Introduction
Osteosarcoma is the most common primary bone tumor
that occurs mainly in children and adolescents. Twenty percent of
osteosarcoma patients succumb to this disease as a result of tumor
metastasis or an unresectable tumor. The remaining 80% of patients
generally present with small metastases when diagnosed; a number of
these patients develop pulmonary metastasis within a year and the
5-year survival rate is only 15% (1,2).
Recently, various chemotherapies and new diagnostic techniques have
been developed. These developments have dramatically improved the
5-year survival rate for osteosarcoma patients up to 55–70%
(1). Chemotherapeutic regimens used
in the treatment of osteosarcoma, such as adriamycin, methotrexate
and/or cisplatin, result in significant morbidity, including
cardiac toxicity, infertility and renal dysfunction. In order to
improve the long-term survival rate of this type of cancer, it is
crucial for research efforts to identify new targets and
techniques, especially for gene therapy.
It has been suggested that protein phosphorylation
is a critical step in protein structural change. PIN1 is composed
of 2 functional domains, an amino-terminal WW domain (amino acids
1–39) involved in protein-protein interaction which mediates PIN1
binding to phosphorylated protein and a COOH-terminal
peptidyl-prolyl cis-trans isomerase (PPIase) domain (amino
acids 45–163) that functions in catalysis (3). Phosphorylation of Ser/Thr-Pro residues
plays an important role in several cellular events. An anomalous
change in the protein during the process of signal transduction
usually leads to unwanted cell proliferation and culminates in the
development of cancer (4,5).
PIN1 was originally identified by a yeast 2-hybrid
screen as a protein that interacts with NIMA (never in mitosis gene
A). It consists of 163 amino acid residues and is located at 19p13
(6,7). It has been reported that PIN1 is
involved in several aspects of signal transduction, such as cell
growth, proliferation, differentiation and apoptosis and is
upregulated in various types of tumor tissues. Currently,
approximately 20 types of PIN1 targets are known, such as p53,
β-catenin and cyclin D1, and most are related to the development of
tumors (8,9). PIN1 has important effects in a number
of carcinogenic signaling pathways. It catalyzes the composite of a
carcinogenic substance and prevents the degradation of the
carcinogenic substance. Thus, PIN1 as an amplifier of carcinogenic
signals, plays a crucial role in the process of transforming an
oncogene to a signal of cell proliferation and differentiation.
Cyclin D1 is one of the important targets of PIN1.
PIN1 affects not only the Ras/JNK pathway to accelerate the
synthesis of cyclin D1 during transcription (10), but also prevents the degradation of
cyclin D1 by changing the structure of cyclin D1 to block its
movement from the nucleolus to the cytoplasm (11). β-catenin is a crucial downstream
transcriptional factor of the Wnt signaling pathway, and also
improves cyclin D1 synthesis. PIN1 specifically binds to the
phosphorylated motif of β-catenin, altering its configuration and
thus inhibiting the combination of β-catenin and APC. PIN1 also
improves the expression of cyclin D1 by increasing the stability of
β-catenin and its accumulation in the nucleolus (9). Cyclin D1 is a member of the cyclin
family, and is known to catalyze the G1-S phase
transition. Cyclin D1 phosphorylates Rb protein by activating CDK4
and CDK6. The phosphorylated Rb protein releases E2F, which
initiates DNA transcription and induces the transition from the
G1 to S phase. The overexpression of cyclin D1 leads
mammary glandular cells to transform into inocytes (12,13)
and promotes breast cancer formation (14). Together, these reports strongly
suggest that PIN1 and cyclin D1 play an important role in the
occurrence and development of tumors.
The expression of PIN1 in human breast and oral
cancers is closely correlated with the cyclin D1 level (15,16).
However, the relationship between PIN1 and osteosarcoma has not
been previously studied. Therefore, we studied the expression
patterns of PIN1 in human osteosarcoma tissues. We further
investigated the effect of the overexpression of PIN1 in
osteosarcoma cells on cell proliferation and cycle. Our data
demonstrated that PIN1 traverses the G0-G1
and/or G1-S transitions through a coordinated mechanism
involving the upregulation of cyclins and CDKs and the
downregulation of p21.
Materials and methods
Cell culture and reagents
Human osteosarcoma MG-63 and U2-OS cells were
purchased from the American Type Culture Collection (ATCC,
Rockville, MD, USA) and HEK293T cells were maintained in our
laboratory. The cells were grown at 37°C in a humidified atmosphere
of 5% CO2 in DMEM medium supplemented with 10% fetal
bovine serum and 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml
streptomycin and 2.5 μg/ml amphotericin B. All reagents were
purchased from Sigma (St. Louis, MO, USA) unless otherwise
noted.
Tissue preparation
Eight paraffin samples of human osteosarcoma and
normal tissues were obtained from Chonbuk National University
Hospital after Ethics Committee approval of the use of human
tissue. Tissues were fixed in a 4% formalin solution and embedded
in wax blocks. Each block was cut into 5-μm sections and stained
with specific antibodies. Eight cases of fresh human osteosarcoma
and normal tissues (5–8 mm3) were acquired during
surgery and stored at −80°C until use.
