Introduction
Colon cancer is a tumor of the large intestine,
which accounts for >9% of all the cancer cases (1), and is the third most common cancer
worldwide (2). Surgery is the
standard treatment strategy for patients in the early stages of
colon cancer. However, recurrence and metastasis often occur
following surgery. Therefore, identifying the predictors of early
recurrence and metastasis in patients with colon cancer is
important (3). Currently, proteomics
is at the frontier of life sciences. Protein fingerprinting, which
is one of the technologies used to study the proteome, includes
surface-enhanced laser desorption/ionization time-of-flight mass
spectrometry (SELDI-TOF-MS) and protein chip techniques.
SELDI-TOF-MS is a novel and comparative technology used in
proteomics (4–6). The application of comparative technology
for proteomics, which screens for markers of tumorigenesis, tumor
development and metastasis, is extremely promising (7). Our previous studies have found that
certain protein peaks may represent specific biomarkers for
colorectal adenomas usign SELDI-TOF-MS technology (8). However, data regarding how to predict
early tumor recurrence in situ following colon cancer
surgery is limited. The present study used SELDI-TOF-MS technology
to analyze differences in the serum protein fingerprints of
patients with and without tumor recurrence in situ following
colon cancer surgery. In addition, specific proteins were screened,
with the aim of identifying potential biomarkers of tumor
recurrence in situ.
Materials and methods
Participants
In total, 50 patients with colon cancer recurrence
and 50 patients without colon cancer recurrence were selected
between January 2012 and January 2014 at the Department of
Gastroenterology, Renmin Hospital of Wuhan University (Wuhan,
China). The group presenting in situ colon cancer recurrence
following surgery included patients with anastomotic recurrence of
a malignant tumor within five years after the radical resection of
a colon tumor. By contrast, the group without colon cancer
recurrence following surgery included patients without cancer
recurrence within five years after surgery. Overall, 52 patients
were male and 48 were female. The mean age was 66.1±5.5 years. The
clinical presentation of the patients was confirmed by colonoscopy
and histopathological biopsies. The patients did not suffer from
any other diseases that affect the serum protein content, including
liver or kidney disease, or an endocrine disorder. All the
participants were of Chinese ethnicity. The study was approved by
the Ethics Committee of Renmin Hospital of Wuhan University. All
the patients accepted and signed informed consent.
Patient samples and protein
profiling
Following fasting, 5 ml of venous blood was obtained
from each of the 100 patients from the recurrence and
non-recurrence groups. The samples were then placed in drying tubes
and centrifuged at 2,504 × g for 15 min at 4°C. Next, the upper
serum was collected and transferred into Eppendorf tubes
(Sigma-Aldrich, St. Louis, MO, USA) which were placed in a −80°C
refrigerator and reserved.
The serum samples were then removed from the −80°C
refrigerator, thawed and centrifuged at 626 × g for 10 min at 4°C.
Next, 10 µl was collected from each sample and placed in a 1.5-ml
centrifuge tube with 20 µl U9 buffer (Ciphergen Biosystems, Inc.,
Fremont, CA, USA). The tubes were then centrifuged at 626 × g for
30 min at 4°C, which lead to protein denaturation.
Subsequently, 100 µl WCX2 beads (Ciphergen
Biosystems, Inc.) were added into a 200-µl Eppendorf tube and fixed
onto a magnetic rack. The supernatant was then discarded after 1.5
min. In total, 100 µl WCX buffer solution (Ciphergen Biosystems,
Inc.) was added for 5 min in order to pre-activate the beads. Next,
10 µl processed serum sample was added to the activated beads,
mixed well and incubated at room temperature for 30 min. The
samples were then set onto the magnetic rack, and the supernatant
was discarded after 1.5 min. In total, 100 µl WCX buffer solution
was added to eluate the WCX2 beads. Eluant (10 µL 1%
trifluoroacetic acid) was added and 1 µl eluent that was rich in
proteins was loaded onto an Au/steel chip (Ciphergen Biosystems,
Inc.) along with 1 µl sinapic acid saturated solution
(Sigma-Aldrich), air-dried and placed into a chip reader for
analysis (8).
