Introduction
An immediate reaction to hypoxia is a reduction in
the rate of global protein synthesis, which is thought to reduce
energy demands when oxygen and adenosine triphosphate levels are
low (1). This response triggers
stressors that alter the environment of the endoplasmic reticulum
(ER) by impairing nascent ER protein glycosylation, disulfide bond
formation or calcium level interference with protein folding,
resulting in the ER stress response (2–4). To
prevent the deleterious effects of ER stress, cells have evolved
numerous protective strategies, including activating signal
transduction pathways that trigger complex transcriptional and
translational responses, which is known as the unfolded protein
response (UPR) (5). The UPR is
mediated by three distinct ER-transmembrane signaling proteins that
act as major proximal sensors of the ER stress response, consisting
of inositol-requiring transmembrane kinase and endonuclease 1α,
activation of transcription factor 6 (ATF6) and protein kinase-like
ER kinase (6–8). Full-length p90-ATF6 is located in the ER
membrane, and in the absence of ER stressors, the protein is
maintained in an inactive form by binding to glucose-regulated
protein (GRP)78 (9). When activated,
cleaved ATF6, or p50-ATF6, then translocates into the nucleus,
where it activates the transcription of ER stress
response-associated genes, including GRP78 (10,11).
Cyclophilin B (CypB) is a 21-kDa protein belonging
to the cyclophilin family of peptidyl-prolyl cis-trans
isomerases (12). CypB has been
identified in the ER and nucleus and is also present in notable
levels in the blood and breast milk (13). It has previously been revealed that
CypB plays important roles in the response to ER stress-mediated
cell death (14). Although it has
been reported that the expression of CypB is regulated under
physiological and pathophysiological states, the mechanism of the
regulation is largely unknown.
The aim of the present study was to explore the
possible functional mechanisms underlying the induction of CypB
under hypoxic conditions. In addition, the transcription factors
involved in the increase of CypB expression under hypoxia have yet
to be reported in the literature. In the present study, it was
demonstrated that hypoxia transcriptionally upregulates CypB
through ATF6. Therefore, understanding the molecular basis of the
regulation of CypB under hypoxic conditions is of clinical
importance for the removal of gastric adenocarcinoma cells. These
results indicate that CypB may be a key molecule in the adaptation
of cells to hypoxia.
Meterials and methods
Reagents and antibodies
Fetal bovine serum (FBS) and RPMI-1640 medium were
purchased from Lonza (Walkersville, MD, USA). X-tremeGENE DNA
transfection reagents were purchased from Roche Applied Science
(Mannheim, Germany). The cyclophilin B antibody was purchased from
Abcam (Cambridge, UK). Actin, human influenza hemagglutinin
(HA)-tag and an enhanced chemiluminescence western blotting kit
were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA). Cyclophilin B, calnexin, ATF6 and ATF4 were purchased from
Abcam. All other chemical reagents were obtained from Sigma-Aldrich
(St. Louis, MO, USA).
Plasmid constructions
cDNA encoding the C-terminal 670 amino acids of ATF6
was subcloned into the pcDNA vector (Invitrogen, Carlsbad, CA,
USA), between the BamHI and XhoI sites for protein
expression, which tagged the protein with an HA-tag at the
C-terminal end. The human CypB promoter reporter constructs (−974,
−800, −350, −250 and −100) were kindly provided by Dr Sung Soo Kim
(Kyung Hee University, Seoul, Korea). All constructs were verified
by sequencing.
Cell culture and in vitro hypoxia
model
The human gastric carcinoma AGS cells (American Type
Culture Collection, Manassas, VA, USA) were maintained in RPMI-1640
medium supplemented with 10% FBS, in humidified air containing 5%
CO2 at 37°C. The cells were incubated under either
normoxic (20% O2) or hypoxic conditions (0.1%
O2). Cultured AGS cells were subjected to hypoxic
conditions at 37°C for the time periods indicated, while controls
were maintained in normoxic conditions at 37°C for the same time
periods.