Transfection of cells
Preparation and amplification of the adenovirus were
performed as previously described (17). Adenoviruses encoding PIN1 were
created using the Virapower Adenoviral Expression System according
to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA).
Site-specific recombination between entry vectors (PIN1-pENTR) and
the adenoviral destination vector (pAd/PL-DEST) were established
with LR Clonase II (Invitrogen). The control adenovirus (Ad-LacZ,
S36A) was obtained from R.H. Unger (University of Texas
Southwestern Medical Center, Dallas, TX, USA). The virus (100 MOI)
was added to the cells and incubated for 4 h at 37°C. After
conventional culturing of the cells for 48 h, the cells were
collected to perform the experiment.
MTT assay for cell proliferation
The viability of cultured cells was determined by
assaying the reduction of
3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
to formazan. After transfection with the PIN adenovirus (Ad-PIN1)
or control adenovirus (Ad-LacZ), the cells were washed twice in
96-well plates with phosphate-buffered saline (PBS), and MTT (100
μg/100 μl of PBS) was added to each well. The cells were then
incubated at 37°C for 2 h and DMSO (100 μl) was added to dissolve
the formazan crystals. Absorbance was measured at 570 nm with a
Spectra Max Plus model (Molecular Devices, Sunnyvale, CA, USA). In
the juglone treatment group, cells were incubated with different
doses of juglone (1, 10, 20 or 50 μM) for 6, 12 or 24 h after virus
treatment. Other processes were the same as the ones described
above.
Cell cycle analysis
Cells (1–5×105) in 6-well plates were
incubated with Ad-PIN1 or AD-LacZ for 4 h. Cells were treated with
or without juglone for 48 h. After trypsinization and collection,
cells were washed 3 times with PBS and fixed in 70% ethanol
overnight at 4°C. Cells were then washed 3 times in PBS with 0.1%
BSA. Cells were incubated with 5 mg/ml of RNase A (DNase free) and
50 mg/ml of PI for 90 min at 4°C. The percentages of cells in
different phases of the cell cycle were measured with a FACStar
flow cytometer (Becton-Dickinson, San Jose, CA, USA).
Histology and immunohistochemistry
Tissue sections (5 μm) were stained with hematoxylin
and eosin (H&E) for analysis under a light microscopy.
Immunohistochemical staining was performed with the Dako Envision
system (Dako, Carpinteria, CA, USA), which uses dextran polymers
conjugated with horseradish peroxidase to avoid contamination with
endogenous biotin. The paraffin samples were warmed at 55°C for 30
min. After deparaffinization, tissue sections were treated using a
microwave antigen retrieval procedure in 0.01 M sodium citrate
buffer (pH 6.0) and immunostained with antibody against PIN1.
Western blot analysis
After collection, cells were placed in lysis buffer
(M-PER; Thermo Scientific) for 30 min and then centrifuged at
13,200 × g for 30 min at 4°C. The supernatant was stored at −80°C
until use. The fresh osteosarcoma tissues (200 mg) were cut into
small pieces and put in lysis buffer (M-PER), homogenized, then
centrifuged at 13,200 × g for 15 min at 4°C. The supernatant was
stored at −80°C until use. Protein concentration was determined
using the Bradford method. The lysates (20 μg/lane) were resolved
in a 10% SDS-PAGE gel depending on the target protein sizes and the
resolved proteins were transferred onto Immobilon-PVDF membranes
(Millipore, Billerica, MA, USA) by electroblotting. Immunoblotting
was performed using a primary antibody directed specifically
against PIN1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a
dilution of 1:2,000. A β-actin monoclonal antibody (Santa Cruz
Biotechnology) was used as a control to demonstrate equal loading
and transfer. The membranes were incubated with secondary
antibodies at a dilution of 1:2,000. The membrane blots were
developed using an Amersham ECL™ Advance Western Blotting Detection
kit (Amersham, Arlington Heights, IL, USA).
Statistical analysis
Statistical analysis of the data was performed using
ANOVA and Duncan’s test. P-value <0.05 was considered to
indicate a statistically significant difference.
