Introduction
Glioma is the most common primary intracranial tumor
in both adults and children. The World Health Organization (WHO)
classification scheme divides gliomas into grades I through IV,
based on increasing levels of malignancy (1). The prognosis of patients with glioma
is poor and is closely related to WHO grade. Glioblastoma
multiforme (GBM) WHO grade IV is the most malignant variant with a
median survival time of 1 year (2).
Efforts to better understand the biological basis of glioma
progression may yield important, clinically relevant insights into
disease management. Many aggressive treatment approaches, such as
postoperative chemotherapy and radiation therapy, have been used
clinically. Yet, these approaches do not benefit all patients
equally. Adverse effects associated with these approaches
dramatically deteriorate the quality of life of certain patients.
Therefore, individualized therapy should be considered as a
valuable approach for patients with high-grade gliomas. Recently,
molecular diagnostics has emerged as a powerful tool to discover
new biomarkers, network and therapeutic targets, realizing the
proof of principle that personalized medicine can increase survival
and cure cancer patients. Thus, elucidation of these critical
molecular events may improve therapy and individualize therapeutic
interventions for patients with gliomas.
Deubiquitylating enzymes (DUBs) antagonize the
ubiquitylation of substrates by cleaving polyubiquitin and
monoubiquitin and thus afford an additional level of protein
post-translational regulation (3).
Herpesvirus-associated ubiquitin-specific protease (HAUSP, also
known as ubiquitin specific protease 7, USP7) is a cysteine
protease that was originally identified as a binding partner for
the Herpes simplex viral (HSV) protein infected cell protein 0
(ICP0/Vmw110) (4). Subsequently,
numerous proteins have been identified as potential
substrate/binding partners of HAUSP (5,6). Some
of the better characterized substrates of HAUSP play crucial roles
in tumor suppression, DNA repair, immune responses, viral
replication and epigenetic control (7). HAUSP has been shown to inactivate
several tumor suppressors by nuclear export forkhead box O
transcription factor (FOXO4) and phosphatase and tensin homologue
deleted on chromosome 10 (PTEN) inactivation (8,9) or
degradation [p53 degradation following murine double minute 2
(Mdm2) stabilization] (10,11).
HAUSP is overexpressed in human prostate cancer, and
more importantly, high levels of HAUSP are directly correlated with
tumor aggressiveness (8). In
contrast, Masuya et al(12)
found that a reduction in HAUSP gene expression may play an
important role in NSCLC carcinogenesis, particularly in
adenocarcinomas, through p53-dependent pathways. These data suggest
that HAUSP may in some instances act as a tumor suppressor (by
stabilizing p53) or an oncogene (by stabilizing Mdm2 and
redistributing PTEN). Thus, this regulation may be dependent on
genetic context and also tissue or cell type due to differential
expression of other proteins.
In order to gain further insight into the role of
HAUSP in the progression of glioma, we used immunohistochemical
assay, quantitative real-time PCR and western blot analysis to
investigate the expression pattern of HAUSP in glioma specimens and
normal control brain tissues. Next, we analyzed the relationship
between HAUSP expression and the glioma stage as well as the
survival of patients.
Materials and methods
Patients and clinical specimens
This study was approved by the Research Ethics
Committee of Anhui Provincial Hospital of Anhui Medical University.
Written informed consent was obtained from all of the patients. All
specimens were handled and anonymized according to ethical and
legal standards.
Fresh glioma specimens were obtained from 80
patients who underwent surgical treatment at the Department of
Neurosurgery, Anhui Provincial Hospital, Anhui Medical University
between January 2008 and October 2008. None of the patients had
received radiotherapy, immunotherapy and chemotherapy prior to
surgery. Additionally, normal brain tissue samples were obtained
from 10 patients who underwent surgery for reasons other than
malignancy, such as cerebral trauma. These normal control samples
were collected during partial resection of normal brain tissue for
decompression treatment following severe head injury. Parts of each
specimen were snap-frozen in liquid N2 for 10 min and
stored in a −80°C ultra-freezer for mRNA and protein isolation;
other parts of each specimen were fixed in 10% formalin and
paraffin-embedded for histological sectioning. All of the glioma
samples were verified by pathological analysis and classified
according to the WHO 2007 classification standard. There were 27
low-grade (WHO grades I and II) and 53 high-grade tumors (WHO
grades III and IV) (Table I).
