Introduction
Cervical cancer is one of the leading causes of
cancer-associated mortalities among women worldwide, accounting for
>270,000 mortalities annually (1,2). Although
certain advances have been achieved in the early detection and
systemic treatment of patients receiving chemotherapy, hormonal
therapy and immunotherapy, the precise mechanisms underlying
cervical carcinogenesis have remained elusive. Previous studies
have suggested that carcinogenesis is closely associated with the
aberrant expression of certain proteins associated with cell
signaling regulation (3,4). These genetic and molecular events
ultimately contribute to the initiation and progression of tumors.
Therefore, the identification of alterations to specific genes
associated with cervical cancer may provide a conceptual framework
for the future analysis of this complex disease.
The Notch signaling pathway is an important
regulator of embryonic cell fate decisions, proliferation and
patterning (5). Notch receptors are
large, transmembrane epidermal growth factor-like repeat-containing
proteins that contain an extracellular domain responsible for
ligand binding, and a cytoplasmic domain involved in signal
transduction (6,7). In total, mammals express four Notch
receptors: Notch1, 2, 3 and 4, which are activated by Serrate
(Jagged) and Delta (Delta-like) ligands expressed on the surface of
adjacent cells. This activation results in a sequence of
proteolytic cleavage events in the receptor, initially by a
disintegrin and metalloproteinase enzyme and then by the
γ-secretase complex, which releases the Notch intracellular domain
(NICD) from the membrane (8). The
NICD is subsequently translocated to the nucleus where it interacts
with CSL transcription factors and activates the expression of
target genes (9).
In addition to its central role in the developmental
process, Notch1 has been reported to be deregulated in a variety of
types of cancer (10). Hyperactivated
Notch1 signaling has been implicated in the development of T-cell
lymphoblastic leukemia (11), colon
cancer (12) and breast cancer
(13). In a further study,
accumulative events revealed that Notch1 was a major factor in
tumorigenesis due to its involvement in the regulation of cell
growth, proliferation, differentiation and apoptosis (14). By contrast, Nicolas et al
(15) reported that Notch1 was able
to function as a tumor-suppressor gene in mammalian skin, which
indicated that the role of Notch1 in tumorigenesis remains
controversial. At present, the precise association between Notch1
and cervical cancer is yet to be elucidated. A better understanding
of the role of Notch1 in the development of cervical cancer may
provide novel insights into the process of tumorigenesis.
The present study used immunohistochemical staining
to examine the expression of Notch1 in human cervical cancer
tissues and normal cervical tissues. In addition, the association
between Notch1 and various clinicopathological characteristics was
investigated. Furthermore, the effect of Notch1 on the
proliferation of cervical cancer cells was evaluated by small
interfering (si)RNA-mediated Notch1 knockdown.
Materials and methods
Cell lines and cell culture
Human cervical cancer cell lines HeLa, SiHa, C33A,
HT-3 and Caski were purchased from the American Type Culture
Collection (Manassas, VA, USA). The HeLa, SiHa and C33A cells were
maintained in Dulbecco's modified Eagle's medium (DMEM;
Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum (FBS; Invitrogen Life Technologies, Carlsbad, CA,
USA). The HT-3 cells were maintained in McCoy's 5A medium
(Sigma-Aldrich) with 15% FBS, and the Caski cells were maintained
in RPMI-1640 medium (Invitrogen Life Technologies) with 10% FBS.
All cell lines were incubated at 37°C in an atmosphere of 5%
CO2.
Immunohistochemical analysis
A total of 84 samples, including 59 cervical cancer
tissues and 25 adjacent normal cervical tissues, were obtained via
surgical resection from patients previously diagnosed with cervical
cancer who had not received chemotherapy, immunotherapy or
radiotherapy at the Zaozhuang Group Hospital (Zaozhuang, China)
between July 2011 and June 2013. This study was approved by the
ethics committee of the Zaozhuang Group Hospital, and patients
provided informed consent prior to sample collection.
Immunohistochemistry (ICH) procedures were performed
on 5-µm sections prepared from 10% formalin (Luoyang Haohua
Chemical Reagent Co., Ltd., Luoyang, China)-fixed, paraffin (Shanghai
Hua Lingkang Complex Equipment Plant, Shanghai, China)-embedded
tissues. Following deparaffinization with xylene (Tianjin Tianli
Chemical Reagent Co., Ltd., Tianjin, China) and rehydration,
sections were retrieved in 10 mM citrate buffer (Tianjin Tianli
Chemical Reagent Co., Ltd.) (pH 6.0) and blocked with 3%
H2O2 (Tianjin Tianli Chemical Reagent Co.,
Ltd.). The sections were then washed three times with
phosphate-buffered saline (PBS; Tianjin Tianli Chemical Reagent
Co., Ltd.) and incubated with primary polyclonal goat anti-human
Notch1 (dilution, 1:100; cat. no. sc-6014; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) antibody overnight at 4°C.
Next, the sections were incubated with horseradish
peroxidase-conjugated secondary rabbit anti-goat IgG antibodies
(dilution, 1:1,000; cat. no. ZB-2306; Beijing Zhongshan Golden
Bridge Biotechnology Co., Ltd., Beijing, China) for 30 min at room
temperature, visualized using diaminobenzidine (Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd.) and counterstained with
hematoxylin (Sigma-Aldrich).