Results
PIN1 is overexpressed in human
osteosarcoma tissues
To assess the effect of PIN1 on osteosarcoma
formation and development, PIN1 expression was assessed in human
osteosarcoma and normal tissues by immunohistochemistry and western
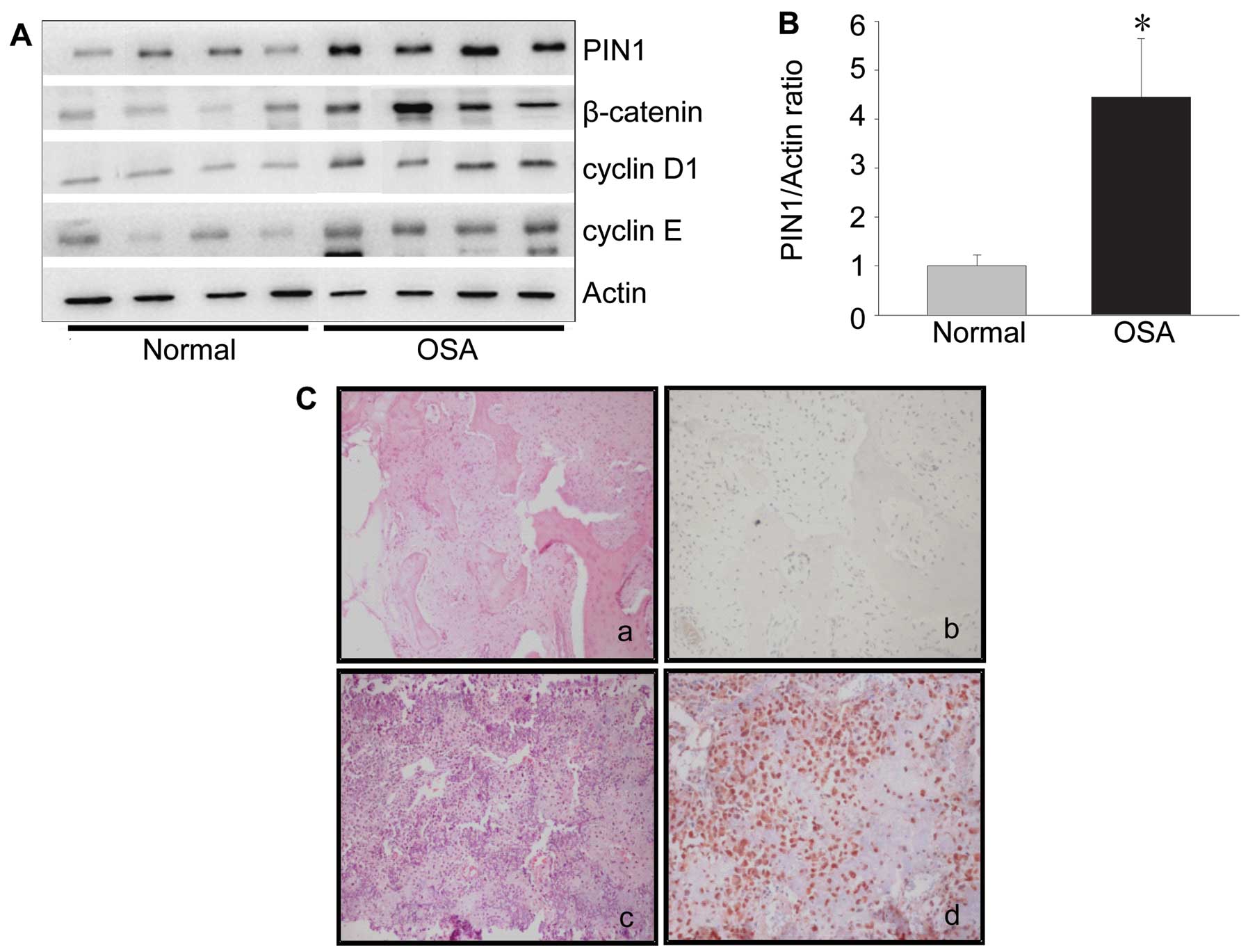
blotting. In the western blotting study, the expression levels of
PIN1 protein were 4-fold higher in osteosarcoma tissues compared
with normal tissues (Fig. 1A and
B). The cyclin D1 level was also significantly increased in
osteosarcoma tissues (Fig. 1A).
Meanwhile, the expression of proteins upstream of cyclin D1,
including β-catenin and cyclin E were also significantly increased
in osteosarcoma tissues (Fig. 1A).
In the immunohistochemical study, the PIN1 expression was observed
in the nucleus and cytoplasm of human osteosarcoma tissues
(Fig. 1C).