Patient data included age, gender, date and type of initial
operation, and details of the follow-up. Clinical information was
obtained by reviewing medical records and radiographic images, by
interview in the clinic or by telephone, and by review of death
certificate. A patient was considered to have recurrent disease
when revealed either by magnetic resonance imaging or the
occurrence of new neurologic symptoms. In the follow-up period,
overall survival was calculated from diagnosis to death or last
follow-up; the total period of follow-up was 16–60 months.
 | Table ILevel of expression of HAUSP protein
in glioma specimens as determined using immunohistochemical
analysis, and the comparison with clinicopathological
variables. |
Table I
Level of expression of HAUSP protein
in glioma specimens as determined using immunohistochemical
analysis, and the comparison with clinicopathological
variables.
| | Expression of HAUSP
protein | |
|---|
| |
| |
|---|
| n | − | + | ++ | +++ | P-value |
|---|
| Total | 80 | 11 | 15 | 22 | 32 | |
| Gender | | | | | | 0.925a |
| Male | 48 | 7 | 9 | 15 | 17 | |
| Female | 32 | 4 | 6 | 7 | 15 | |
| Age (years) | | | | | | 0.378a |
| <60 | 54 | 6 | 10 | 16 | 21 | |
| ≥60 | 26 | 5 | 5 | 6 | 10 | |
| KPS | | | | | | <0.05a |
| <80 | 47 | 4 | 8 | 14 | 21 | |
| ≥80 | 33 | 7 | 7 | 8 | 11 | |
| WHO grade | | | | | | <0.05b |
| I | 8 | 3 | 4 | 1 | 0 | |
| II | 19 | 4 | 5 | 7 | 3 | |
| III | 25 | 3 | 3 | 8 | 11 | |
| IV | 28 | 1 | 3 | 6 | 18 | |
Immunohistochemical assays
The formalin-fixed, paraffin-embedded tissue
sections (4 μm) were deparaffinized in xylene and dehydrated
through a graduated alcohol series. Endogenous peroxidase activity
was blocked with 3% H2O2 in methanol for 20
min. Antigen retrieval was performed by microwaving sections in
0.01 M sodium citrate (pH 6.0). Non-specific binding was blocked by
incubating sections with 5% BSA in phosphate-buffered saline (PBS)
for 30 min at room temperature. Without being washed, these
sections were incubated with polyclonal anti-HAUSP antibody
(sc-30164; 1:400; Santa Cruz Biotechnology Inc., CA, USA) in PBS at
4°C overnight in a moist box. Wash steps the sections were
incubated with biotinylated goat anti-rabbit immunoglobulin G
(1:400; Sigma) for 1 h at room temperature wash steps, and
expression was detected using the streptavidin-peroxidase complex.
The brown color indicative of peroxidase activity was developed by
incubation with 0.1% 3,3-diaminobenzi-dine (Sigma) in PBS with
0.05% H2O2 for 5 min at room temperature. The
sections were lightly counterstained with hematoxylin. Two
pathologists scored the immunohistochemical staining under
double-blinded conditions without prior knowledge of the clinical
or clinicopathological status of the specimens. Images captured for
all sections were acquired using an Olympus BX51. The expression of
HAUSP in glioma tissue was evaluated by scanning the entire tissue
specimen under low magnification (x40), followed by confirmation by
scanning under high magnification (×200 and ×400). Positive cells
were indicated by the presence of a distinct brown color in the
nuclei or cytoplasm. Normal brain tissues were used as control
tissues, and non-immune IgG was also used as a negative control
antibody for immunohistochemical staining.
Real-time polymerase chain reaction
Total RNA was purified from all 80 glioma specimens
and 10 control normal brain tissues using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA). The concentration and purity of
RNA were determined spectrophotometrically at 260/280 nm using a
Nanodrop spectrophotometer (Ocean Optics, Dunedin, FL, USA). Then
cDNA was synthesized from ~5 μg RNA/20 μl using a cDNA reverse
transcription kit (Qiagen, Germany). Real-time polymerase chain
reaction (real-time PCR) amplification was performed using a 7500
Fast System Real-Time PCR cycler (Applied Biosystems, Foster City,
CA, USA). Primers were designed using Primer Express v3.0 software
(Applied Biosystems). The HAUSP primers were
5′-ATGAACCACCAGCAGCAGCAGC-3′ (forward) and
5′-GCGTGGCATCACCATAATCTTCC-3′ (reverse), while the internal control
β-actin primers were 5′-ATGGATGATGATATCGCCGCGCTC-3′ (forward) and
5′-TTTCTCCATGTCGTCCAGTTGG-3′ (reverse). After first-strand
synthesis, an equivalent of 50 ng of starting total cellular RNA
(1/10 of the cDNA reaction) was added to two duplicate PCR reaction
tubes, each containing 12.5 μl SYBR-Green mix, 0.5 μl SYBR-Green
Rox (both from Takara, Dalian, Liaoning, China), 100 nmol/l forward
primer, and 100 nmol/l reverse primer in a final volume of 25 μl.
