutations were found at cysteine 176
and 238, in conserved regions III and IV, respectively. These
cysteines are also directly involved in the binding of the zinc
molecule (Table II). Six (12.8%)
mutations were identified within the zinc-binding loop domains L2
and L3 (codons 163–195 and 236–251, respectively) (Table II). No mutation was detected at the
3 hot-spot codons 248, 273 and 175, nor at the highly mutagenic
codons 245, 249 and 282 (14).
Furthermore, only one mutation was identified at codon 176 and 2 at
codon 220.
Identification of novel mutations in the
TP53 gene
In the present study, 16 new mutations were
identified in the TP53 gene. These mutations were found in
14 different patients. One of these patients harbored 3 mutations.
The majority of these mutations were transitional (10 transitions
vs. 6 transversions) (Table II).
In addition, 7 novel changes were identified in the TP53
gene (not previously reported in breast cancer or any other tumor
type, IARC database, 2008). These changes (5 base substitutions and
2 deletions) were found in 6 different patients, since the tumor
from one patient had 2 of these novel mutations at codons 58 and 64
(Table II). All of the 5 base
substitutions were located in exon 4 at codons 58, 64, 55, 78 and
107 (3 transitions and 2 transversions) (Table II). The 2 novel deletions of 3 and
15 bp were identified in exon 5 (Table
II). The second deletion did not lead to a premature stop
codon, whereas the first one did following the addition of 15 new
amino acids.
The frequency of the 7 novel changes was <1%.
Therefore, they were considered as mutations. To verify this, we
sequenced exon 4 which encompassed the 5 base substitutions from
108 DNA blood samples from healthy Arab female controls. No
substitutions were identified at these sites, confirming that the
substitutions identified in the breast cancer tissues were novel
mutations. Therefore, the frequency of p53 mutations in the Arab
breast cancer patients was 39.49%.
Association between TP53 mutations and
the age of Arab breast cancer patients
The potential link between TP53 mutations and
the age of breast cancer patients was investigated. The patients
were divided into two subgroups depending on their age; the first
group included patients younger than 50 years of age (young
patients), and the second included patients of 50 years or older
(‘old’ patients). As expected, most of the Arab breast cancer
patients (68%) were under 50 years of age, confirming the early
onset of breast cancer in this population. Notably, among 33
patients that harbored TP53 mutations in their tumors, 24
(73%) were young patients, whereas only 9 (27%) were considered
older patients. In each subgroup, patients with tumors harboring
TP53 mutations were compared with those patients with tumors
without TP53 mutations. Table
III shows that the TP53 gene mutations were more
frequent in tumors from younger patients with a prevalence of 35%,
whereas in the older patients the TP53 mutations were only
27%. However, the difference was not statistically significant
(p=0.45).
 | Table IIIAssociation of TP53 gene
mutations with the clinicopathological characteristics of Arab
breast cancer patients. |
Table III
Association of TP53 gene
mutations with the clinicopathological characteristics of Arab
breast cancer patients.
| Total (n) | Positive n (%) | Negative n (%) | P-value |
|---|
| Age |
| <50 | 69 | 24 (34.8) | 45 (65.2) | 0.4480 |
| ≥50 | 33 | 9 (27.3) | 24 (72.0) | |
| Menopausal
status |
| Premenopausal | 68 | 24 (23.08) | 44 (42.31) | 0.1690 |
|
Postmenopausal | 36 | 8 (7.69) | 28 (26.92) | |
| ER status |
| Positive | 48 | 13 (18.31) | 35 (49.30) | 0.7690 |
| Negative | 23 | 7 (9.86) | 16 (22.54) | |
| PR status |
| Positive | 1 | 0 (0.00) | 1 (7.69) | 0.7640 |
| Negative | 12 | 1 (7.69) | 11 (84.62) | |
| ErbB2 status |
| Positive | 45 | 18 (17.48) | 27 (26.21) | 0.0850 |
| Negative | 58 | 14 (13.59) | 44 (42.72) | |
| Involvement of
lymph nodes |
| Positive | 46 | 16 (17.20) | 30 (32.26) | 0.4590 |
| Negative | 47 | 13 (13.98) | 34 (36.56) | |
| Clinical stage of
tumors |
| I | 16 | 3 (3.19) | 13 (13.83) | |
| II | 35 | 11 (11.70) | 24 (25.53) | 0.0447 |
| III | 22 | 12 (12.77) | 10 (10.64) | |
| IV | 21 | 4 (4.26) | 17 (18/09) | |
| Histopathological
grade of tumors |
| I | 9 | 3 (2.88) | 6 (5.77) | |
| II | 53 | 14 (13.46) | 39 (37.50) | 0.6120 |
| III | 42 | 15 (14.42) | 27 (25.96) | |
Association between TP53 mutations and
the clinocopathological characteristics of Arab breast cancer
patients
To investigate the potential role of p53 in the
development and progression of primary breast tumors, the
clinicopathological characteristics of the patients with tumors
harboring p53 mutations were compared with those of patients that
had tumors without p53 mutations. A statistically significant
correlation between the presence of p53 mutations and the clinical
stage of the tumors was found (p=0.0447). Patients with locally
advanced breast cancer stage III A+B showed the highest proportion
of p53 mutations. On the other hand, no statistically significant
correlation was found with the other characteristics, such as the
menopausal status, the histopathological grade, the presence or
absence of lymphatic or vascular invasion, ER/PR status and
Her2neu.
