Introduction
Glioma is one of the most common and lethal cancers
due to the high incidence, mortality rate and disability rate
associated with the disease, which accounts for 32–45% of all
primary brain tumours (1,2) and 70–80% of all malignant brain tumours
(2–4).
Although microsurgery, chemotherapy and radiotherapy have been
developed as treatments for glioma, the majority of patients
succumb to the disease less than two years subsequent to diagnosis
(5). Therefore, there is an urgent
requirement for novel efficient glioma therapies. To achieve this
aim, it is necessary to explore the molecular abnormalities
underlying glioma formation and malignant progression. In addition,
reversing such aberrant changes to evaluate their influence on
glioma development in vitro and in vivo may provide
notable insights into the functions of these glioma-associated
molecules, paving the way for their potential clinical
applications.
Protein synthesis is a major step in gene expression
and translational regulation is critical for homeostasis and normal
cell physiology (6). It is not
unexpected that aberrant protein synthesis markedly contributes to
tumorigenesis and malignant progression, as abnormalities in
protein synthesis lead to the interrupted translation of either
general or specific mRNAs, which are essential for cell survival,
migration or angiogenesis (7).
Numerous studies have described the abnormalities of translational
processes underlying various types of cancers, and these results
indicated the involvement of translation factors in tumorigenesis
(6,8–10). Out of
these processes, translation initiation is the rate-limiting step
of protein translation, and close associations have been identified
between multiple cancer types and eukaryotic translation initiation
factors (eIFs), which are critical components in translation
initiation (8). Therefore, probing
the changes and functions of eIFs in glioma progression is
necessary, and may enable the development of novel therapies for
the treatment of glioma. Among eIFs, the eIF3 complex is
particularly important as it serves as a scaffold to mediate
translation initiation and start codon recognition. Additionally,
eukaryotic translation initiation factor 3, subunit C (eIF3c), a
component of the eIF3 complex, is essential for the assembly of the
eIF3 complex and the translational initiation complex (11). Although the association between eIF3c
and tumorigenesis has been investigated in several studies
(11–14), the expression pattern and function of
eIF3c in glioma has yet to be elucidated.
The present study aimed to clarify the association
between eIF3c and glioma. The expression profiles of eIF3c in human
glioma tissues were analysed, and the expression of eIF3c was then
manipulated using RNA interference (RNAi) in glioma U-87 MG cells
to investigate the function of eIF3c in tumour proliferation and
apoptosis. A glioma xenograft model was also established in nude
mice and the impact of eIF3c inhibition on tumorigenesis was
investigated, suggesting that eIF3c is a potential target for
glioma treatment.
Materials and methods
Preparation of clinical specimens and
immunohistochemical (IHC) analysis
Written informed consent was obtained from all
patients involved in the present study or from the patient's
family, and the study was approved by the Ethics Committee of The
Second Hospital of Hebei Medical University (Shijiazhuang, Hebei,
China). A total of 83 formalin-fixed paraffin-embedded (FFPE)
glioma samples were obtained from patients who underwent surgery at
The Second Hospital of Hebei Medical University from January 2008
to December 2013 with a protocol approved by the same institution,
and the detailed information is summarised in Table II. In total, 25 FFPE normal brain
tissues from traumatic brain injury patients were collected and
used as control samples. To probe the eIF3c protein expression
pattern, eIF3c IHC staining was performed in glioma tissues and
normal brain tissues as previously described (15). eIF3c immunoreactivity was then
determined independently by two pathologists, and the eIF3c
staining was scored as follows: absent, <5% eIF3c-positive
nuclei; and present, 6–100% eIF3c-positive nuclei.
 | Table II.Assessment of the association between
eukaryotic translation initiation factor 3, subunit C expression
and clinical pathological parameters by immunohistochemical
staining. |
Table II.
Assessment of the association between
eukaryotic translation initiation factor 3, subunit C expression
and clinical pathological parameters by immunohistochemical
staining.
| Clinical
pathological parameters | n | eIF3c
expression | Positive
percentage, % | χ2 | P-value |
|---|
|
|---|
| No | Yes |
|---|
| Age, years |
|
<50 | 49 | 8 | 41 | 83.67 | 0.0250 | 0.8745 |
|
≥50 | 34 | 6 | 28 | 82.35 |
|
|
| Gender |
|
Male | 44 | 7 | 37 | 84.09 | 0.0613 | 0.8044 |
|
Female | 39 | 7 | 32 | 82.05 |
|
|
| Tumour diameter,
cm |
| ≥3 | 50 | 9 | 41 | 82.00 | 0.1150 | 0.7332 |
|
<3 | 33 | 5 | 28 | 84.85 |
| Glioma grades |
|
I+II | 27 | 9 | 18 | 66.67 | 6.0950 | 0.0136 |
|
III+IV | 56 | 5 | 51 | 91.07 |
|
|
Preparation of eIF3c small interfering
RNA (si)RNA lentivirus
A lentiviral system and siRNA were employed to
manipulate eIF3c expression, as previously described (16). Briefly, candidate siRNA specifically
targeting human eIF3c was designed for the target sequence
GACCATCCGTAATGC CATGAA using online tools, and the target sequence
for the negative control siRNA was TTCTCCGAACGTGTCACGT. Stem-loop
DNA oligonucleotides were then synthesised and inserted into the
lentiviral pGCSIL-GFP vector (GeneChem Co., Ltd., Shanghai, China).