The amplification protocol consisted of denaturation at 95°C for 10
min (to activate the enzyme), followed by 45 cycles of denaturation
at 95°C for 10 sec and annealing and extension at 60°C for 34 sec,
using an ABI SDS 7500 system (Applied Biosystems). Expression of
HAUSP was analyzed using the 2−ΔΔCt method. HAUSP mRNA
expression was normalized relative to expression of β-actin in the
same sample.
Western blot analysis
Total tissue proteins were purified from all 80
glioma tissue samples and 10 normal brain tissue specimens and were
lysed in SDS lysis buffer (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 1 mM
EDTA, 0.5% SDS), followed by centrifugation at 12,000 × g for 10
min at 4°C. The supernatants were collected and protein
concentrations were determined using a Bio-Rad protein assay dye
reagent (Bio-Rad, Hercules, CA, USA). For electrophoresis, aliquots
containing equal amounts of whole protein lysate (50 μg) were
separated by size on 4–20% polyacrylamide gel (Invitrogen) under
SDS denaturing conditions. Separated proteins were transferred to
PVDF membranes at 200 mA for 2 h. The membranes were blocked with
5% non-fat milk in 1X Tris-buffered saline containing 0.1% Tween-20
and incubated with primary antibody against HAUSP (sc-30164;
1:1,000; Santa Cruz Biotechnology Inc.), or actin (1:1,000;
Beyotime, China) overnight at 4°C. Finally, blots were incubated
with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or
goat anti-mouse IgG antibody (ZSGB-BIO) for 1 h at room
temperature. Immunoblots were visualized by chemiluminescence using
an ECL detection system (BeyoEcl Plus; Beyotime) and the intensity
of the bands was determined using the Image-Pro Plus 6.0 software
(Japan).
Statistical analysis
All statistical analyses were performed using SPSS
software (version 16.0; SPSS Inc., Chicago, IL, USA). The rank-sum
test was used to analyze ranked data. Measured data were analyzed
using one-way analysis of variance (ANOVA). Randomized block design
ANOVA was used to analyze the differences between different tissue
types. In the analysis of glioma morbidity for all patients, we
used the Kaplan-Meier estimator and univariate Cox regression
analysis to assess the marginal effect of each factor. The
differences between groups were tested by log-rank analyses. The
joint effect of different factors was assessed using multivariate
Cox regression. Spearman’s analysis was carried out to analyze the
correlation between HAUSP mRNA and protein expression levels. A
P-value <0.05 was considered to indicate a statistically
significant result.
Results
Expression levels of HAUSP in patients