Discussion
In the present study, the frequency of TP53
mutations in Arab breast cancer patients living in Saudi Arabia was
found to be 39.49%. This frequency is considered to be relatively
high, since it is significantly higher than the previously reported
mean proportion of 25% (range 15–71%; examined in 1425 breast tumor
samples worldwide) (19). It is
also higher than the prevalence of p53 mutations in breast tumors
determined in a meta-analysis (18%) (20) and in the IARC mutation prevalence
database on all breast cancers, R9 release (28%) (21). Therefore, the frequency of
TP53 mutations in the KSA is one of the highest in the
world. It is similar to the frequency found in Kashmir (44%)
(22), the USA (45%) (21), Japan (47.5%) (21), the UK (34.5%) (21), and in African-Americans (34.5%)
(23). However, it is higher than
the prevalence reported in patients from Delhi, India (3%)
(24), France (19%) (25), Tokyo (25%) (26) and US midwestern Caucasians (30%)
(27). This variation in p53
mutations in breast cancers may be due to factors such as the
ethno-geographically diverse populations studied, exposure to
various carcinogens, size of the studied population, life-style and
dietary habits. Notably, 7 novel mutations (not previously reported
in the TP53 gene) were identified during this study; 5 of
the 7 mutations were found in exon 4. Therefore, tumors from Arab
breast cancer patients have a high prevalence (28.57%) of
TP53 mutations in exons 4 and 5, whereas the smallest
proportion of TP53 mutations (9.52%) was found in exon 7.
However, in the IARC database, exon 5 has the highest proportion of
TP53 mutations in breast cancer (30.6%) followed by exon 7
(23.5%), while exon 4 represents only 4.2% of mutations (IARC
TP53 Database, R14 release, November 2009, http://www.iarc.fr/p53/homepage.htm/).
Therefore, even the distribution of TP53 mutations in the
various exons of the gene appears to be population-dependent.
Brazilian women of African descent have a higher proportion of
mutations in exons 5 and 7, whereas Brazilian women of Caucasian
descent have more mutations in exon 8. No mutations were found in
Brazilian patients of African descent in exon 4 (29). In the Kashmiri population, no
mutation was found in exon 5, and 52.9% of mutations were
identified in exon 6 (22). To the
best of our knowledge, this study is the first to report a high
proportion of mutations in exon 4 of the TP53 gene.