The sections were observed under a light microscope
(CX21; Olympus Corporation, Tokyo, Japan) and independently scored
by two investigators. The final score was determined by multiplying
the staining intensity (scored as: 1, no staining; 2, weak
staining; and 3, strong staining) by the percentage of positive
cells (scored as: 0, 0–10% positive cells; 1, 10–25% positive
cells; 2, 26–50% positive cells; 3, 51–75% positive cells; and 4,
76–100% positive cells). Samples with a score of >3 were
considered positive.
Western blot analysis
All tumor tissues and cells were lysed on ice with
lysis buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 2 mM EDTA; 1%
NP-40; and 0.1% SDS; Sigma-Aldrich) containing a protease inhibitor
(Complete Mini; Roche Diagnostics, Branchburg, NJ, USA). The
lysates were then separated by 10% SDS-PAGE (Sigma-Aldrich) and
transferred onto PVDF membranes (EMD Millipore, Billerica, MA,
USA). Subsequent to blocking with 5% fat-free milk, the membranes
were incubated with primary polyclonal goat anti-human Notch1
(dilution, 1:500; cat. no. sc-6014; Santa Cruz Biotechnology Inc.),
polyclonal rabbit anti-human Ki67 (dilution, 1:500; cat. no.
sc-15402; Santa Cruz Biotechnology, Inc.) and monoclonal mouse
anti-human β-actin (dilution, 1:1,000; cat. no. sc-47778; Santa
Cruz Biotechnology, Inc.) antibodies at 4°C overnight. This was
followed by incubation with horseradish peroxidase-conjugated
rabbit anti-goat (dilution, 1:5,000; ZB-2306; ZSGB-Bio Technology
Co., Ltd, Beijing, China), goat anti-rabbit (dilution, 1:5,000;
cat. no. A6124; Thermo Fisher Scientific, Waltham, MA, USA) or goat
anti-mouse (dilution, 1:5,000; cat. no. A16090; Thermo Fisher
Scientific) IgG secondary antibodies at room temperature for 1 h.
The proteins were detected by enhanced chemiluminescence (ECL; EMD
Millipore) and visualized on X-ray film. The relative expression of
Notch1 and Ki67 was determined by the Quantity One system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), and normalized to β-actin
for quantification. The intensity of protein expression was scored
according to the ratio of Notch1/Ki67 to β-actin, respectively, as
follows: Notch1, negative expression, <0.25; weak expression,
0.25–0.5; and strong expression, >0.5; and Ki67, negative
expression, <0.5; weak expression, 0.5–1; and strong expression,
>1.
cDNA microarray database analysis
A cDNA microarray database (GSE5787) from an
established human cervical cancer study was retrieved using GEO
profiles (http://www.ncbi.nlm.nih.gov./geoprofiles/?term=GSE5787)
and Pearson's correlation was used to analyze the association
between Notch1 and Ki67 mRNA expression.
RNA interference
A Notch1-targeting siRNA oligonucleotide was
obtained from GenePharma Co., Ltd. (Shanghai, China). The sequences
of the Notch1-targeting siRNA nucleotides were as follows:
siNotch1–1, 5′-GATCCTGGCGGGAAGTGTGAAGCGT-3′; siNotch1–2,
5′-AGCTTAATGGCGGGAAGTGTGAAGC-3′; and siNotch1–3,
5′-AGACGCTTCACACTTCCCGCCATTA-3′. The negative control was designed
as random sequence. A total of 1×105 cells/well were
seeded into six-well plates and transfected with siRNA
oligonucleotides using Lipofectamine® 2000 (Invitrogen Life
Technologies) according to the manufacturers' instructions.
Cell proliferation
The cells were seeded into six-well plates at a
density of 5×104 cells/well and incubated in DMEM
supplemented with 10% FBS at 37°C in an atmosphere of 5%
CO2. The cells were harvested and counted on days 1, 3,
5, and 7 using a hemocytometer (Shanghai Qiujing Biochemical
Reagent Instrument Co., Ltd., Shanghai, China). Cell proliferation
was then assessed following the construction of cell growth
curves.
In order to analyze the cell viability, cells were
seeded into 96-well plates at a density of 1×103
cells/well and cultured at 37°C for 7 days, as aforementioned. MTT
assays were then performed according to standard protocols
(16). In brief, 20 µl MTT (5 mg/ml)
was added to each well and incubated for 4 h at 37°C, followed by
150 µl dimethyl sulfoxide (Sigma-Aldrich). The absorbance was read
at 540 nm using a Bio-Rad 3350 microplate reader (Bio-Rad
Laboratories, Inc.).