eIF3c siRNA-expressing lentivirus was prepared using Lentivector
Expression Systems (GeneChem Co., Ltd., Shanghai, China), and the
U-87 MG cell line was infected with associated lentivirus to
determine the knockdown efficiency using western blot analysis and
reverse transcription polymerase chain reaction (PCR).
Cell proliferation assay using
5-bromo-2-deoxyuridine (BrdU)
U-87 MG cells infected with eIF3c or scrambled
siRNA-expressing lentivirus were cultured for 48 h. These cells
were then resuspended and seeded at the proper density into 96-well
plates and cultured in a 5% CO2 incubator at 37°C for
another one or four days. During the final 2–24 h, the BrdU reagent
was diluted at a ratio of 1:100 and then added to 96-well plates
(10 µl/well). Subsequent to 2–24 h of incubation, BrdU signals and
cell proliferation were analysed using a BrdU cell proliferation
ELISA (catalogue number, 11647229001; Roche Applied Science,
Penzberg, Germany), according to the manufacturer's instructions.
Briefly, the cells were fixed using 200 µl FixDenat for each well
for 30 min, and the cells were then blocked with 5–10% bovine serum
albumin for an additional 30 min at room temperature. Anti-BrdU
monoclonal antibody from mouse-mouse hybrid cells (clone BMG
6H8,Fab fragment) conjugated with peroxidase; this antibody was a
component of BrdU cell proliferation ELISA kit (catalogue no.
11647229001; Roche Applied Sciences, Penzberg, Germany) was
diluted, added and incubated for 90 min at room temperature.
Subsequent to washing three times using washing buffer, substrate
solution was added and the cells were incubated for 5–30 min in the
dark. Colour was developed with 10% H2SO4 for
30 min, and the BrdU density was determined at 450 nm.
Cell cycle analysis
Cell cycle analysis was performed according to
previously described methods (17).
Briefly, U-87 MG cells were infected with eIF3c or scrambled
siRNA-expressing lentivirus and incubated for 96 h. Then, the cells
were resuspended and seeded in 6 cm dishes until ∼80% coverage was
achieved. The cells were then harvested and fixed with ice-cold 70%
ethanol for at least 1 h. Subsequent to washing with
phosphate-buffered saline (PBS) buffer, the cells were treated with
staining solution that was prepared using 2 mg/ml 40X propidium
iodide stock solution, 10 mg/ml 100X RNase stock solution and 1X
PBS buffer at a dilution of 25:10:1,000. The cell cycle profiles
were then analysed with FACSCalibur (Becton-Dickinson, Franklin
Lakes, NJ, USA). In total, ≥1×106 cells per sample were
prepared for cell cycle analysis, and the experiments were
performed in triplicate.
Apoptosis assay
Cell apoptosis was analysed using Annexin
V-allophycocyanin (APC) staining. Briefly, U-87 MG cells were
infected with eIF3c or scrambled siRNA-expressing lentivirus and
incubated for 96 h. These cells were collected and washed with PBS
buffer. The cells were resuspended at a final density of
1×106-1×107/ml in staining buffer and 100 µl
cell suspension was mixed with 5 µl Annexin V-APC and incubated at
room temperature in the dark for 10–15 min. Cell apoptosis was
analysed using FACSCalibur (Becton-Dickinson, Franklin Lakes, NJ,
USA).
Colony formation assay
As previously described (18), the U-87 MG cells transfected with
eIF3c or scrambled siRNA-expressing lentivirus were cultured for 48
h and were collected in the logarithmic phase of growth. Subsequent
to the counting of the cells using a haemocytometer, the cells were
seeded in triplicate into six-well plates at a density of 800
cells/well and cultured for 14 days at 37°C in an incubator with a
5% CO2 atmosphere. The cells were then fixed with
paraformaldehyde for 30–60 min and stained with Giemsa stain for 20
min. Subsequent to washing with double-distilled H2O
several times, images of the cell plates were obtained using
Micropublisher 3.3RTV (Olympus, Tokyo, Japan) and the cell colonies
were counted.