Using a PBS Type II protein chip reader (Ciphergen
Biosystems, Inc.) to analyze the Au/steel chip, the computer
converted the raw data into protein fingerprinting at a speed of
1×109/sec. Ciphergen protein chip 3.0 software
(Ciphergen Biosystems, Inc., Fremont, CA, USA) was used to
calibrate the data. The abscissa of the mass spectrum represents
the mass to charge ratio (M/Z) of proteins, whilst the ordinate
represents the relative value.
Statistical analysis
Statistical analysis was performed using Biomarker
Wizard version 3.1.0 software (Ciphergen Biosystems, Inc.). In
addition, the data were processed using the SPSS software for
Windows (version 17.0; SPSS Inc., Chicago, IL, USA). Differences in
data between the two groups were analyzed using the t-test, while
the χ2 test was used to quantify the data. P<0.05 was
considered to indicate a statistically significant difference.
Results
Comparison of age and gender
In the recurrence (mean age, 67.3±4.5 years; male,
28; female, 22) and non-recurrence groups (mean age, 65.9±5.7
years; male, 24; female, 26), the differences in the mean age and
gender were not found to be statistically significant
(P>0.05).
Screening the differences of serum
protein peaks from patients with and without colon cancer
recurrence in situ following surgery
The protein molecular masses were set at
2,000–20,000 Da. A protein peak of <2,000 Da was automatically
cleared in order to exclude the impact of certain chemical
compositions, such as sinapic acid, sodium acetate and
trifluoroacetate, on the experimental results. Using the Biomarker
Wizard software, nine protein peaks were identified, which
exhibited statistically significant differences between the
recurrence and non-recurrence groups (P<0.05), including the
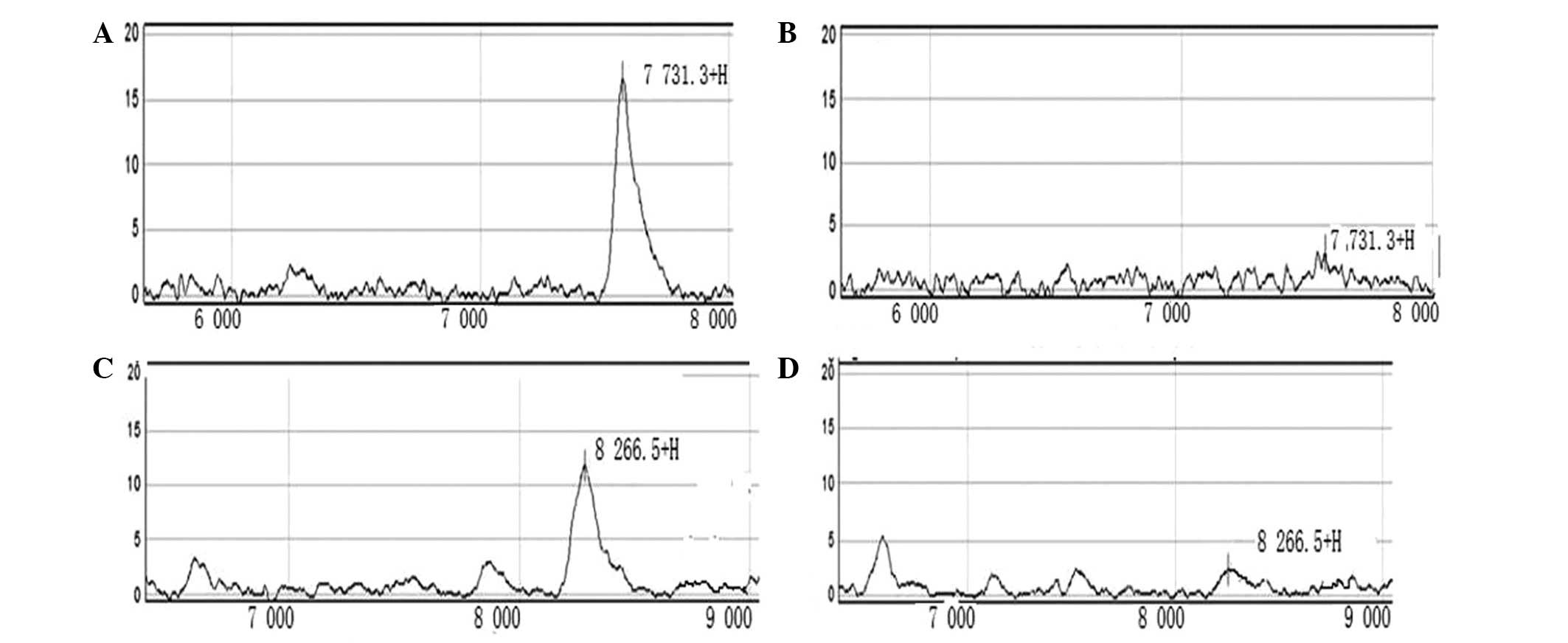
2,687.5, 4,506.1, 5,571.2, 6,879.3, 7,731.3, 8,266.5, 8,969.7,
12,408.1 and 18,257.6 Da proteins. Upon comparing the nine protein
peaks, two protein peaks, 7,731.3 Da and 8,266.5 Da, were found to
have a value of P<0.001 (Table I;
Fig. 1).
 | Table I.Differences in protein expression
levels following serum protein fingerprinting in the colon cancer
recurrence and non-recurrence groups. |
Table I.
Differences in protein expression