Colony formation assay
The cells were plated in 60-mm culture dishes at a
density of 300 cells/well and cultured in DMEM supplemented with
10% FBS at 37°C for 3 weeks. Next, the colonies were rinsed with
PBS, fixed with absolute methanol (Xi'an Chemical Reagent Factory,
Xi'an, China) for 15 min and stained with Giemsa solution (Beijing
DingGuo ChangSheng Biotechnology Co., Ltd., Beijing, China) for 30
min. Colonies containing ≥50 cells were considered to be
positive.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Statistical analysis was performed using SPSS software version 16.0
(SPSS Inc., Chicago, IL, USA). Differences between groups were
analyzed using the χ2 test or Student's t-test,
depending on the data type. Correlations between Notch1 and Ki67
expression were evaluated using the Pearson's correlation test. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
Notch1 expression is enhanced in
cervical cancer tissues
In order to determine the role of Notch1 in cervical
carcinogenesis, its endogenous expression in human cervical cancer
and normal cervical tissues was examined by immunohistochemical
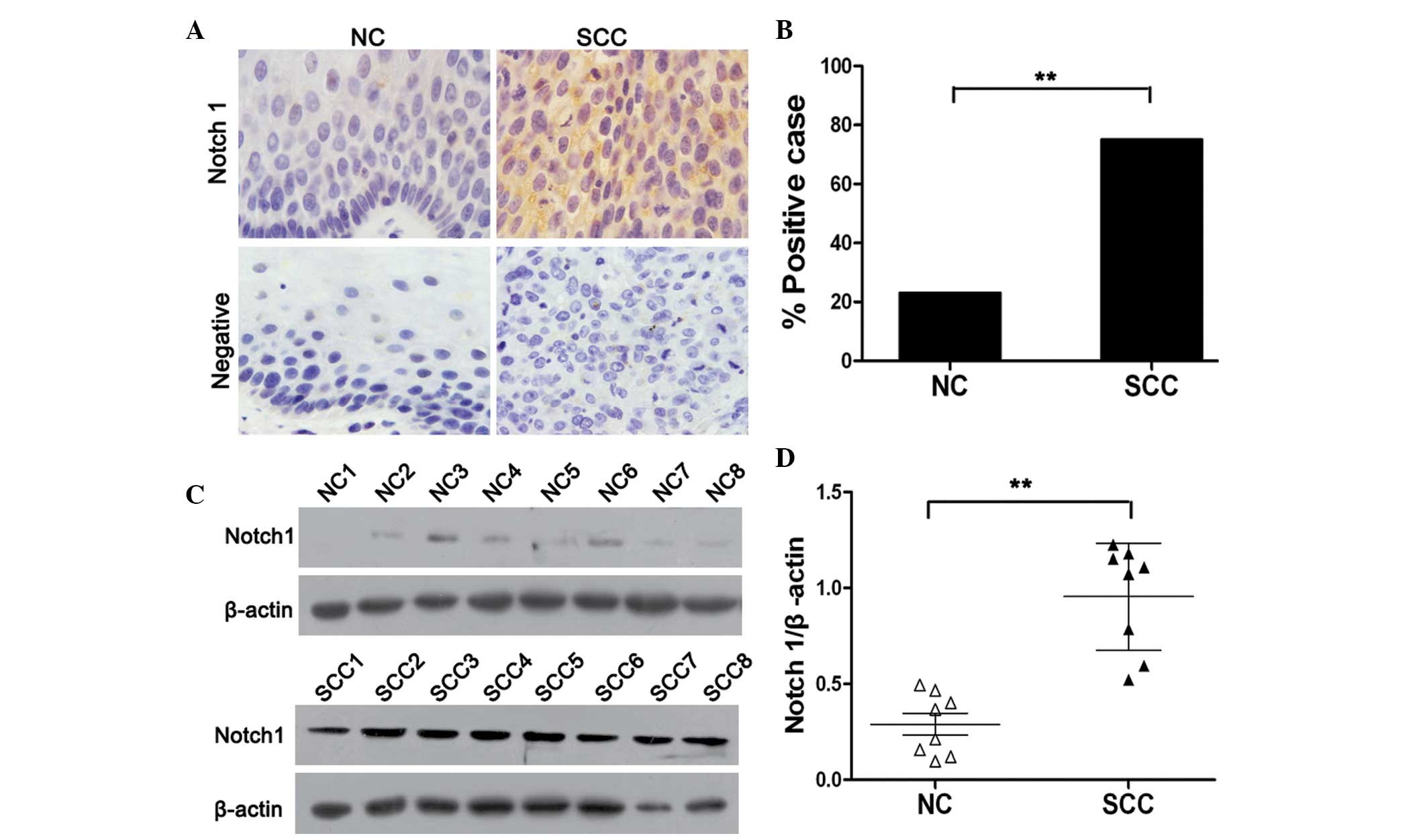
staining. As shown in Fig. 1A, the
expression of Notch1 was primarily identified in the cytoplasm and
membrane of cancer cells. Notch1 expression was significantly
higher in cervical cancer tissues compared with that of normal
cervical tissues (P<0.01). Notch1 protein was positively
expressed in 23% (6/25) of the normal cervical tissue samples, and
75% (44/59) of the squamous cervical cancer tissue specimens
(Fig. 1B). In order to further
investigate the expression of Notch1 in cervical cancer, a western
blot assay was performed using eight randomly selected normal
cervical and cervical cancer tissues. In accordance with the IHC
findings, it was revealed that the levels of Notch1 in cervical
cancer tissues were significantly higher than those in normal
cervical tissues (P<0.01). Together, these results indicated
that Notch1 is highly expressed in cervical cancer, and is
therefore likely to be associated with the progression of cervical
cancer.
Notch1 expression is associated with
tumor differentiation
Correlations between Notch1 expression and the
clinicopathological characteristics of patients with cervical
cancer were investigated. As shown in Table I, a significant positive correlation
was observed between Notch1 expression and tumor differentiation
(P<0.01), but not between Notch1 expression and patient age or
lymph node metastasis status (P>0.05). The percentage of
positive cases in the poorly-differentiated group was markedly
higher than that in the well/moderately-differentiated group. This
finding confirmed that Notch1 was closely associated with the
progression of cervical cancer.
 | Table I.Correlation between Notch1 expression
and clinicopathological characteristics. |
Table I.