PIN1 induces cell proliferation in human
osteosarcoma cells
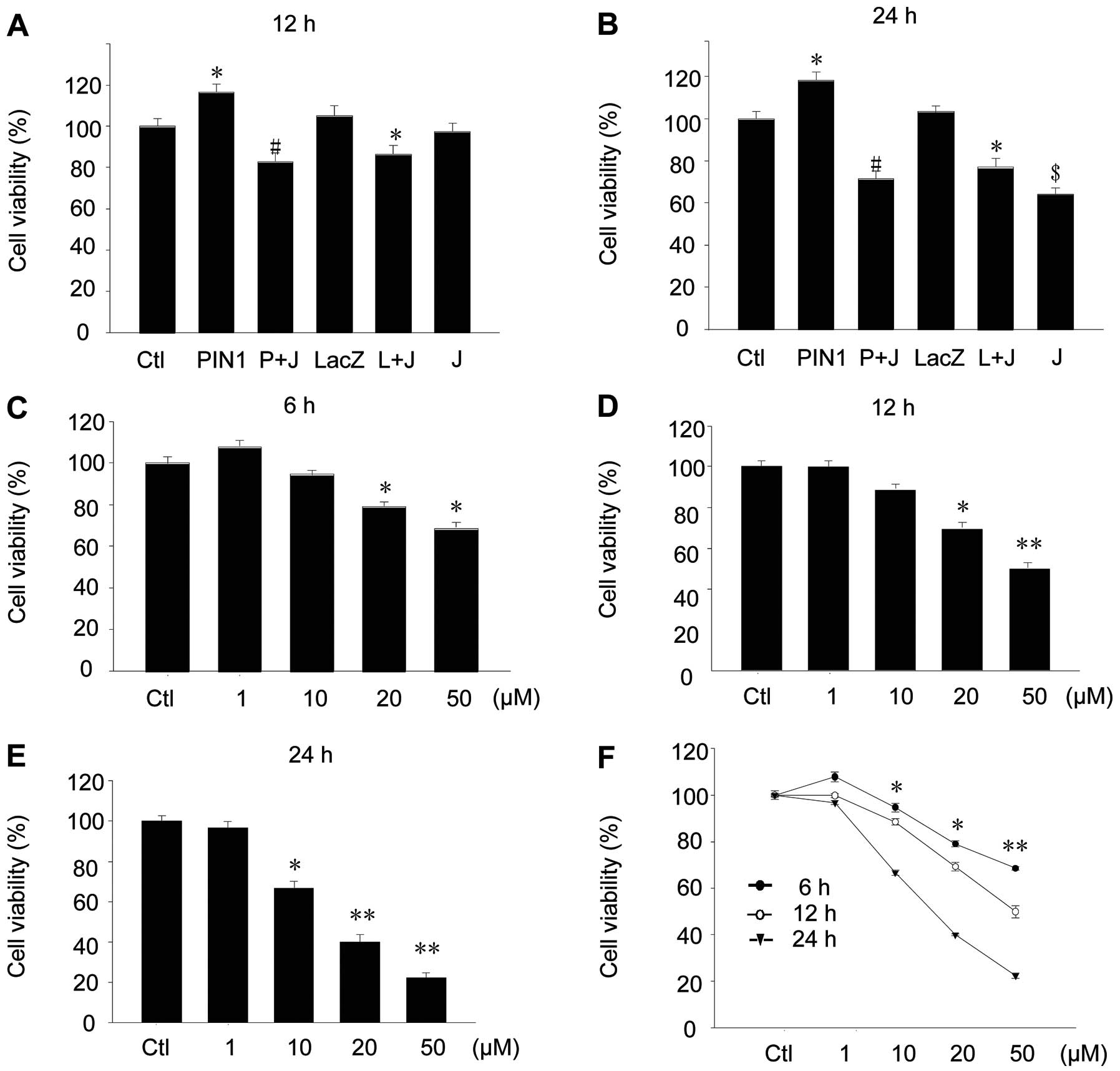
After transfection of the PIN1 adenovirus for 24 h,
cellular proliferation was assessed using human osteosarcoma cells,
U2-OS and MG63. MTT analysis found that PIN1 overexpression
increased the cellular proliferation by ~118±5.5% in 12 h and
121±7.5% in 24 h (Fig. 2A and B).
When the PIN1 inhibitor juglone was added, the effects of PIN1
overexpression were abolished (Fig. 2A
and B). Juglone also induced cell death of the U2-OS and MG63
cells in a concentration- and time-dependent manner (Fig. 2C-F).