Reverse transcription-polymerase chain
reaction (RT-PCR) and quantitative PCR analysis
Total cellular RNA was extracted from cells using
TRIzol reagent (Invitrogen). The cDNA was synthesized from 1 µg of
total RNA using M-MLV reverse transcriptase (Fermentas, Hanover,
MD, USA). The specific primers used for RT-PCR are as follows: CypB
forward, 5′-AATTCCATCGTGTAATCAAGGACTT-3′ and reverse,
5′-TCTTGACTGTCGTGATGAAGAACT-3′; and β-actin forward,
5′-GTACTTGCGCTCAGGAGGAG-3′ and reverse, 5′-TCGTGCGTGACATTAAGGGG-3′.
The PCR products were analyzed by agarose gel electrophoresis and
visualized using ethidium bromide. The signals were quantitated
using ImageJ software (version 1.45; National Institutes of Health,
Bethesda, MD, USA). Quantitative PCR was performed using an ABI
prism 7300 Sequence Detection System (Applied Biosystems Life
Technologies, Foster City, CA, USA) with SYBRGreen PCR Master Mix
(Applied Biosystems). The PCR reaction was carried out for 40
thermal cycles of denaturation at 94°C for 30 sec, annealing at
55°C for 30 sec and extension at 72°C for 30 sec. Expression of the
target gene was analyzed by an absolute quantification method and
normalized using β-actin levels.
Luciferase assay
The CypB promoter sequence was analyzed using
Genomatix MatInspector (Genomatix Software GmbH, Munich, Germany).
One ER stress element (ERSE) candidate was located at 222 bp, ∼209
bp from the CypB open reading frame. The AGS cells were transfected
with 1.0 µg of the pGL3 basic-derived plasmids together with the
pCMV-β-galactosidase internal control plasmid (Promega, Madison,
WI, USA). The activity of luciferase and β-galactosidase was
measured using 50 µl of each cell lysate and a fluorescence
microplate reader (Victor 1420 Multilabel Counter; Wallac, Turku,
Finland). The luciferase activity was normalized on the basis of
the β-galactosidase values. The transfection experiments were
performed in triplicate and repeated at least three times.
Western blot analysis
The cells were washed twice with cold PBS on ice and
harvested by scraping with a rubber cell scraper. The cells were
pelleted by centrifugation at 4°C and resuspended directly into a
lysis buffer, which consisted of 50 mM Tris-HCl (pH 7.4), 150 mM
NaCl, 1% Triton X-100, 0.5% Igepal CA-630, 2 mM EDTA, 10 mM NaF, 2
mM Na3VO4 and 0.01% protease inhibitor
cocktail. The cell lysates were subjected to SDS-PAGE and
transferred to a nitrocellulose membrane. Subsequent to blocking in
5% skim milk and Tris-buffered saline with 0.1% Tween-20, signals
were detected and analyzed by a Kodak X-OMAT 2000 image analyzer.
Densitometric analysis was performed using ImageJ software.
Statistical analysis
The results were expressed as the mean ± standard
deviation, which was obtained from at least three independent
experiments. Statistical analyses were conducted using a Student's
t-test. By convention, P<0.05 was considered to indicate
a statistically significant difference.