When we compared the TP53 mutational pattern
in the Arab breast cancer population to the patterns of 15 other
populations from low and high breast cancer-risk countries, we
found that the Saudi population is characterized by a low frequency
(2.1%) of the G:C→A:T transition (at CpG sites) and a high
frequency (48.9%) of the mutational type G:C→A:T transition (at
non-CpG sites). Thus, the Arab population living in Saudi Arabia
possesses the second highest frequency of G:C→A:T transitions at
non-CpG sites after a New Orleans population of African or
Caucasian descent (57%) (23). On
the other hand, the frequency of G:C→A:T transitions at CpG sites
in the KSA is the lowest in the world. IARC mutation spectrum data
on all breast cancer cases reported frequencies of 17.7% at the
non-CpG sites and 21.3% at the CpG sites (21). This variation in the TP53
mutation pattern among different populations may be due to exposure
to various environmental mutagens (23). The association between mutations and
specific exogenous mutagens has been observed in the TP53
gene. The best example is the CC→TT tandem dipyrimidine transition
associated with UV light and G→T transversions associated with
benzo(a)pyrene (14). In the Saudi
breast cancer patients, the most distinguishing feature of the
TP53 mutation pattern was the excess of G:C→A:T transition
at the non-CpG sites, which was rarely found at the CpG sites. The
transition of cytosine to thymine at the CpG sites may result from
spontaneous deamination of methylated cytosine (29). Therefore, the low frequency of this
transition in the Saudi breast cancer patients is likely to be due
to the low cytosine methylation at the CpG sites. On the other
hand, the G:C→A:T transitions at the non-CpG sites is induced by
various carcinogens, in particular oxidizing agents and alkylating
agents such as N-nitroso compounds (e.g., nitrosoamines and
N-nitrosodimethylamine ‘NDMA’) (14). The carcinogenic effect of the
N-nitroso compounds on the mammary gland of laboratory animals is
well established, suggesting that human mammary epithelial cells
contain DNA adducts due to exposure to these chemicals (30,31).
N-nitroso compounds (e.g., N-nitrosdimethylamines) are
procarcinogenic agents that are bioactivated by enzymatic
metabolism (32,33). These agents lead to guanine
alkylation generating o6-alkylguanine (e.g.,
o6-methylguanine), which typically results in G:C→A:T
transitions (34). This adduct can
be directly repaired by alkylguanine alkyltransferase enzymes (e.g.
o6-methylguanine DNA methyl transferase enzymes)
(35). This enzyme has been
detected in breast tissue with large inter-individual variations in
activity (36). Zaidi et al
demonstrated that the presence of estrogen increased the amount of
o6-methylguanine in the DNA of breast xenografts
(34). Therefore, high exposure to
nitrosamines (or NDMA) with insufficient capacity for DNA repair or
high levels of estrogen may lead to the accumulation of DNA damage
and the formation of mutations that trigger cellular transformation
and then breast carcinogenesis. These mutagens and the type of
mutations they induce have been shown to play a role in the
etiopathogenesis of oesophageal and gastric carcinomas (37–39).
Findings of our study showed that among the 33
patients with tumors harboring TP53 mutations, 24 (73%) were
young patients (<50 years of age), while only 9 (27%) were older
patients (≥50 years of age). Furthermore, TP53 mutations
occurred more frequently in tumors from young patients with a
prevalence of 34.8% than in the older patients with a prevalence of
27.3%. However, this difference was not statistically significant
(p=0.45). Studies have reported the presence of an association
between TP53 mutations and the age of breast cancer onset
(13). However, Nagai et al
who reported on the Brazilian population, found no significant
correlation between the age of breast cancer patients and p53
mutations (28).
In the present study, the frequency of p53 mutations
in the Arab breast cancer patients was found to be among the
highest in the world (40%), with a high proportion of these
mutations localized in exon 4 of the gene. Five out of these 12
mutations were identified for the first time. We also identified 2
novel deletions in exon 5. In addition, 16 mutations were
identified for the first time in these breast cancer patients. A
total of 70% of the patients harboring p53 mutations in their
tumors were younger than 50 years of age. Therefore, it can be
concluded that the TP53 gene plays a signficant role in
breast carcinogenesis and the early onset of the disease among Arab
female individuals.
Acknowledgements
We are very thankful to KACST for their financial
help. We also thank the KFSH & RC administration as well as the
Training and Education and ORA offices for their continuous
assistance. This study was performed under the RAC proposal
#2040037 and KACST #LPG 10-9.
References
|
1
|
Parkin DM: International variation.