Correlation between Notch1 expression
and clinicopathological characteristics.
| Characteristic | All cases |
Notch1-negativea cases |
Notch1-positiveb cases | P-valuec |
|---|
| Total, n | 59 | 15 | 44 |
|
| Age, n (%) |
|
|
| 0.480 |
| ≥55
years | 23 (39) | 7 (47) | 16 (36) |
|
| <55
years | 36 (61) | 8 (53) | 28 (64) |
|
| Tumor
differentiation, n (%) |
|
|
| <0.01 |
|
Well/moderate | 28 (47) | 13 (87) | 15 (34) |
|
|
Poor | 31 (53) | 2 (13) | 29 (66) |
|
| Tumor stage, n
(%) |
|
|
| 0.201 |
|
I–II | 35 (59) | 11 (73) | 24 (55) |
|
|
III–IV | 24 (41) | 4 (27) | 20 (45) |
|
| Lymph node
metastasis, n (%) |
|
|
| 0.714 |
|
Negative | 37 (63) | 10 (67) | 27 (61) |
|
|
Positive | 22 (37) | 5 (33) | 17 (39) |
|
Notch1 expression is positively
correlated with Ki67 expression in cervical cancer tissues
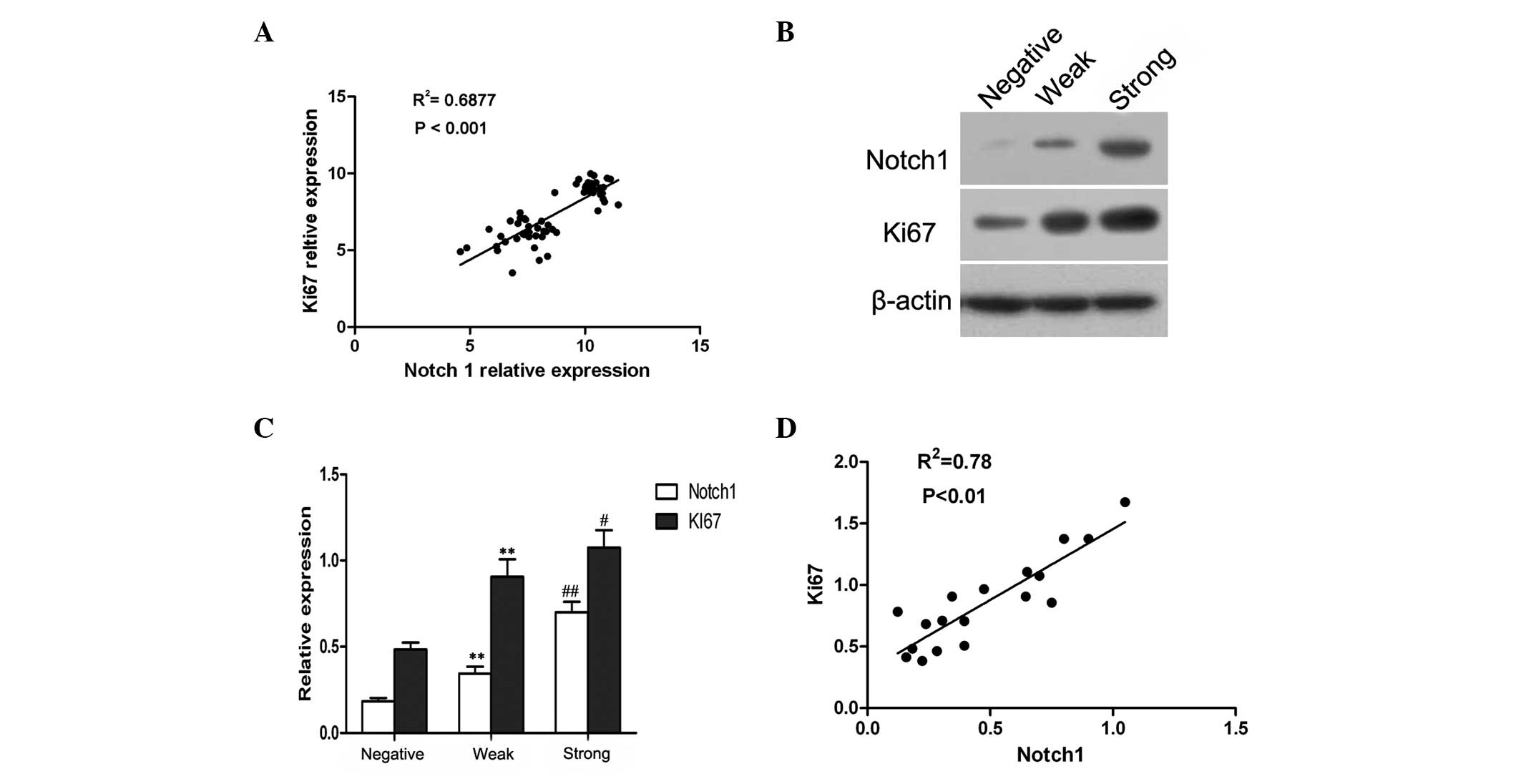
Through analyzing a complementary (c)DNA microarray
database (GSE5758) from an established human cervical cancer study,
a significant, positive correlation was identified between Notch1
and Ki67 expression in cervical cancer (Fig. 2A). It is known that Ki67 is a marker
of cell proliferation (17),
therefore, these observations implied that Notch1 may also be
involved in the proliferation of cervical cancer cells.