Cell proliferation assay using cell
counting methods
Briefly, U-87 MG cells infected with eIF3c or
scrambled siRNA-expressing lentivirus were cultured for 48 h and
were collected in the logarithmic phase of growth. The cells were
then resuspended and seeded in triplicate in 96-well plates, at a
density of 2,000 cells/well, and then cultured at 37°C in an
incubator with a 5% CO2 atmosphere. After 24 h, images
of the cell plates were captured and the images were quantified
using Cellomics ArrayScan VTI (Thermo Fisher Scientific, Waltham,
MA, USA) once a day for five days, and then a cell growth curve was
generated based on data obtained using the Cellomics ArrayScan
VTI.
RNA extraction and reverse
transcription PCR
Total RNA from cultured tumour cells was extracted
with TRIzol (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer's instructions. In total, 2 µg RNA was
reverse-transcribed using M-MLV reverse transcriptase (Promega,
Madison, WI, USA) and Oligo(dT) primers (Sangon Biotech Co., Ltd.,
Shanghai, China) to produce cDNA. To quantify the expression of
eIF3c in the U-87 MG cells infected with eIF3c siRNA-expressing
lentivirus and reverse transcription PCR was performed using the
TP800 Real-Time PCR system (Takara Bio, Inc., Otsu, Japan), with
GAPDH acting as an internal control. The primers used for reverse
transcription PCR were as follows: GAPDH forward, 5′-TGACTTCAACAG
CGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCC AAA-3′; and eIF3c
forward, 5′-AGATGAGGATGAGGATGA GGAC-3′ and reverse,
5′-GGAATCGGAAGATGTGGA ACC-3′. The expression of eIF3c was
normalised to GAPDH expression, and data analysis was conducted
using the comparative CT method.
Western blot analysis
To determine the knockdown efficiency of the eIF3c
siRNA-expressing lentivirus in U-87 MG cells, western blot analysis
was performed to analyse the protein levels of eIF3c. Briefly, the
U-87 MG cells transfected with eIF3c or scrambled siRNA-expressing
lentivirus were cultured for 96 h. The cells were then harvested,
lysed with modified RIPA buffer, boiled in 1X SDS loading buffer
and resolved by 10% SDS-PAGE. The gel was transferred to
polyvinylidene fluoride membranes (Amersham, Uppsala, Sweden), and
the membranes were blocked with 5% milk in Tris-buffered saline
with Tween-20 (TBST) for 1 h. The membranes were then incubated
overnight at 4°C with primary rabbit polyclonal anti-eIF3c antibody
(catalog no. ab170841,Abcam, Cambridge, MA, USA) and Synthetic
peptide within human eIF3s8 aa 126–155 (N-terminal) conjugated to
keyhole limpet haemocyanin (Immunogen, Waltham, MA, USA) at a 1:50
dilution, washed three times in TBST, and the signals from a horse
radish peroxidase reaction were detected using the SuperSignal
chemiluminescent substrate (Pierce Biotechnology, Inc., Rockford,
IL, USA).
Tumorigenicity analysis in nude
mice
The protocols that required nude mice were approved
by the Institutional Animal Care and Use Committee of The Second
Hospital of Hebei Medical University (Shijiazhuang, Hebei, China)
and the animals were fed according to institution guidelines. A
glioma xenograft model in nude mice was established as previously
described, with certain modifications (19), as the untreated U-87 MG cells and the
U-87 MG cells transfected with eIF3c or scrambled siRNA-expressing
lentivirus were cultured for 48 h. The cells were then collected
and resuspended with PBS buffer at a density of 1–2×107
cells/ml, and the nude mice were subcutaneously injected with
∼2×105 cells. The mice were allocated to three groups,
as follows: Negative control group injected with untreated U-87 MG
cells; control group injected with scrambled siRNA-treated U-87 MG
cells; and knockdown group injected with eIF3 siRNA-treated U-87 MG
cells. The nude mice were fed and the diameter and weight of the
tumours were measured every other day from day 10 for 18 days.
Statistical analysis
The present data were analysed with GraphPad Prism 6
software, and all experiments were repeated at least three times.
Differences between the groups were analysed for statistical
significance using Student's two-tailed t-test. P<0.05
was considered to indicate a statistically significant difference.
The data reported in Tables I and
II was analysed using
Cochran-Mantel-Haenszel statistics and SAS software (SAS Institute,
Inc., Cary, NC, USA).
 | Table I.Assessment of the expression of
eukaryotic translation initiation factor 3, subunit C in human
glioma and normal brain samples by immunohistochemical
staining. |
Table I.