levels following serum protein fingerprinting in the colon cancer
recurrence and non-recurrence groups.
|
|
| Average intensity of
protein peak (mean ± SD) |
|---|
|
|
|
|
|---|
| Protein peak, Da | P-value | Recurrence group | Non-recurrence
group |
|---|
| 2,687.5 | 0.0300 | 19.63±4.32 | 14.09±3.34 |
| 4,506.1 | 0.0400 | 14.23±5.16 | 10.55±4.76 |
| 5,571.2 | 0.0300 | 1.37±0.33 | 0.96±0.29 |
| 6,879.3 | 0.0200 | 10.02±3.18 | 7.11±3.02 |
| 7,731.3 | 0.0002 | 17.26±5.39 | 3.66±1.12 |
| 8,266.5 | 0.0003 | 12.97±3.77 | 3.56±1.11 |
| 8,969.7 | 0.0300 | 10.50±3.56 | 15.12±5.78 |
| 12,408.1 | 0.0300 | 3.76±1.09 | 1.39±0.27 |
| 18,257.6 | 0.0300 | 7.22±2.22 | 4.42±1.57 |
Discussion
Each year ~930,000 new cases of colorectal cancer
are diagnosed worldwide (9). With
regard to digestive tract tumors, the incidence of colorectal
cancer is next only to gastric cancer. Colorectal cancer has become
one of the three major types of cancer in China, with its incidence
growing at a rate of 4.2% per year, far exceeding that of the
international average (10). The
principle of colon cancer treatment involves surgical excision
combined with post-surgical chemotherapy and radiotherapy to reduce
recurrence and metastasis, as well as to improve survival. However,
a number of colon cancer patients experience in situ
recurrence at different times following surgery, resulting in
significant reductions in survival rates (11). Identifying an early index for in
situ recurrence may aid with early treatment and, therefore,
prolong survival. Therefore, the development of a simple, quick and
minimally invasive method to accurately detect early recurrence
following colon cancer surgery is essential.
In recent years, proteomic technology has developed
rapidly. Proteomic technology is considered to be a potentially
powerful mean of identifying the mechanisms involved in cancer and
provide effective control measures (12). SELDI-TOF-MS is widely used for the
early diagnosis of cancer and the early prediction of coronary
heart disease and other disorders (8,13,14). It is able to detect the small proteins
that are closely associated with tumorigenesis. A previous study
indicated that, at present, SELDI-TOF-MS is the most promising
method for the early detection of tumors (15).