In order to determine whether increased Notch1
expression was associated with the proliferation of cervical cancer
cells, 18 cervical cancer cases were randomly selected, and the
expression of Notch1 and Ki67 was assessed by western blot
analysis. A representative blot is shown in Fig. 2B, and the relative quantitative
expression of Notch1 and Ki67 is summarized in Fig. 2C. Based on the intensity of protein
expression, the cases were categorized into three grades; negative,
weak and strong. It was established that the expression of Ki67 in
cervical cancer was similar to that of Notch1 (Fig. 2C). Furthermore, the correlation
analysis confirmed that Notch1 expression was significantly
associated with Ki67 expression in cervical cancer (r=0.88;
P<0.01; Fig. 2B). These data
suggested that Notch1 was likely to be associated with the
proliferation of cervical cancer cells.
Knockdown of Notch1 inhibits the
proliferation and colony formation of cervical cancer cells in
vitro
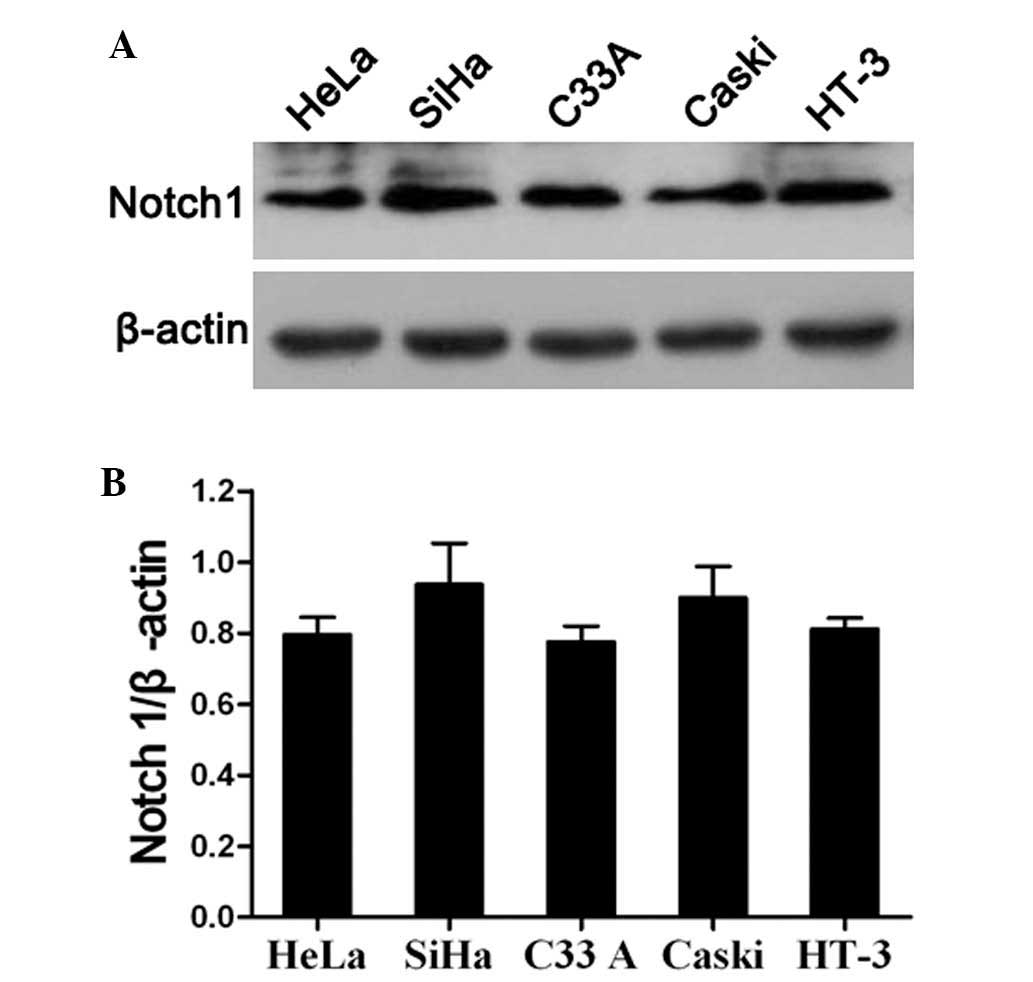
The present study also analyzed the expression of
Notch1 in the HeLa, SiHa, C33A, Caski and HT-3 cervical cancer cell
lines by western blotting. The results revealed that Notch1 was
highly expressed in all cervical cancer cell lines (Fig. 3A and B). This indicated that Notch
signaling is likely to be activated in cervical cancer cell lines.