Assessment of the expression of
eukaryotic translation initiation factor 3, subunit C in human
glioma and normal brain samples by immunohistochemical
staining.
| Group | n | eIF3c expression | Positive percentage,
% | χ2 | P-value |
|---|
|
|---|
| No | Yes |
|---|
| Glioma tissues | 83 | 14 | 69 | 83.13 | 55.0385 | <0.0001 |
| Normal brain
tissue | 27 | 26 | 1 | 3.70 |
Results
High eIF3c expression is correlated
with malignant phenotypes in human glioma
Aberrant eIF3c expression has been observed in
several human cancer types. In the present study, the expression
pattern of eIF3c was evaluated in 83 FFPE glioma tissues and 27
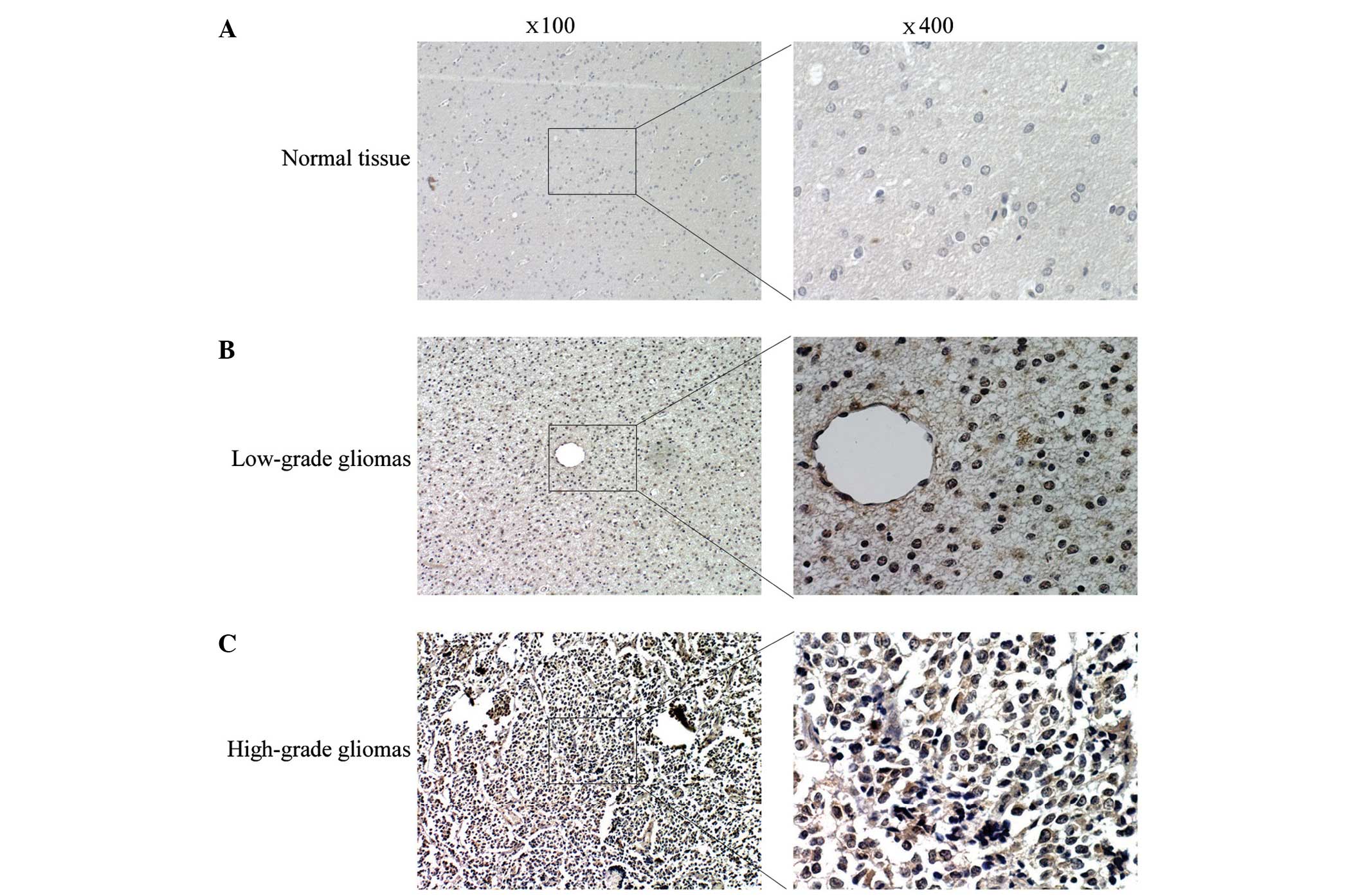
normal brain tissues using IHC staining. The IHC results revealed
that eIF3c expression was significantly increased in glioma tissues
compared with normal brain samples, with positive staining of eIF3c
in 69 out of 83 (83.13%) glioma tissues and in 1 out of 27 (3.70%)
normal brain tissues (Table I).
Furthermore, the association between eIF3c expression and
clinicopathological parameters were analysed in the present study.