Oncogene. 23:6329–6340. 2004. View Article : Google Scholar
|
|
2
|
Garfinkel L, Boring CC and Heath CW:
Changing trends. An overview of breast cancer incidence and
mortality. Cancer. 74:222–227. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Polyak K, Porter DA, Krop IE, Nasser S,
Sgroi D, Kaelin CM, Marks JR and Riggins G: On the birth of breast
cancer. Biochim Biophys Acta. 1552:1–13. 2001.PubMed/NCBI
|
|
4
|
Ezzat AA, Ibrahim EM, Raja MA, Al-Sobhi S,
Rostom A, Stuart RK, al-Mulhim FA, al-Amri A, al-Muhanna FA and
Ajarim D: Locally advanced breast cancer in Saudi Arabia: high
frequency of stage III in a young population. Breast cancer in the
eastern province of Saudi Arabia. Med Oncol. 16:95–103. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Neuhausen SL: Ethnic differences in cancer
risk resulting from genetic variation. Cancer. 86:2575–2582. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perera NM and Gui GP: Multi-ethnic
differences in breast cancer: current concepts and future
directions. Int J Cancer. 106:463–467. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polyak K: Molecular alterations in ductal
carcinoma in situ of the breast. Curr Opin Oncol. 14:92–96. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
9
|
Bargonetti J and Manfredi JJ: Multiple
roles of the tumor suppressor p53. Curr Opin Oncol. 14:86–91. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Soussi T, Legros Y, Lubin R, Ory K and
Schlichtholz B: Multifactorial analysis of p53 alteration in human
cancer: a review. Int J Cancer. 57:1–9. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Olivier M, Langerod A, Carrieri P, Bergh
J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche
I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A,
Niederacher D, Kato S, Ishioka C, Hainaut P and Borresen-Dale AL:
The clinical value of somatic TP53 gene mutations in 1,794 patients
with breast cancer. Clin Cancer Res. 12:1157–1167. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hartmann A, Blaszyk H, McGovern RM,
Schroeder JJ, Cunningham J, De Vries EM, Kovach JS and Sommer SS:
p53 gene mutations inside and outside of exons 5–8: the patterns
differ in breast and other cancers. Oncogene. 10:681–688.
1995.PubMed/NCBI
|
|
13
|
Berns EM, Foekens JA, Vossen R, Look MP,
Devilee P, Henzen-Logmans SC, van Staveren IL, van Putten WL,
Inganas M, Meijer-van Gelder ME, Cornelisse C, Claassen CJ,
Portengen H, Bakker B and Klijn JG: Complete sequencing of TP53
predicts poor response to systemic therapy of advanced breast
cancer. Cancer Res. 60:2155–2162. 2000.PubMed/NCBI
|
|
14
|
Olivier M and Hainaut P: TP53 mutation
patterns in breast cancers: searching for clues of environmental
carcinogenesis. Semin Cancer Biol. 11:353–360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lowe SW, Ruley HE, Jacks T and Housman DE:
p53-dependent apoptosis modulates the cytotoxicity of anticancer
agents. Cell. 74:957–967. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Borresen-Dale AL: TP53 and breast cancer.
Hum Mutat. 21:292–300. 2003. View Article : Google Scholar
|
|
17
|
Geisler S, Borresen-Dale AL, Johnsen H,
Aas T, Geisler J, Akslen LA, Anker G and Lonning PE: TP53 gene
mutations predict the response to neoadjuvant treatment with
5-fluorouracil and mitomycin in locally advanced breast cancer.
Clin Cancer Res. 9:5582–5588. 2003.PubMed/NCBI
|
|
18
|
Langerod A, Zhao H, Borgan O, Nesland JM,
Bukholm IR, Ikdahl T, Karesen R, Borresen-Dale AL and Jeffrey SS:
TP53 mutation status and gene expression profiles are powerful
prognostic markers of breast cancer. Breast Cancer Res. 9:R302007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hartmann A, Blaszyk H, Kovach JS and
Sommer SS: The molecular epidemiology of p53 gene mutations in
human breast cancer. Trends Genet. 13:27–33. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pharoah PD, Day NE and Caldas C: Somatic
mutations in the p53 gene and prognosis in breast cancer: a
meta-analysis. Br J Cancer. 80:1968–1973. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olivier M, Eeles R, Hollstein M, Khan MA,
Harris CC and Hainaut P: The IARC TP53 database: new online
mutation analysis and recommendations to users. Hum Mutat.