PIN1 induces the transition of the
G0/G1 to the G2/M phase in human
osteosarcoma cells
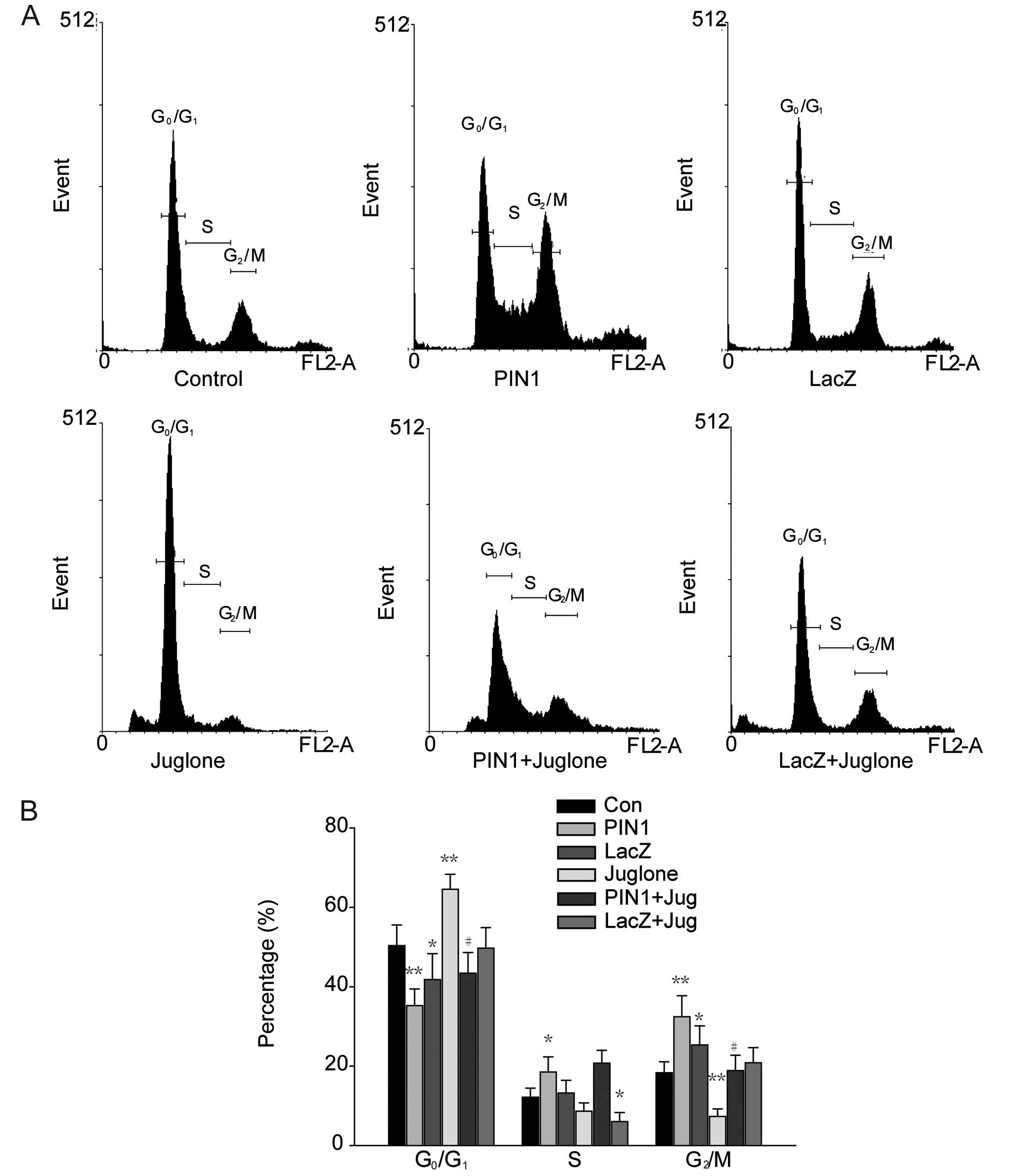
Following the overexpression of PIN1, cell cycle
distribution was determined by FACS analysis (Fig. 3A). PIN1 overexpression decreased the
distribution of G0/G1 phase cells to
35.26±3.41%, whereas G0/G1 phase cells of the
control group accounted for 50.36±3.27% of the total population.
The percentages of cells at the S and G2/M phases were
18.52±2.52 and 32.44±3.83%, respectively, in PIN1-overexpressed
cells, while the same cell populations comprised 12.17±2.31 and
18.33±1.96%, respectively, in the control cells. There was no
change in the control virus-transfected cells:
G0/G1 (41.81±3.75%), S (13.22±3.09%) and
G2/M (25.35±3.18%). Juglone treatment abolished all
effects of PIN1 overexpression.
PIN1 modulates the protein levels of
G2/M phase regulators
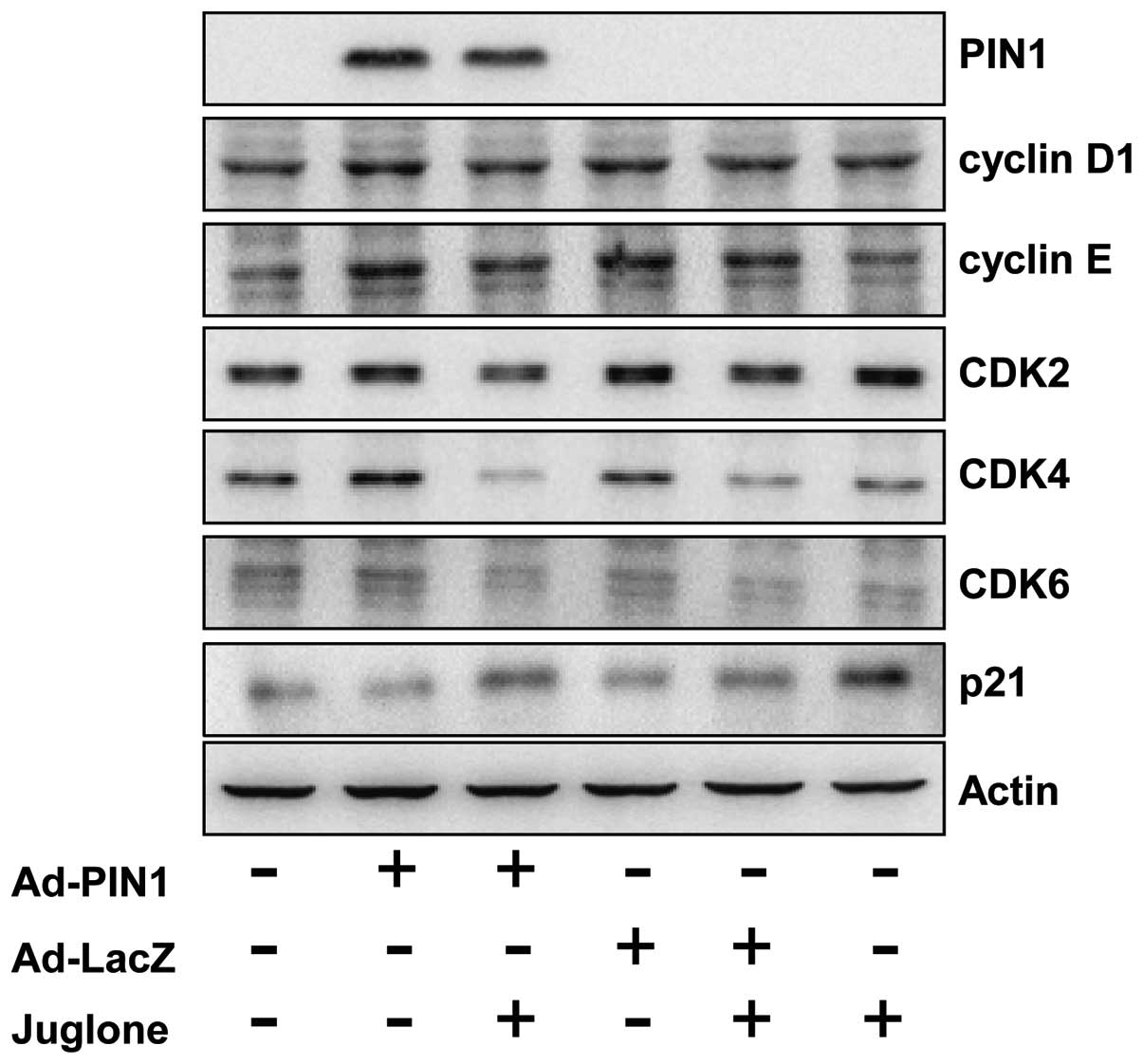
Consistent with increased cell growth, PIN1 induced
the transition from G0/G1 to G2/M
in the U2-OS cells. We assessed the effect of PIN1 on cell cycle
regulatory molecules that play important roles in the cell cycle
transition. The levels of cyclins D1 and E and their associated
cyclin-dependent kinases, CDK4 and CDK6, were increased in
PIN1-overexpressed cells (Fig. 4).
However, the expression of the cell cycle inhibitor, p21, was
decreased in PIN1 overexpressed cells. Juglone abolished
PIN1-mediated changes in cell cycle protein expression.
Discussion
PIN1, a peptidyl-prolyl cis-trans isomerase
(PPIase), binds to phosphorylated Ser/Thr-Pro motifs and catalyzes
a conformational change in its target, thereby altering the
conformation of its substrates (2).