Results
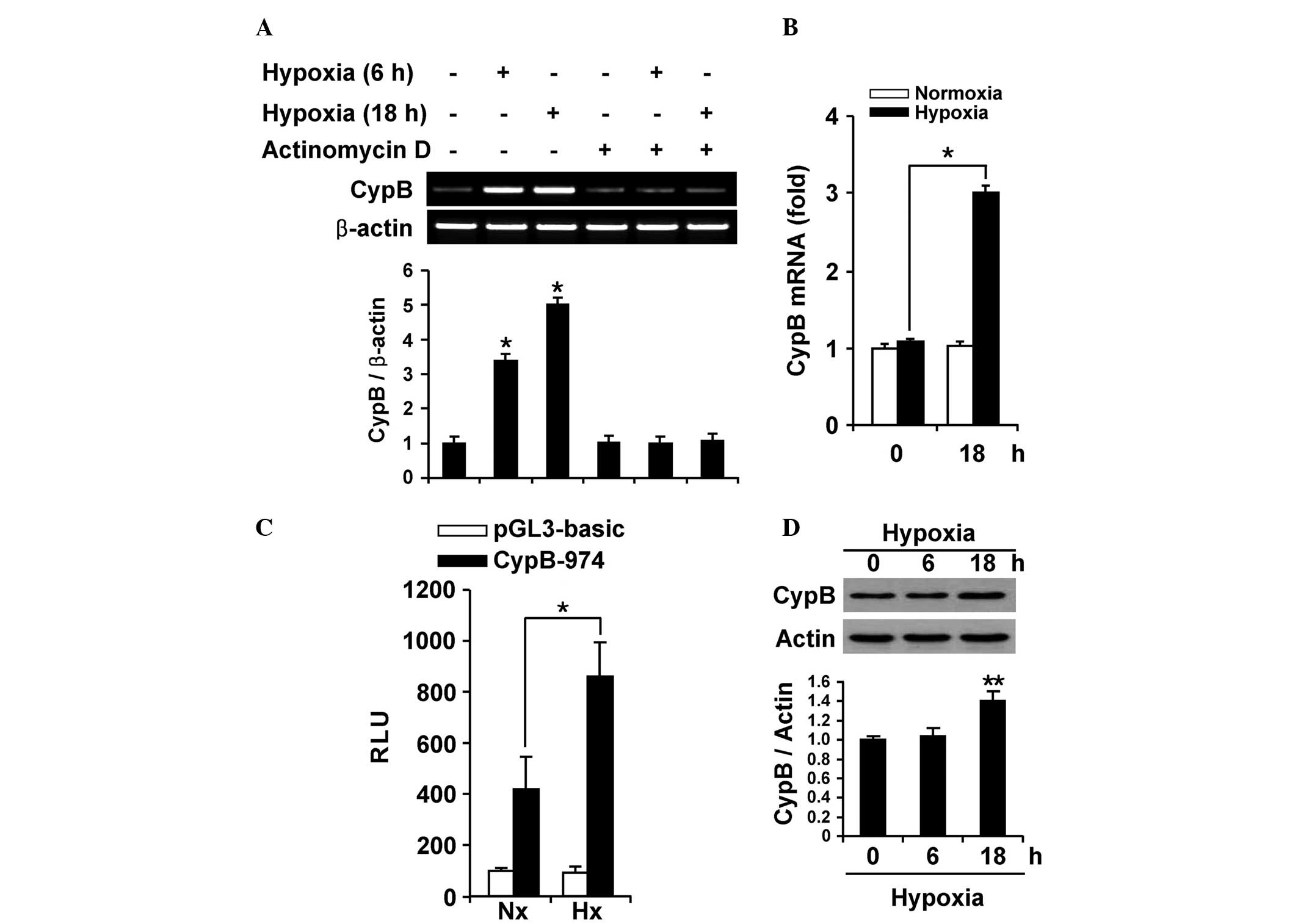
Hypoxia results in the transcriptional
upregulation of CypB expression
Since overexpression of CypB has been demonstrated
in numerous types of cancer cells, the present study investigated
whether the elevated CypB level is caused by transcriptional
induction. RT-PCR was performed using AGS cells that were subjected
to hypoxic conditions for 0, 6 and 18 h (Fig. 1A). Upregulated CypB mRNA expression
was observed during a 6 h exposure to hypoxia, and up to a
five-fold increase in CypB mRNA expression was identified in the
cells exposed to an 18 h period of hypoxia, compared with the
negative control. In addition, the treatment of AGS cells with 5
µg/ml actinomycin D, an mRNA synthesis inhibitor, abolished the
hypoxia-mediated induction of CypB mRNA expression, indicating that
upregulated CypB mRNA is mainly induced, not by mRNA stability, but
by the de novo synthesis of mRNA under hypoxia. The
quantitative PCR also demonstrated upregulated CypB mRNA levels
under hypoxia, consistent with the results shown in Fig. 1A and B. To further understand the
molecular mechanisms by which CypB mRNA is induced by hypoxia, the
CypB promoter activity was monitored using the wild-type CypB-974
promoter in AGS cells (Fig. 1C). As
expected, the luciferase activity of the wild-type CypB-974
promoter also demonstrated a significant increase under hypoxic
conditions compared with normoxic conditions. These results
indicate that the CypB gene is transcriptionally induced under
hypoxic conditions in AGS cells.