In order to further investigate the specific role of Notch1 in the
progression of cervical cancer, SiHa and C33A cells were randomly
selected for use as an in vitro model for the assessment of
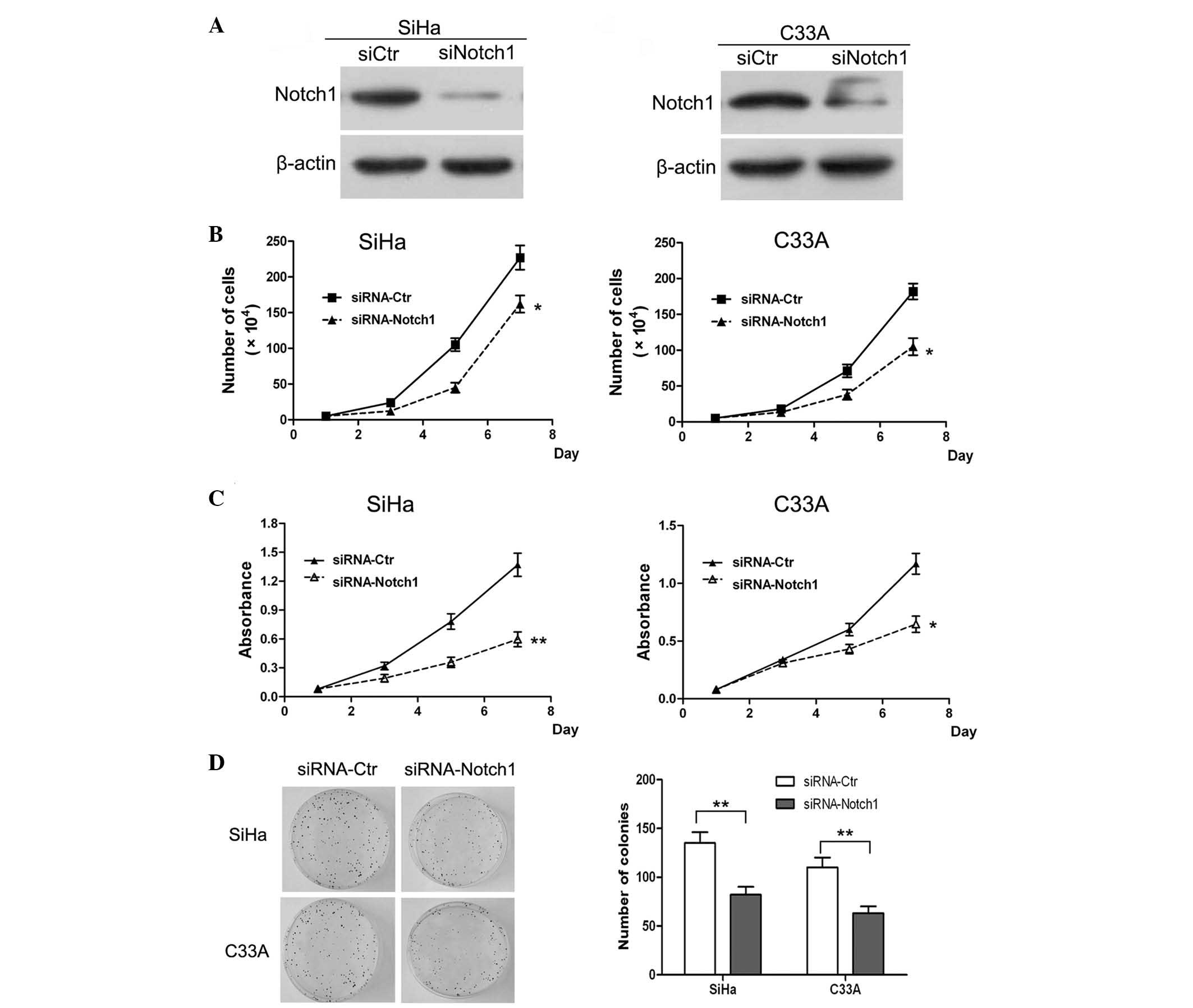
Notch1 function in the subsequent experiments. First, the
expression of Notch1 was knocked down in SiHa and C33A cells by
siRNA. The western blot results of the knockdown are shown in
Fig. 4A. It was revealed that Notch1
was significantly downregulated in the siRNA knockdown SiHa and
C33A cells. Subsequently, a cell growth curve assay and MTT assay
were performed. The cell growth curves demonstrated that Notch1
knockdown in SiHa and C33A cells resulted in a significant
inhibition of cell proliferation (P<0.05; Fig. 4B). In addition, the MTT assay analysis
of SiHa and C33A cells revealed that Notch1 knockdown resulted in a
significant decrease in cell viability (Fig. 4C). These findings confirmed the role
of Notch1 in the promotion of cell proliferation.
A colony formation assay was also performed in order
to determine whether Notch1 enhanced the proliferation ability of
the cells. It was revealed that the number of formed colonies in
Notch1 knockdown SiHa cells was significantly lower than that in
the control cells (P<0.01; Fig.
4D). Similar results were observed following Notch1 knockdown
in C33A cells. Therefore, it was concluded that knockdown of Notch1
expression significantly inhibited the proliferation of SiHa and
C33A cells in vitro. Previous studies have indicated that
Notch1 regulates cell transformation and functions as an oncogene
to promote tumor growth (12,14). Collectively, the results of the
present study indicated that Notch1 is a critical regulator of
cervical cancer cell proliferation in vitro.
Discussion
Cervical cancer is the third most common type of
malignant tumor and the fourth leading cause of cancer-associated
mortalities among women worldwide (18). Particularly in certain developing
countries, the incidence and mortality rates of cervical cancer
remain high due to a lack of screening and appropriate therapeutic
facilities and drugs (19).