No significant association was observed between eIF3c expression
and the patient age or gender, or the tumour volume (Table II). However, a strong correlation has
been observed between eIF3c expression and glioma grades, with
signals for eIF3c expression being detected in 91.07% of high-grade
glioma samples (grades III and IV; n=56) and in only 66.67% of
low-grade glioma tissue samples (n=27) (Table II). Similarly, Fig. 1 demonstrates that eIF3c expression was
increased in high-grade glioma compared with low-grade glioma and
was barely detected in normal tissues. These results suggested that
increased eIF3c expression is associated with a high grade in
glioma.
Lentiviral-mediated siRNA inhibits
eIF3c expression efficiently in human glioma cell lines
The significantly increased expression of eIF3c in
human malignant glioma tissues compared with normal brain tissues
prompted the exploration of its biological roles in glioma in
vitro and in vivo. An RNA interference (RNAi) strategy
based on lentivirus-mediated siRNA was used to deplete eIF3c
expression. To determine the knockdown efficiency, the malignant
human glioma cell line U-87 MG was infected with eIF3c or scrambled
siRNA-expressing lentivirus. The eIF3c mRNA and protein levels were
determined using quantitative fluorescent PCR and western blot
analysis, respectively. Treatment with eIF3c siRNA led to a ∼95%
reduction in the expression of eIF3c mRNA and evident depletion of
the expression of eIF3c protein (Fig. 2A
and B). Therefore, the present results demonstrated that a
highly efficient knockdown of eIF3c expression at the mRNA and
protein levels was achieved using the lentivirus-mediated RNAi
strategy.
eIF3c knockdown inhibits the
proliferation of U-87 MG cells
Tumour cells are characterised by uncontrolled
division and proliferation. To evaluate the role of eIF3c in the
proliferation of U-87 MG cells, the malignant human glioma cell
line U-87 MG was infected with eIF3c or scrambled siRNA-expressing
lentivirus. Cell numbers were monitored using Cellomics ArrayScan
VTI every day for five days, and significant proliferation
inhibition was observed in U-87 MG cells after 24 h of infection.
Furthermore, the inhibitory effect of eIF3c knockdown on cell
proliferation had increased four days subsequent to transfection,
indicating a positive association between proliferation suppression
and the duration of cell culture (Fig. 3A
and B). To further confirm the suppressive effect of eIF3c RNAi
on the proliferation of U-87 MG cells, a BrdU incorporation assay
was performed to assess cell proliferation states in various
treatment conditions. In this assay, cell proliferation was
evaluated by DNA synthesis, which is quantified by the BrdU
incorporation ratio. Initially, the BrdU incorporation ratio was
determined 24 h after the cells were plated and normalised to one.
Consistent with the result of cell counting, BrdU incorporation
after four days of cell plating was significantly suppressed in
U-87 MG cells by eIF3c knockdown compared with controls, with
average incorporation changes of 8.5-fold and 4.4-fold,
respectively (Fig. 3C). These data
suggested that eIF3c knockdown markedly suppressed the
proliferation of the malignant human glioma U-87 MG cell line.
eIF3c knockdown impairs the ability of
U-87 MG cells to form colonies
In addition to uncontrolled cell proliferation,
increased clonogenic capacity is a characteristic of tumour cells
that underlines malignant tumour progression. Therefore, a colony
formation assay was employed to determine the impact of eIF3c
knockdown on the colony formation ability of the malignant human
glioma U-87 MG cell line. The results revealed that a reduction in
the expression of eIF3c in U-87 MG cells impaired colony formation
ability, leading to a ∼34.8% reduction in colony number formation,
with an average of 23 clones in cells treated with scrambled
lentiviral siRNA, and only 15 clones in cells treated with eIF3c
lentiviral siRNA.
eIF3c knockdown induces cell cycle
arrest
To explore the underlying mechanisms of the
inhibitory effect of eIF3c knockdown on U-87 MG cell growth, the
impact of eIF3c knockdown on cell cycle distribution of U-87 MG
cells was analysed using fluorescence-activated cell sorting. It
was revealed that eIF3c knockdown induced a significant
G0/G1 phase arrest in the U-87 MG cells, with
a corresponding reduction in the percentage of S-phase cells. The
proportion of cells in the G0/G1 phase
increased between 71.42 and 79.05% in the eIF3c siRNA treatment
group, while the proportion in the S phase decreased between 17.09
and 7.83% (Fig. 5A and B). Therefore,
the present results indicated that eIF3c knockdown led to
inhibition of the G1-S transition.
eIF3c knockdown promotes apoptosis in
U-87 MG cells
In addition to unlimited proliferation and elevated
clonogenic capacity, evading apoptosis is also essential for tumour
initiation and development (20,21). As
eIF3c knockdown led to a reduction in cell number (Fig. 3B), the present study attempted to
investigate the involvement of apoptosis in this process. U-87 MG
cells were treated with eIF3c lentiviral siRNA or scrambled
lentiviral siRNA and an Annexin V-APC assay was employed to
evaluate the status of cell apoptosis. Apoptosis was observed in
5.83% of U-87 MG cells treated with eIF3c siRNA, while only being
observed in 2.78% of U-87 MG cells treated with scrambled siRNA
(Fig. 5C and D). It was suggested
that eIF3c knockdown may induce cell apoptosis in the malignant
human glioma U-87 MG cell line.