The CypB protein level was subsequently monitored in
AGS cells under hypoxic conditions, and it was found that CypB
protein expression was significantly increased (Fig. 1D). A time-course experiment
demonstrated that CypB protein expression under hypoxic conditions
reached a maximum level after 6 h and was sustained until the cells
had been exposed to hypoxia for 18 h. Densitometric analysis
revealed that the CypB protein expression was higher in AGS cells
exposed to hypoxic conditions when compared with the control,
suggesting a crucial role of CypB in hypoxia. These results
indicate that CypB expression is upregulated by hypoxia in AGS
cells.
Hypoxia-mediated induction of CypB
requires an ATF6 transcription factor
The promoters of ER chaperones contain ERSEs
(10,15) and previous studies have revealed that
the human CypB promoter contains ER stress-responsive elements that
are induced by ATF6-dependent pathways (16). To identify the region of the CypB
promoter that is responsible for hypoxia-induced transcriptional
upregulation, the luciferase activity was measured using
incrementally truncated reporter plasmids, comprising
pGL3-CypB-974, −800, −350, −250 and −100, that were transfected
into AGS cells (Fig. 2A). The
luciferase activity in each assay was normalized to that of a
co-transfected pGL3-basic plasmid, pCMV-β-galactosidase luciferase.
The pCMV-β-galactosidase luciferase plasmid did not respond to
hypoxia, but the pGL3-CypB-250, −350, −800 and −974 reporter
plasmids demonstrated significantly increased luciferase activity
under hypoxic conditions. The empty pGL3-basic vector and
pGL3-CypB-100 plasmid did not demonstrate hypoxia-induced
luciferase activity. These results suggest that the
cis-element responsible for the response to hypoxic
conditions is within the region 250 bp upstream of the
transcription start site. Within this region, the ERSE element was
located using a promoter analysis program. Therefore, it was
hypothesized that the ERSE element is associated with
hypoxia-induced transcription activity. In the following promoter
experiments, pGL3-CypB-250, a plasmid containing the ERSE element
of the CypB promoter, was used to test this hypothesis in
detail.