Increasing evidence has revealed that aberrant cell proliferation
is associated with the dysregulation of signaling pathways that
link co-regulated genes, which are required for cell homeostasis
during malignant transformation (20). The canonical Notch1 signaling pathway
is believed to be a fundamental transduction pathway involved in
directly transmitting signals from the cell surface to the nucleus
(21). Notch1 has been demonstrated
to have essential roles in the regulation of tumor growth,
invasion, metastasis and angiogenesis (11,22–24).
Furthermore, the overexpression of Notch1 has been observed in
numerous types of human cancer (25,26), and
Notch signaling has been reported to have an oncogenic role in
breast (27), colorectal (28), ovarian (29), pancreatic and prostate cancer
(30), as well as leukemia and
lymphoma (31). However, the function
of Notch1 in cervical cancer progression remains poorly
understood.
In the present study, immunohistochemical staining
and western blotting revealed that Notch1 expression was
significantly higher in cervical cancer tissues than that in normal
cervical tissues. This finding revealed a potential role for Notch1
in cervical carcinogenesis. However, the expression of Notch1 in
cervical cancer remains controversial. Zagouras et al
(32) reported that the expression of
Notch1 in situ and in invasive squamous cancers of the
cervix was higher than in normal cervical tissues (32). However, Talora et al (33) reported that the expression of Notch1
was highly cell- and context-specific. The present study found that
elevated Notch1 expression was significantly associated with tumor
differentiation, but not with age and lymph node metastasis status,
which suggested that Notch signaling may be involved in the
progression of cervical cancer. Taken together, these results
indicate that Notch1 has an essential role in the development and
progression of cervical cancer.
It has previously been reported that the Notch
signaling pathway has a fundamental role in modulating the balance
between cell proliferation, differentiation and apoptosis (5), and that continuous activated Notch1
signaling promotes cell proliferation (34) and survival (35).
In the present study, the reanalysis of cervical
cancer cDNA from a microarray dataset (GSE5787) indicated that Ki67
expression was significantly associated with Notch1 expression in
cervical cancer. Further investigation revealed that the expression
of Ki67 was positively correlated with the expression of Notch1 in
cervical cancer. Ki67 is a marker of cell proliferation.
Accordingly, the results of the present study suggested that Notch1
expression was associated with cell proliferation, and that
cervical cancer cells with high Notch1 expression may have a higher
proliferative activity. As cell proliferation is regulated by
multiple extracellular signals (36),
the role of Notch1 in cervical carcinogenesis has remained elusive.
In order to determine the precise role of Notch1 in tumor
progression, its expression in the SiHa and C33A cervical cancer
cell lines was knocked down by siRNA transfection. The cell growth
curves and MTT assay revealed that the knockdown of Notch1
significantly inhibited the proliferation of cervical cancer cells
in vitro. In addition, an inhibition of colony formation
induced by the knockdown of Notch1 was observed in SiHa and C33A
cells. These findings supported the hypothesis that activated
Notch1 signaling promotes cervical cancer progression. These
results are consistent with those of a previous study, which
demonstrated that Notch1 had a pro-oncogenic role in the
progression of cervical cancer (37).
In conclusion, the present study demonstrated that
Notch1 was highly expressed in cervical cancer, and that the
increased expression of Notch1 promoted the progression of cervical
cancer through enhancing cell proliferation. Collectively, these
results suggested that Notch1 has an important role in cervical
carcinogenesis, and is therefore a candidate therapeutic target for
the treatment of cervical cancer. However, the function of Notch1
in regulating cancer progression is complex, and further studies
are required in order to elucidate the mechanisms underlying the
role of Notch1 in promoting the progression of cervical cancer.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saavedra KP, Brebi PM and Roa JC:
Epigenetic alterations in preneoplastic and neoplastic lesions of
the cervix. Clin Epigenetics. 4:132012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan LH, Wang W, Yeung W, Deng Y, Yuan P
and Mak KK: Hedgehog signaling induces osteosarcoma development
through Yap1 and H19 overexpression. Oncogene. 33:4857–4866. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burgess AW, Faux MC, Layton MJ and Ramsay
RG: Wnt signaling and colon tumorigenesis - a view from the
periphery. Exp Cell Res. 317:2748–2758. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Egan S, St-Pierre B and Leow C: Notch
receptors, partners and regulators: From conserved domains to
powerful functionsIn: Protein Modules in Signal Transduction.
Pawson AJ: Springer; New York, NY: pp. 273–324. 1998
|
|
7
|
Callahan R and Egan SE: Notch signaling in
mammary development and oncogenesis. J Mammary Gland Biol
Neoplasia. 9:145–163. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kopan R: Notch: A membrane-bound
transcription factor. J Cell Sci. 115:1095–1097. 2002.PubMed/NCBI
|
|
9
|
Lai EC: Keeping a good pathway down:
Transcriptional repression of Notch pathway target genes by CSL
proteins. EMBO Rep. 3:840–845. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harrison H, Farnie G, Brennan KR and
Clarke RB: Breast cancer stem cells: Something out of notching?