At the molecular level, protein-protein interactions are mediated
by specialized molecules consisting of dedicated signaling domains
and their cognate binding partners. By interacting with these
domains, PIN1 has the ability to modify protein function. For this
reason, PIN1 has been implicated as a key regulator in a number of
cell events, including cell cycle progression, transcription, cell
proliferation and apoptosis (10,11).
PIN1 was originally identified as a binding partner for cell cycle
proteins, several transcriptional factors and cell cycle regulatory
proteins. Given its ability to regulate these proteins, PIN1 is
suggested to act as a ‘timer’ in cell cycle progression (4).
Previous studies indicated that PIN1 is
overexpressed in a number of diseases. Subsequent studies confirmed
that the expression levels of PIN1 are increased in tissues of
various carcinoma and cancer cell lines (9,18–24). A
study using 60 different types of human tumors indicated that tumor
tissues have higher PIN1 protein levels compared with corresponding
normal tissues (19). PIN1
overexpression has been considered an indicator of prognosis in
several types of cancers, including lung (25) and colon cancers (24). In some cases, the overexpression of
PIN1 indicates a short tumor recurrence period following treatment
(18). In the current study, we
performed immunohistochemistry to examine PIN1 expression in normal
and osteosarcoma human tissues. In 8 human tissue samples, PIN1
expression was very low in the normal tissues, whereas high levels
of PIN1 were observed in osteosarcoma tissues. Consistent with this
finding, western blot data demonstrated that PIN1 levels were
4-fold higher in osteosarcoma tissues compared to the normal
tissues.