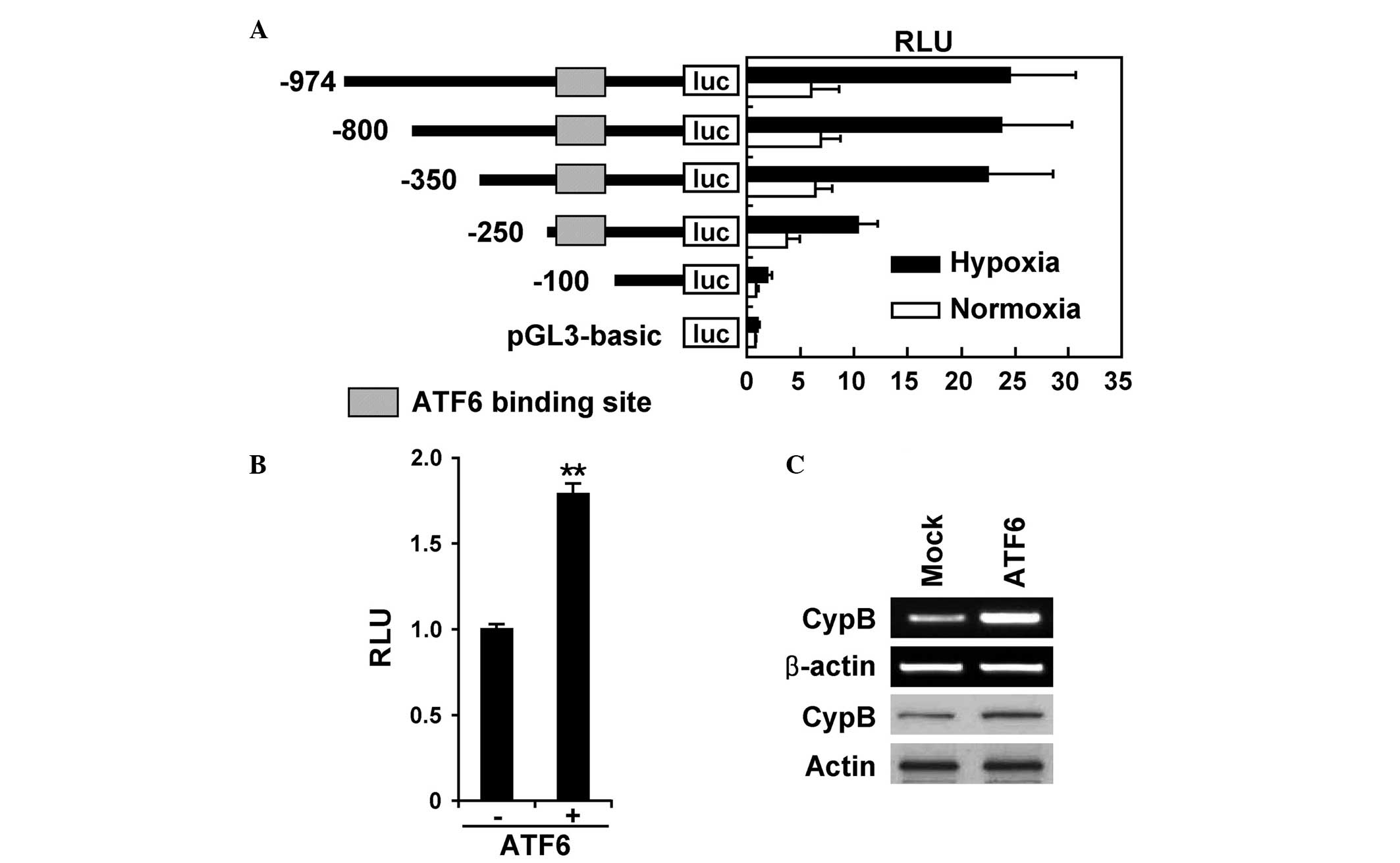 | Figure 2.ATF6 mediates hypoxia-induced
transcriptional upregulation of CypB. (A) Schematic representation
of deletion constructs for the human CypB promoter, revealing the
location of ATF6. The AGS cells were transfected with various
deletion constructs of pGL3-CypB, comprising pGL3-CypB-974, −800,
−350, −250 and −100, together with 0.4 mg pCMV-β-galactosidase
vector. The cells were incubated under normoxic or hypoxic
conditions for 24 h, and assayed for luciferase activity. (B) The
AGS cells were transfected with the pGL3-CypB-250 plasmid and the
active form-ATF6 (p50ATF6) expression plasmid. Subsequent to
transfection, the cells were incubated under normoxic or hypoxic
conditions for 24 h and assayed for luciferase activity. The data
are reported as the mean ± standard deviation from three
independent experiments. (C) The AGS cells were transfected with
p50ATF6 in a dose-dependent manner. The expression of the CypB
protein was analyzed by western blot analysis. ATF6, activation of
transcription factor 6; CypB, cyclophilin B; luc, luciferase; RLU,
relative light units. |
ATF6 is a UPR transducer that binds to the CCACG
region of ERSE elements in the promoter region of UPR-responsive
genes (15). ATF6 is constitutively
synthesized as a 90-kDa protein, termed p90ATF6, that is converted
to a 50-kDa protein, termed p50ATF6, particularly in ER-stressed
cells prior to the induction of GRP78. p50ATF6 translocates into
the nucleus, where it specifically activates the transcription of
ER stress response-associated genes, including GRP78 (17). To determine whether CypB is regulated
by p50ATF6 at the transcriptional level under hypoxic conditions,
the AGS cells were transiently co-transfected with ATF6
transcription factor expression plasmids and pGL3-CypB-250
plasmids. The luciferase activity of these transfected cells was
subsequently analyzed (Fig. 2B). It
was found that p50ATF6 expression plasmids increased the luciferase
activity in the transfected cells compared with cells transfected
with the pcDNA3 empty vector. RT-PCR and western blot analysis
revealed that CypB protein expression is increased by the
expression of p50ATF6 in AGS cells (Fig.
2C). These results indicate that the hypoxia-mediated
activation of CypB transcription requires ATF6.
Discussion
In the present study, the ER stress-mediated ATF6
pathway of CypB expression was demonstrated to be one of the
mechanisms induced by hypoxia in AGS cells. It has previously been
reported that CypB activation was found to protect AGS cells