Depletion of eIF3c delays tumoursphere
formation in vivo
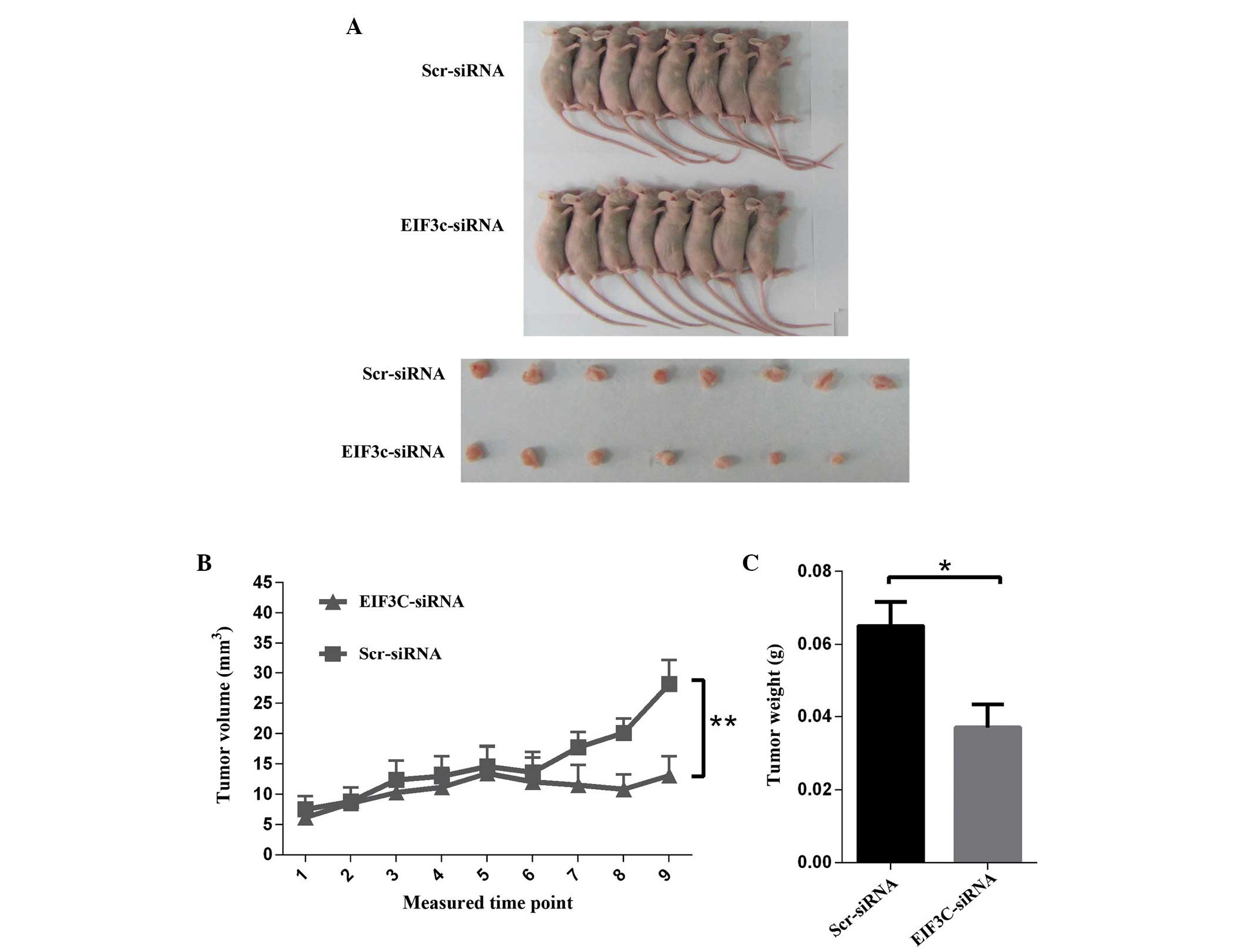
To investigate the role of eIF3c in glioma in
vivo, a glioma xenograft model was established in nude mice.
The mice were injected subcutaneously with ∼2×106 U-87
MG cells pre-transfected with eIF3c or scrambled siRNA-expressing
lentivirus for 48 h. The tumour growth in nude mice was
subsequently estimated by measuring tumour volume every other day
after day 10 for 18 days. In the initial 11 days of measurement, no
significant difference was observed between the various groups.