The present study used SELDI-TOF-MS to compare and
analyze the serum protein fingerprints of patients with (n=50) or
without (n=50) colon cancer recurrence in situ following
surgery. The Biomarker Wizard software was used to conduct
statistical analysis. The results demonstrated the presence of nine
protein peaks that exhibited statistically significant differences
between the two study groups (P<0.05). In particular, two
protein peaks, corresponding to 7,731.3 Da and 8,266.5 Da,
exhibited a value of P<0.001. The identification of the two
protein peaks is important for predicting the likelihood of
recurrence in situ of colon cancer following surgery. In
addition, these peaks provide information regarding the early
prediction of post-operative recurrence and may assist the
identification of specific biomarkers for the prediction of in
situ recurrence following colon cancer surgery.
In order to conduct SELDI-TOF-MS, only a blood
sample is required; therefore, it is a relatively simple technique
compared with other clinical examinations (16). Furthermore, SELDI-TOF-MS is able to
detect highly-specific serum proteins in patients with colon cancer
recurrence in situ following surgery. Using this technology
as an early diagnostic method has broad prospects. SELDI-TOF-MS is
therefore believed to be an effective method for the early
detection and diagnosis of colon cancer recurrence following
surgery.
Acknowledgements
The present study was supported, in part, by grants
from the Natural Science Foundation of Hubei Province (nos.
302-132139 and 2013BKB013).
References
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: incidence, mortality, survival and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Q and Chen H: Epigenetic modifications
of metastasis suppressor genes in colon cancer metastasis.
Epigenetics. 6:849–852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sirop S, Kanaan M, Korant A, et al:
Detection and prognostic impact of micrometastasis in colorectal
cancer. J Surg Oncol. 103:534–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van den Boom D, Wjst M and Everts RE:
MALDI-TOF mass spectrometry. Methods Mol Biol. 1015:71–85. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyer K and Ueland PM: Use of
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry for multiplex genotyping. Adv Clin Chem. 53:1–29.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dingle TC and Butler-Wu SM: Maldi-tof mass
spectrometry for microorganism identification. Clin Lab Med.
33:589–609. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rodrigo MA, Zitka O, Krizkova S, et al:
MALDI-TOF MS as evolving cancer diagnostic tool: a review. J Pharm
Biomed Anal. 95:245–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou ZY, Tao DD, Cao JW and Luo HS:
Application of surface-enhanced laser desorption/ionization
time-of-flight mass spectrometry technology for the diagnosis of
colorectal adenoma. Oncol Lett. 5:1935–1938. 2013.PubMed/NCBI
|
|
9
|
Herszényi L and Tulassay Z: Epidemiology
of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci.
14:249–258. 2010.PubMed/NCBI
|
|
10
|
Cui J, Wang JP, Peng JS, et al: Proteomic
analysis of colon cancer. Chin J Pathophysi. 24:1013–1017.
2008.
|
|
11
|
Sampieri K and Fodde R: Cancer stem cells
and metastasis. Semin Cancer Biol. 22:187–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu SY, Liu Z, Ma WJ, et al: New potential
biomarkers in the diagnosis of esophageal squamous cell carcinoma.
Biomarkers. 14:340–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grizzle WE, Semmes OJ, Basler J, et al:
The early detection research network surface-enhanced laser
desorption and ionization prostate cancer detection study: A study
in biomarker validation in genitourinary oncology. Urol Oncol.
22:337–343. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bakry R, Rainer M, Huck CW and Bonn GK:
Protein profiling for cancer biomarker discovery using
matrix-assisted laser desorption/ionization time-of-flight mass
spectrometry and infrared imaging: a review. Anal Chim Acta.
690:26–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Henderson NA and Steele RJ: SELDI-TOF
proteomic analysis and cancer detection. Surgeon. 3:383–390. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boggio KJ, Obasuyi E, Sugino K, et al:
Recent advances in single-cell MALDI mass spectrometry imaging and
potential clinical impact. Expert Rev Proteomics. 8:591–604. 2011.
View Article : Google Scholar : PubMed/NCBI
|