against hypoxia through the attenuation of ER stress-mediated cell
death. However, the mechanism for the induction of the ER stress
pathways following exposure to hypoxia have not been clearly
identified.
In the present study, ATF6, an ER stress-associated
transcription factor, was found to mediate the upregulation of CypB
under hypoxic conditions (Fig. 1).
ERSE, ERSE-II and ATF6-binding cis-acting element have been
reported as consensus sequences for the binding of p50ATF6
(15,18,19). The
ER stress response element CCAAT-N9-gtaaCGTGG (ERSE-III), which is
located within the CypB promoter region and is similar to the
conventional ERSE-I motif of CCAAT-N9-CCACG (19), was consistently used for ATF6 binding
under hypoxic conditions, as demonstrated by the luciferase assay.
Hypoxia was found to significantly upregulate CypB mRNA expression
in a time-dependent and ATF6-dependent manner. The stimulation of
ERSE-mediated transcription activity by ATF6 requires the integrity
of the tripartite structure of the ERSE, a high-affinity CCAAT
binding site for NF-Y/CBF and a functional NF-Y complex, which is
consistent with the present results.
It has been established that ER stress favors
survival and induces apoptotic signaling in various types of cells.
Therefore, the decision between survival and apoptosis may depend
on the balance between survival signaling and apoptotic signaling.
In future studies, the involvement of ER stress-specific apoptotic
signaling in hypoxia in AGS cells may be examined.
In summary, ATF6 upregulates CypB gene expression
through an increase in the promoter activity of CypB in AGS cells,
which may demonstrate anti-apoptotic properties under hypoxic
conditions. Future studies investigating the mechanism through
which CypB prevents ER stress may aid the present understanding of
the nature of hypoxia-associated diseases, including solid
tumors.
Acknowledgements
This study was supported by a grant from the Kyung
Hee University in 2012 (grant no., KHU-20121733), and a grant from
the Basic Science Research Program through the National Research
Foundation of Korea (NRF), which is funded by the Ministry of
Education (grant no., NRF-2013R1A1A2060694).
References
|
1
|
Hochachka PW, Buck LT, Doll CJ and Land
SC: Unifying theory of hypoxia tolerance: Molecular/metabolic
defense and rescue mechanisms for surviving oxygen lack. Proc Natl
Acad Sci USA. 93:9493–9498. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee AS: The glucose-regulated proteins:
Stress induction and clinical applications. Trends Biochem Sci.