PIN1 expression is closely correlated with cyclin
D1, β-catenin and cyclin E levels, which are intrinsically involved
in oncogenesis (16,21). The correlation between PIN1 and
cyclin D1 expression suggests that PIN1 expression is not only
highly regulated, but that it also plays specific roles in
oncogenesis. To examine this possibility, we transiently
overexpressed PIN1 in 2 osteosarcoma cell lines, MG-63 and U2-OS
and then examined its effects on cell proliferation. Results
demonstrated that PIN1 overexpression increased cell proliferation
and that juglone decreased cell proliferation. These results
revealed that PIN1 causes cell cycle progression. This conclusion
is further supported by our observation of increased cell numbers
in the G2/M phase of the cell cycle.
Cell proliferation is strictly governed by a family
of protein kinase complexes that are controlled, in part, by CDKs
and their binding partners, the cyclins (26,27).
Normally, these complexes are activated by the passage of a
specific checkpoint and then cells progress through the phases of
the cell cycle. Any defect in this machinery causes an alteration
in cell cycle regulation that may result in unwanted cellular
proliferation culminating in the development of cancer (28,29).
Mechanistically, PIN1 increases cyclin D1 gene expression by
activating β-catenin/T-cell transcription factor (TCF) (9) and directly binds to cyclin D1 to
increase its stability (8).
Furthermore, PIN1 regulates Ki67, cyclin E and c-MYC (30,31).
Cyclins D1 and E, along with CDKs 2, 4 and 6, are the driving
forces behind the G1-S transition in the cell cycle,
whereas p21 induces the G1 and G2/M phase
cell cycle arrest (32). In our
study, significant increases in the expression of cyclins D1 and E
and CDKs 2, 4 and 6 were observed in the PIN1-transfected U2-OS
cells. Increases in these checkpoint proteins lead to uncontrolled
cell growth and aberrant cell function, with the cells quickly
traversing the G0-G1 and/or G1-S
transitions. Thus, PIN1, by regulating the expression of cyclin D1,
cyclin E and CDKs, has the ability to promote tumor growth.
Recently, PIN1 has been suggested to promote the
epithelial-mesenchymal transition (33), often regarded as the initiation of a
metastatic and aggressive cancer. However, more detailed studies
are required to elucidate the mechanism underlying PIN1-induced
metastasis in osteosarcoma cell lines.
Osteosarcoma continues to be the leading cause of
bone cancer that mainly occurs in adolescent and young adults.
Delayed diagnosis and lack of effective therapeutics cause
osteosarcoma to recur and spread to distant sites (17). Although surgery offers the best
treatment option, this cancer is often not detected early enough
for surgery to be effective. The possibility of reducing the
incidence and the burden of this cancer is what makes the search
for promising advances in diagnosis and treatment of the utmost
importance. The present study focused on the prognostic marker that
contributes to osteosarcoma carcinogenesis and the effects of PIN1
on tumorigenesis. This study demonstrates that PIN1 may be a
suitable prognostic marker for osteosarcoma and that it markedly
induces cell proliferation through the upregulation of the
CDK/cyclin complex and the downregulation of p21 expression. Taken
together, these results suggest the possible application of PIN1 as
a target for cancer prevention or therapy in osteosarcoma.
Acknowledgements
This study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education, Science and Technology
(2010-0021514).
References
|
1
|
Galat A: Peptidylprolyl cis/trans
isomerases (immunophilins): biological diversity - targets -
functions. Curr Top Med Chem. 3:1315–1347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu KP, Suizu F, Zhou XZ, Finn G, Lam P and
Wulf G: Targeting carcinogenesis: a role for the prolyl isomerase
Pin1? Mol Carcinog. 45:397–402. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu KP, Hanes SD and Hunter T: A human
peptidyl-prolyl isomerase essential for regulation of mitosis.
Nature. 380:544–547. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blume-Jensen P and Hunter T: Oncogenic
kinase signalling. Nature. 411:355–365. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hunter T: Prolyl isomerases and nuclear
function. Cell. 92:141–143. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berger M, Stahl N, Del Sal G and Haupt Y:
Mutations in proline 82 of p53 impair its activation by Pin1 and
Chk2 in response to DNA damage. Mol Cell Biol. 25:5380–5388. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Albert A, Lavoie S and Vincent M: A
hyperphosphorylated form of RNA polymerase II is the major
interphase antigen of the phosphoprotein antibody MPM-2 and
interacts with the peptidyl-prolyl isomerase Pin1. J Cell Sci.
112:2493–2500. 1999.PubMed/NCBI
|
|
8
|
Liou YC, Ryo A, Huang HK, et al: Loss of
Pin1 function in the mouse causes phenotypes resembling cyclin
D1-null phenotypes. Proc Natl Acad Sci USA. 99:1335–1340. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryo A, Nakamura M, Wulf G, Liou YC and Lu
KP: Pin1 regulates turnover and subcellular localization of
beta-catenin by inhibiting its interaction with APC. Nat Cell Biol.
3:793–801. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yaffe MB, Schutkowski M, Shen M, et al:
Sequence-specific and phosphorylation-dependent proline
isomerization: a potential mitotic regulatory mechanism. Science.