However, after day 13, the tumour growth was markedly suppressed in
the group treated with lentiviral eIF3c siRNA, and the suppression
effect was more discernible with an increasing time after tumor
implantation (Fig. 6A and B). Nude
mice were sacrificed after 27 days of feeding, and the isolated
tumours were weighed. These results demonstrated that tumour weight
was significantly reduced by 43% subsequent to treatment with eIF3c
siRNA (Fig. 6B and C).
Discussion
Overexpression of eIFs is common in numerous types
of cancer and the aberrant expression of several eIFs, including
eIF3b (22), eIF4e (23) and eIF5a (24), has also been observed in tissues
obtained from patients with glioma, emphasizing the critical role
of eIFs in glioma. The upregulation of eIF3c, a component of the
eIF3 complex, has been reported in several tumour types, including
colon cancer (14), testicular
seminoma (12) and meningioma
(25). Furthermore, the
overexpression of individual eIF3 subunits, including eIF3a, −3b,
−3c, −3h and −3i, may promote the malignant transformation of
NIH3T3 cells. These findings suggest a causal association between
the expression of components of the eIF3 complex and tumour
progression (13). However, prior to
the current study, the role of eIF3c in glioma remained elusive. To
the best of our knowledge, the present study is the first to
uncover the specific overexpression of eIF3c in human glioma. In
normal brain tissue, eIF3c expression was barely detected, while
eIF3c expression was markedly increased in glioma tissue. These
results suggested that the role of eIF3c in glioma is specific and
essential. Additionally, an increased expression of eIF3c is
strongly associated with an increased glioma grade, which provides
additional evidence for the involvement of eIF3c in the development
of glioma.
In support of the critical functions of eIF3c in
glioma development and progression, it was found that eIF3c
knockdown inhibited the proliferation and colony formation ability
(Figs. 3 and 4) of the human malignant glioma U-87 MG cell
line. The present study also demonstrated that such suppression may
be due to cell cycle arrest in the G0/G1
phase and apoptosis induced by eIF3c depletion.
As a component of the eIF3 complex, eIF3c mainly
functions through the eIF3 complex, which is composed of 13
subunits, with an aggregate molecular weight of ∼700 kDa, and the
eIFs that orchestrate translation initiation, the rate-limiting
step of protein translation (26).
Thus, the upregulation of translational initiation factors may
enhance the initiation of protein translation, leading to
acceleration of the protein synthesis involved in tumour initiation
and progression (27). It has been
demonstrated that the eIF3 complex serves as a dynamic scaffold for
mammalian target of rapamycin complex 1 (mTORC1) recruitment and
that eIF3c is critical for the assembly of the eIF3 complex
(28). Therefore, eIF3c upregulation
may enhance mTORC1-mediated translation upregulation, ultimately
leading to aberrant increased protein synthesis and tumour
progression, while eIF3c depletion may impair the assembly of the
eIF3 complex, leading to robust reduction of protein synthesis.
Consistent with this hypothesis, previous studies have revealed
that eIF3c knockdown induced similar changes in human bladder cell
lines to eIF3b knockdown, leading to the significant inhibition of
novel protein synthesis and cell growth (29,7). It was
also noted that the effect of eIF3c and eIF3b on the cell cycle
process is similar in tumour cells, as depletion of either eIF3c or
eIF3b leads to cell cycle arrest at the G0/G1
phase. It has previously been reported that cell cycle arrest
caused by eIF3b depletion may be due to the suppression of cyclin
expression, the elevation of p27Kip1 levels and the
hypophosphorylation of Rb proteins (23). In consideration of the similarity
between eIF3b and eIF3c, it is possible that eIF3c depletion
induces cell cycle arrest through similar mechanisms.
Overall, the present study focused on the functions
of eIF3c in the development of tumours in vitro and in
vivo. The present results indicated that eIF3c is involved in
multiple processes of glioma growth and survival. Furthermore, the
inhibitory effect of eIF3c knockdown on glioma growth was validated
in a nude mouse xenograft model. The results suggested that eIF3c
may be a potential target for glioma treatment. However, the
underlying mechanisms, including the influence of eIF3c knockdown
on tumour proliferation and apoptosis, remain to be explored. As
eIFs are composed of multiple factors, the systematic evaluation of
the changes of all of the components in various tumour types to
determine their specific association with distinct tumour types and
grades is essential, and manipulating the expression of specific
eIFs in various cell lines to identify potential gene targets may
reveal promising cancer biomarkers and drug targets for tumour
treatment.
References
|
1
|
Wakabayashi T: Clinical trial updates for
malignant brain tumors. Rinsho Shinkeigaku. 51:853–856. 2011.[(In
Japanese)]. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol.
15:(Suppl 2). ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohgaki H: Epidemiology of brain tumors.