26:504–510. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu J and Kaufman RJ: From acute ER stress
to physiological roles of the Unfolded Protein Response. Cell Death
Differ. 13:374–384. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu C, Bailly-Maitre B and Reed JC:
Endoplasmic reticulum stress: Cell life and death decisions. J Clin
Invest. 115:2656–2664. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaufman RJ: Stress signaling from the
lumen of the endoplasmic reticulum: Coordination of gene
transcriptional and translational controls. Genes Dev.
13:1211–1233. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harding HP, Calfon M, Urano F, Novoa I and
Ron D: Transcriptional and translational control in the Mammalian
unfolded protein response. Annu Rev Cell Dev Biol. 18:575–599.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Breckenridge DG, Germain M, Mathai JP,
Nguyen M and Shore GC: Regulation of apoptosis by endoplasmic
reticulum pathways. Oncogene. 22:8608–8618. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schröder M and Kaufman RJ: The mammalian
unfolded protein response. Annu Rev Biochem. 74:739–789. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen J, Chen X, Hendershot L and Prywes R:
ER stress regulation of ATF6 localization by dissociation of
BiP/GRP78 binding and unmasking of Golgi localization signals. Dev
Cell. 3:99–111. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Baumeister P, Roy B, Phan T, Foti D,
Luo S and Lee AS: ATF6 as a transcription activator of the
endoplasmic reticulum stress element: Thapsigargin stress-induced
changes and synergistic interactions with NF-Y and YY1. Mol Cell
Biol. 20:5096–5106. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshida H, Okada T, Haze K, Yanagi H, Yura
T, Negishi M and Mori K: ATF6 activated by proteolysis binds in the
presence of NF-Y (CBF) directly to the cis-acting element
responsible for the mammalian unfolded protein response. Mol Cell
Biol. 20:6755–6767. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Price ER, Zydowsky LD, Jin MJ, Baker CH,
McKeon FD and Walsh CT: Human cyclophilin B: A second cyclophilin
gene encodes a peptidyl-prolyl isomerase with a signal sequence.
Proc Natl Acad Sci USA. 88:1903–1907. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Price ER, Jin M, Lim D, Pati S, Walsh CT
and McKeon FD: Cyclophilin B trafficking through the secretory
pathway is altered by binding of cyclosporin A. Proc Natl Acad Sci
USA. 91:3931–3935. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oh Y, Kim EY, Kim Y, et al:
Neuroprotective effects of overexpressed cyclophilin B against
Aβ-induced neurotoxicity in PC12 cells. Free Radic Biol Med.
51:905–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshida H, Haze K, Yanagi H, Yura T and
Mori K: Identification of the cis-acting endoplasmic reticulum
stress response element responsible for transcriptional induction
of mammalian glucose-regulated proteins. Involvement of basic
leucine zipper transcription factors. J Biol Chem. 273:33741–33749.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim J, Choi TG, Ding Y, et al:
Overexpressed cyclophilin B suppresses apoptosis associated with
ROS and Ca2+ homeostasis after ER stress. J Cell Sci.
121:3636–3648. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye J, Rawson RB, Komuro R, Chen X, Davé
UP, Prywes R, Brown MS and Goldstein JL: ER stress induces cleavage
of membrane-bound ATF6 by the same proteases that process SREBPs.
Mol Cell. 6:1355–1364. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kokame K, Kato H and Miyata T:
Identification of ERSE-II, a new cis-acting element responsible for
the ATF6-dependent mammalian unfolded protein response. J Biol
Chem. 276:9199–9205. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Shen J, Arenzana N, Tirasophon W,
Kaufman RJ and Prywes R: Activation of ATF6 and an ATF6 DNA binding
site by the endoplasmic reticulum stress response. J Biol Chem.
275:27013–27020. 2000.PubMed/NCBI
|