Cancer Res. 70:8973–8976. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Palomero T, Lim WK, Odom DT, et al: NOTCH1
directly regulates c-MYC and activates a feed-forward-loop
transcriptional network promoting leukemic cell growth. Proc Natl
Acad Sci USA. 103:18261–18266. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Li B, Ji ZZ and Zheng PS: Notch1
regulates the growth of human colon cancers. Cancer. 116:5207–5218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolós V, Mira E, Martinez-Poveda B, et al:
Notch activation stimulates migration of breast cancer cells and
promotes tumor growth. Breast Cancer Res. 15:R542013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jundt F, Anagnostopoulos I, Förster R,
Mathas S, Stein H and Dörken B: Activated Notch1 signaling promotes
tumor cell proliferation and survival in Hodgkin and anaplastic
large cell lymphoma. Blood. 99:3398–3403. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nicolas M, Wolfer A, Raj K, et al: Notch1
functions as a tumor suppressor in mouse skin. Nat Genet.
33:416–421. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prasad R, Vaid M and Katiyar SK: Grape
proanthocyanidin inhibit pancreatic cancer cell growth in vitro and
in vivo through induction of apoptosis and by targeting the
PI3K/Akt pathway. PLoS One. 7:e430642012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jaks V, Barker N, Kasper M, et al: Lgr5
marks cycling, yet long-lived, hair follicle stem cells. Nat Genet.
40:1291–1299. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mathew A and George PS: Trends in
incidence and mortality rates of squamous cell carcinoma and
adenocarcinoma of cervix - worldwide. Asian Pac J Cancer Prev.
10:645–650. 2009.PubMed/NCBI
|
|
20
|
Hahn WC and Weinberg RA: Modelling the
molecular circuitry of cancer. Nat Rev Cancer. 2:331–341. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blanpain C, Lowry WE, Pasolli HA and Fuchs
E: Canonical notch signaling functions as a commitment switch in
the epidermal lineage. Genes Dev. 20:3022–3035. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kunnumakkara AB, Anand P and Aggarwal BB:
Curcumin inhibits proliferation, invasion, angiogenesis and
metastasis of different cancers through interaction with multiple
cell signaling proteins. Cancer Lett. 269:199–225. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu C, Li Z, Bi L, et al: NOTCH1 signaling
promotes chemoresistance via regulating ABCC1 expression in
prostate cancer stem cells. Mol Cell Biochem. 393:265–270. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Timmerman LA, Grego-Bessa J, Raya A, et
al: Notch promotes epithelial-mesenchymal transition during cardiac
development and oncogenic transformation. Genes Dev. 18:99–115.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li D, Dong P, Wu C, Cao P and Zhou L:
Notch1 overexpression associates with poor prognosis in human
laryngeal squamous cell carcinoma. Ann Otol Rhinol Laryngol.
123:705–710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu WR, Zhang R, Shi XD, et al: Notch1 is
overexpressed in human intrahepatic cholangiocarcinoma and is
associated with its proliferation, invasiveness and sensitivity to
5-fluorouracil in vitro. Oncol Rep. 31:2515–2524. 2014.PubMed/NCBI
|
|
27
|
Reedijk M, Odorcic S, Chang L, et al:
High-level coexpression of JAG1 and NOTCH1 is observed in human
breast cancer and is associated with poor overall survival. Cancer
Res. 65:8530–8537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bolós V, Blanco M, Medina V, Aparicio G,
Díaz-Prado S and Grande E: Notch signalling in cancer stem cells.
Clin Transl Oncol. 11:11–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kunnimalaiyaan M and Chen H: Tumor
suppressor role of Notch-1 signaling in neuroendocrine tumors.
Oncologist. 12:535–542. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Li Y, Banerjee S, et al:
Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer
cell growth, migration and invasion and induces apoptosis via
inactivation of Akt, mTOR and NF-κB signaling pathways. J Cell
Biochem. 109:726–736. 2010.PubMed/NCBI
|
|
31
|
Weng AP, Millholland JM, Yashiro-Ohtani Y,
et al: c-Myc is an important direct target of Notch1 in T-cell
acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zagouras P, Stifani S, Blaumueller CM,
Carcangiu ML and Artavanis-Tsakonas S: Alterations in Notch
signaling in neoplastic lesions of the human cervix. Proc Natl Acad
Sci USA. 92:6414–6418. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Talora C, Sgroi DC, Crum CP and Dotto GP:
Specific down-modulation of Notch1 signaling in cervical cancer
cells is required for sustained HPV-E6/E7 expression and late steps
of malignant transformation. Genes Dev. 16:2252–2263. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Go MJ, Eastman DS and Artavanis-Tsakonas
S: Cell proliferation control by Notch signaling in Drosophila
development. Development. 125:2031–2040. 1998.PubMed/NCBI
|
|
35
|
Perumalsamy LR, Nagala M and Sarin A:
Notch-activated signaling cascade interacts with mitochondrial
remodeling proteins to regulate cell survival. Proc Natl Acad Sci
USA. 107:6882–6887. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu CY, Liang GB, Du P and Liu YH: Lgr4
promotes glioma cell proliferation through activation of Wnt
signaling. Asian Pac J Cancer Prev. 14:4907–4911. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Srivastava S, Ramdass B, Nagarajan S,
Rehman M, Mukherjee G and Krishna S: Notch1 regulates the
functional contribution of RhoC to cervical carcinoma progression.
Br J Cancer. 102:196–205. 2010. View Article : Google Scholar : PubMed/NCBI
|