Methods Mol Biol. 472:323–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grossman SA, Ye X, Piantadosi S, Desideri
S, Nabors LB, Rosenfeld M and Fisher J: NABTT CNS Consortium:
Survival of patients with newly diagnosed glioblastoma treated with
radiation and temozolomide in research studies in the United
States. Clin Cancer Res. 16:2443–2449. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kong J and Lasko P: Translational control
in cellular and developmental processes. Nature Reviews Genetics.
13:383–394. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Silvera D, Formenti SC and Schneider RJ:
Translational control in cancer. Nature Reviews Cancer. 10:254–266.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sonenberg N and Hinnebusch AG: Regulation
of translation initiation in eukaryotes: Mechanisms and biological
targets. Cell. 136:731–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gandin V, Miluzio A, Barbieri AM, et al:
Eukaryotic initiation factor 6 is rate-limiting in translation,
growth and transformation. Nature. 455:684–688. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wendel HG, Silva RL, Malina A, et al:
Dissecting eIF4E action in tumorigenesis. Genes Dev. 21:3232–3237.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Emmanuel R, Weinstein S, Landesman-Milo D
and Peer D: eIF3c: A potential therapeutic target for cancer.
Cancer Lett. 336:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rothe M, Ko Y, Albers P and Wernert N:
Eukaryotic initiation factor 3 p110 mRNA is overexpressed in
testicular seminomas. Am J Pathol. 157:1597–1604. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Pan X and Hershey JWB: Individual
overexpression of five subunits of human translation initiation
factor eIF3 promotes malignant transformation of immortal
fibroblast cells. J Biol Chem. 282:5790–5800. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song N, Wang Y, Gu XD, Chen ZY and Shi LB:
Effect of siRNA-mediated knockdown of eIF3c gene on survival of
colon cancer cells. J Zhejiang Univ Sci B. 14:451–459. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haapa-Paananen S, Kiviluoto S, et al: HES6
gene is selectively overexpressed in glioma and represents an
important transcriptional regulator of glioma proliferation.
Oncogene. 31:1299–1310. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Zhou GB, Liu P, Song JH, Liang Y,
Yan XJ, Xu F, Wang BS, Mao JH, Shen ZX, et al: Dissection of
mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as
an effective treatment for promyelocytic leukemia. Proc Natl Acad
Sci USA. 105:4826–4831. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasaki K, Tsuno NH, Sunami E, et al:
Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on
colon cancer cells. BMC Cancer. 10:3702010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JF, Xie F, Zhang LJ and Jiang WG:
iASPP is over-expressed in human non-small cell lung cancer and
regulates the proliferation of lung cancer cells through a p53
associated pathway. BMC Cancer. 10:6942010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seely BL, Samimi G and Webster NJ:
Retroviral expression of a kinase-defective IGF-I receptor
suppresses growth and causes apoptosis of CHO and U87 cells in
vivo. BMC Cancer. 2:152002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Enderling H and Hahnfeldt P: Cancer stem
cells in solid tumors: Is ‘evading apoptosis’ a hallmark of cancer?
Prog Biophys Mol Biol. 106:391–399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fernald K and Kurokawa M: Evading
apoptosis in cancer. Trends Cell Biol. 23:620–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang H, Ding X, Zhou C, Zhang Y, Xu M,
Zhang C and Xu L: Knockdown of eukaryotic translation initiation
factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in
glioblastoma cells. Neurol Sci. 33:1057–1062. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tejada S, Lobo MV, Garcia-Villanueva M, et
al: Eukaryotic initiation factors (eIF) 2alpha and 4E expression,
localization and phosphorylation in brain tumors. J Histochem
Cytochem. 57:503–512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Preukschas M, Hagel C, Schulte A, et al:
Expression of eukaryotic initiation factor 5A and hypusine forming
enzymes in glioblastoma patient samples: implications for new
targeted therapies. PloS One. 7:e434682012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scoles DR, Yong WH, Qin Y, Wawrowsky K and
Pulst SM: Schwannomin inhibits tumorigenesis through direct
interaction with the eukaryotic initiation factor subunit c
(eIF3c). Hum Mol Genet. 15:1059–1070. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Emmanuel R, Weinstein S, Landesman-Milo D
and Peer D: eIF3c: A potential therapeutic target for cancer.
Cancer Lett. 336:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Silvera D, Arju R, Darvishian F, et al:
Essential role for eIF4GI overexpression in the pathogenesis of
inflammatory breast cancer. Nature Cell Biol. 11:903–908. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Holz MK, Ballif BA, Gygi SP and Blenis J:
mTOR and S6K1 mediate assembly of the translation preinitiation
complex through dynamic protein interchange and ordered
phosphorylation events. Cell. 123:569–580. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang H, Ru Y, Sanchez-Carbayo M, Wang X,
Kieft JS and Theodorescu D: Translation initiation factor eIF3b
expression in human cancer and its role in tumor growth and lung
colonization. Clin Cancer Res. 19:2850–2860. 2013. View Article : Google Scholar : PubMed/NCBI
|