278:1957–1960. 1997. View Article : Google Scholar
|
|
11
|
Zhou XZ, Kops O, Werner A, et al:
Pin1-dependent prolyl isomerization regulates dephosphorylation of
Cdc25C and tau proteins. Mol Cell. 6:873–883. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujimori F, Takahashi K, Uchida C and
Uchida T: Mice lacking Pin1 develop normally, but are defective in
entering cell cycle from G(0) arrest. Biochem Biophys Res Commun.
265:658–663. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maleszka R, Hanes SD, Hackett RL, de Couet
HG and Miklos GL: The Drosophila melanogaster dodo (dod)
gene, conserved in humans, is functionally interchangeable with the
ESS1 cell division gene of Saccharomyces cerevisiae. Proc
Natl Acad Sci USA. 93:447–451. 1996.
|
|
14
|
Hsu T, McRackan D, Vincent TS and Gert de
Couet H: Drosophila Pin1 prolyl isomerase Dodo is a MAP
kinase signal responder during oogenesis. Nat Cell Biol. 3:538–543.
2001. View
Article : Google Scholar
|
|
15
|
Wulf GM, Ryo A, Wulf GG, et al: Pin1 is
overexpressed in breast cancer and cooperates with Ras signaling in
increasing the transcriptional activity of c-Jun towards cyclin D1.
EMBO J. 20:3459–3472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miyashita H, Uchida T, Mori S, Echigo S
and Motegi K: Expression status of Pin1 and cyclins in oral
squamous cell carcinoma: Pin1 correlates with cyclin D1 mRNA
expression and clinical significance of cyclins. Oncol Rep.
10:1045–1048. 2003.PubMed/NCBI
|
|
17
|
Lee JH, Song MY, Song EK, et al:
Overexpression of SIRT1 protects pancreatic beta-cells against
cytokine toxicity by suppressing the nuclear factor-kappaB
signaling pathway. Diabetes. 58:344–351. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ayala G, Wang D, Wulf G, et al: The prolyl
isomerase Pin1 is a novel prognostic marker in human prostate
cancer. Cancer Res. 63:6244–6251. 2003.PubMed/NCBI
|
|
19
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen SY, Wulf G, Zhou XZ, Rubin MA, Lu KP
and Balk SP: Activation of beta-catenin signaling in prostate
cancer by peptidyl-prolyl isomerase Pin1-mediated abrogation of the
androgen receptor-beta-catenin interaction. Mol Cell Biol.
26:929–939. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fukuchi M, Fukai Y, Kimura H, et al:
Prolyl isomerase Pin1 expression predicts prognosis in patients
with esophageal squamous cell carcinoma and correlates with cyclin
D1 expression. Int J Oncol. 29:329–334. 2006.PubMed/NCBI
|
|
22
|
Kim CJ, Cho YG, Park YG, et al: Pin1
overexpression in colorectal cancer and its correlation with
aberrant beta-catenin expression. World J Gastroenterol.
11:5006–5009. 2005.PubMed/NCBI
|
|
23
|
Kokkinakis DM, Liu X and Neuner RD:
Modulation of cell cycle and gene expression in pancreatic tumor
cell lines by methionine deprivation (methionine stress):
implications to the therapy of pancreatic adenocarcinoma. Mol
Cancer Ther. 4:1338–1348. 2005. View Article : Google Scholar
|
|
24
|
Kuramochi J, Arai T, Ikeda S, Kumagai J,
Uetake H and Sugihara K: High Pin1 expression is associated with
tumor progression in colorectal cancer. J Surg Oncol. 94:155–160.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He J, Zhou F, Shao K, et al:
Overexpression of Pin1 in non-small cell lung cancer (NSCLC) and
its correlation with lymph node metastases. Lung Cancer. 56:51–58.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clurman BE and Roberts JM: Cell cycle and
cancer. J Natl Cancer Inst. 87:1499–1501. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jacks T and Weinberg RA: Cell-cycle
control and its watchman. Nature. 381:643–644. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pestell RG, Albanese C, Reutens AT, Segall
JE, Lee RJ and Arnold A: The cyclins and cyclin-dependent kinase
inhibitors in hormonal regulation of proliferation and
differentiation. Endocr Rev. 20:501–534. 1999.PubMed/NCBI
|
|
30
|
Yeh ES, Lew BO and Means AR: The loss of
PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts
to genomic instability. J Biol Chem. 281:241–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yeh E, Cunningham M, Arnold H, et al: A
signalling pathway controlling c-Myc degradation that impacts
oncogenic transformation of human cells. Nat Cell Biol. 6:308–318.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiong Y, Hannon GJ, Zhang H, Casso D,
Kobayashi R and Beach D: p21 is a universal inhibitor of cyclin
kinases. Nature. 366:701–704. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MR, Choi HK, Cho KB, Kim HS and Kang
KW: Involvement of Pin1 induction in epithelial-mesenchymal
transition of tamoxifen-resistant breast cancer cells. Cancer Sci.
100:1834–1841. 2009. View Article : Google Scholar : PubMed/NCBI
|