19:607–614. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eachkoti R, Hussain I, Afroze D, Aejazaziz
S, Jan M, Shah ZA, Das BC and Siddiqi MA: BRCA1 and TP53 mutation
spectrum of breast carcinoma in an ethnic population of Kashmir, an
emerging high-risk area. Cancer Lett. 248:308–320. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hill KA and Sommer SS: p53 as a mutagen
test in breast cancer. Environ Mol Mutagen. 39:216–227. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hedau S, Jain N, Husain SA, Mandal AK, Ray
G, Shahid M, Kant R, Gupta V, Shukla NK, Deo SS and Das BC: Novel
germline mutations in breast cancer susceptibility genes BRCA1,
BRCA2 and p53 gene in breast cancer patients from India. Breast
Cancer Res Treat. 88:177–186. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Faille A, De Cremoux P, Extra JM, Linares
G, Espie M, Bourstyn E, De Rocquancourt A, Giacchetti S, Marty M
and Calvo F: p53 mutations and overexpression in locally advanced
breast cancers. Br J Cancer. 69:1145–1150. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsuda H, Iwaya K, Fukutomi T and Hirohashi
S: p53 mutations and c-erbB-2 amplification in intraductal and
invasive breast carcinomas of high histologic grade. Jpn J Cancer
Res. 84:394–401. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saitoh S, Cunningham J, De Vries EM, et
al: p53 gene mutations in breast cancers in midwestern US women:
null as well as missense-type mutations are associated with poor
prognosis. Oncogene. 9:2869–2875. 1994.PubMed/NCBI
|
|
28
|
Nagai MA, Schaer Barbosa H, Zago MA,
Araujo Silva W Jr, Nishimoto IN, Salaorni S, Guerreiro Costa LN,
Silva Araujo M, Caldas Oliveira AG, Mourao Neto M and Brentani MM:
TP53 mutations in primary breast carcinomas from white and
African-Brazilian patients. Int J Oncol. 23:189–196. 2003.
|
|
29
|
Kouidou S, Agidou T, Kyrkou A, Andreou A,
Katopodi T, Georgiou E, Krikelis D, Dimitriadou A, Spanos P,
Tsilikas C, Destouni H and Tzimagiorgis G: Non-CpG cytosine
methylation of p53 exon 5 in non-small cell lung carcinoma. Lung
Cancer. 50:299–307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reh BD, DeBord DG, Butler MA, Reid TM,
Mueller C and Fajen JM: O(6)-methylguanine DNA adducts associated
with occupational nitrosamine exposure. Carcinogenesis. 21:29–33.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goldman R and Shields PG: Food mutagens. J
Nutr. 133(Suppl 3): 965S–973S. 2003.PubMed/NCBI
|
|
32
|
Schroeder JC, Conway K, Li Y, Mistry K,
Bell DA and Taylor JA: p53 mutations in bladder cancer: evidence
for exogenous versus endogenous risk factors. Cancer Res.
63:7530–7538. 2003.PubMed/NCBI
|
|
33
|
Hecht SS and Hoffmann D: N-nitroso
compounds and man: sources of exposure, endogenous formation and
occurrence in body fluids. Eur J Cancer Prev. 7:165–166. 1998.
|
|
34
|
Zaidi SN, Laidlaw I, Howell A, Potten CS,
Cooper DP and O’Connor PJ: Normal human breast xenografts activate
N-nitrosodimethylamine: identification of potential target cells
for an environmental nitrosamine. Br J Cancer. 66:79–83. 1992.
View Article : Google Scholar
|
|
35
|
Scharer OD: Chemistry and biology of DNA
repair. Angew Chem Int Ed Engl. 42:2946–2974. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao EH, Fan XJ, Yuan XH, Xin SM, Liu YY
and Yu HT: Levels of O6-methylguanine acceptor protein
in extracts of human breast tumor tissues. Cancer Biochem Biophys.
12:53–58. 1991.
|
|
37
|
Lozano JC, Nakazawa H, Cros MP, Cabral R
and Yamasaki H: G-->A mutations in p53 and Ha-ras genes in
esophageal papillomas induced by N-nitrosomethylbenzylamine in two
strains of rats. Mol Carcinog. 9:33–39. 1994.
|
|
38
|
Mir MM, Dar NA, Gochhait S, Zargar SA,
Ahangar AG and Bamezai RN: p53 mutation profile of squamous cell
carcinomas of the esophagus in Kashmir (India): a high-incidence
area. Int J Cancer. 116:62–68. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Siddiqi M, Kumar R, Fazili Z,
Spiegelhalder B and Preussmann R: Increased exposure to dietary
amines and nitrate in a population at high risk of oesophageal and
gastric cancer in Kashmir (India). Carcinogenesis. 13:1331–1335.
1992. View Article : Google Scholar : PubMed/NCBI
|