with malignant gliomas and normal brain tissue specimens by
immunohistochemical assay and survival analysis
HAUSP expression was assessed in a total of 80
glioma specimens of which 27 were low-grade glioma (grades I and
II) and 53 were high-grade (grades III and IV). Ten specimens
obtained from normal brain tissue served as the control group.
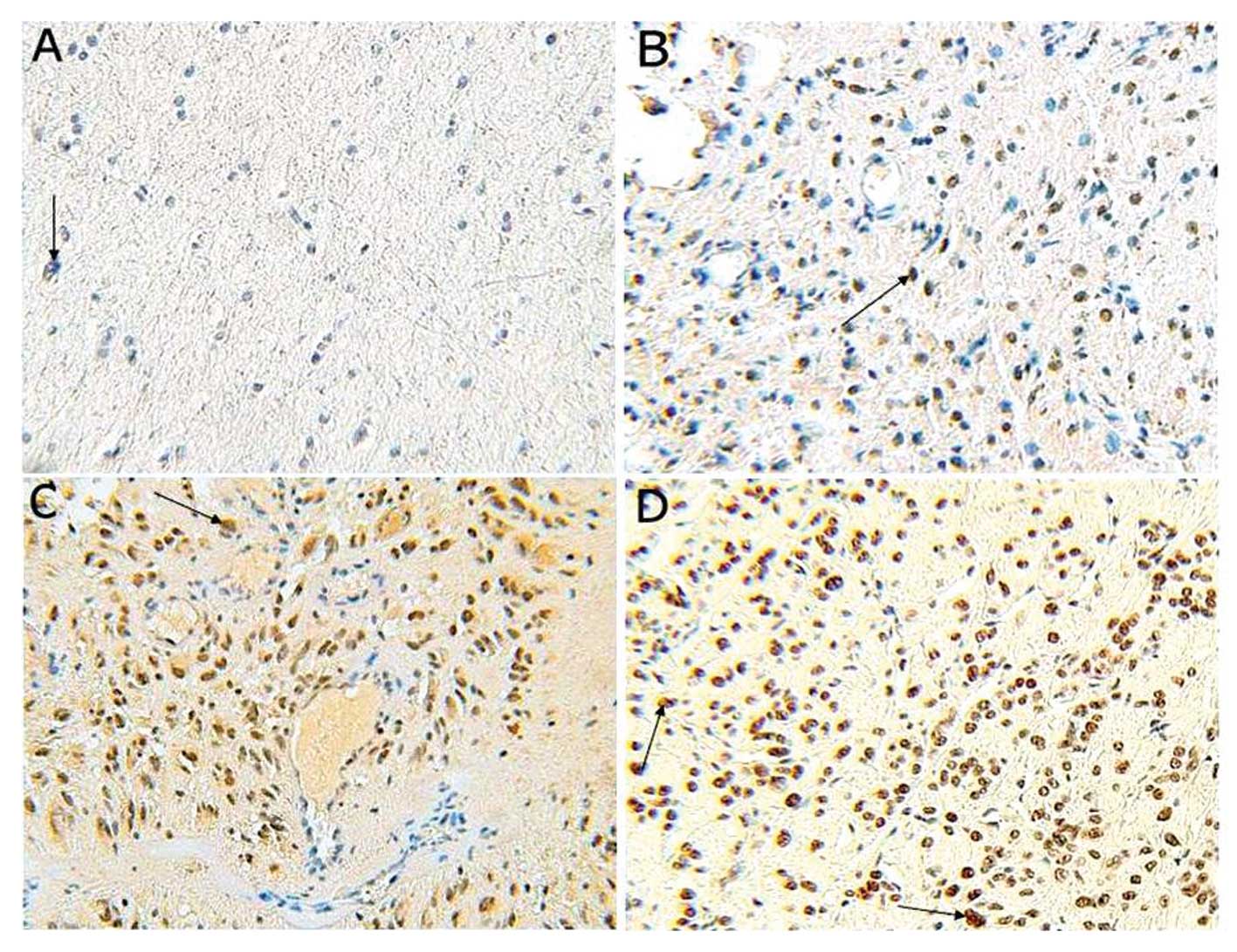
Based on immunohistochemical analysis, positive staining for HAUSP
was predominantly localized in the nuclei of tumor cells, but a
weaker cytoplasmic reaction was noted (Fig. 1). Among the glioma specimens, 69
(86.25%) glioma specimens were positively stained, and 11 (13.75%)
glioma specimens were negatively stained. Among the control
specimens, 6 (60.0%) were positively stained with only weak
staining observed in a few cell nuclei, and 4 (40.0%) were
negatively stained. We also found a significant increase in HAUSP
expression in glioma when compared with the normal brain tissues
(P<0.05).
Based on the hierarchical scores of the
immunohistochemical staining, we analyzed the relationship between
HAUSP staining and clinical factors. HAUSP expression was not
significantly affected by gender and age (both P>0.05) of the
patients. In contrast, the HAUSP expression was closely associated
with WHO grade, with expression increasing from grade I to IV
(P<0.05; Table II). We also
found that HAUSP protein expression was higher in patients with
Karnofsky performance status (KPS) <80 than in those with KPS
≥80 (P<0.05; Table II).
 | Table IIMean values of HAUSP mRNA expression
in clinical glioma samples and normal control tissues, and
comparison with clinicopathological variables. |
Table II
Mean values of HAUSP mRNA expression
in clinical glioma samples and normal control tissues, and
comparison with clinicopathological variables.
| n | HAUSP expression
(mean ± SD) | P-value |
|---|
| Tissue type | | | <0.05a |
| Glioma | 80 | 3.5776±0.6498 | |
| Normal
control | 10 | 1.1928±0.1821 | |
| Gender | | | 0.392a |
| Male | 48 | 3.5152±0.5682 | |
| Female | 32 | 3.6686±0.5147 | |
| Age (years) | | | 0.213a |
| <60 | 54 | 3.4996±0.4941 | |
| ≥60 | 26 | 3.7258±0.5266 | |
| KPS | | | <0.05a |
| <80 | 43 | 3.7128±0.5182 | |
| ≥80 | 37 | 3.4245±0.4814 | |
| WHO grade | | | <0.05b |
| I | 8 | 1.7918±0.3281 | |
| II | 19 | 2.7768±0.4146 | |
| III | 25 | 4.0352±0.5063 | |
| IV | 28 | 5.7928±0.6225 | |
Moreover, we reviewed clinical information of the
glioma patients with HAUSP-positive or -negative tumors. During the
follow-up period, 61 of the 80 glioma patients (76.25%) succumbed
to disease (57 from the HAUSP-positive group and 4 from the
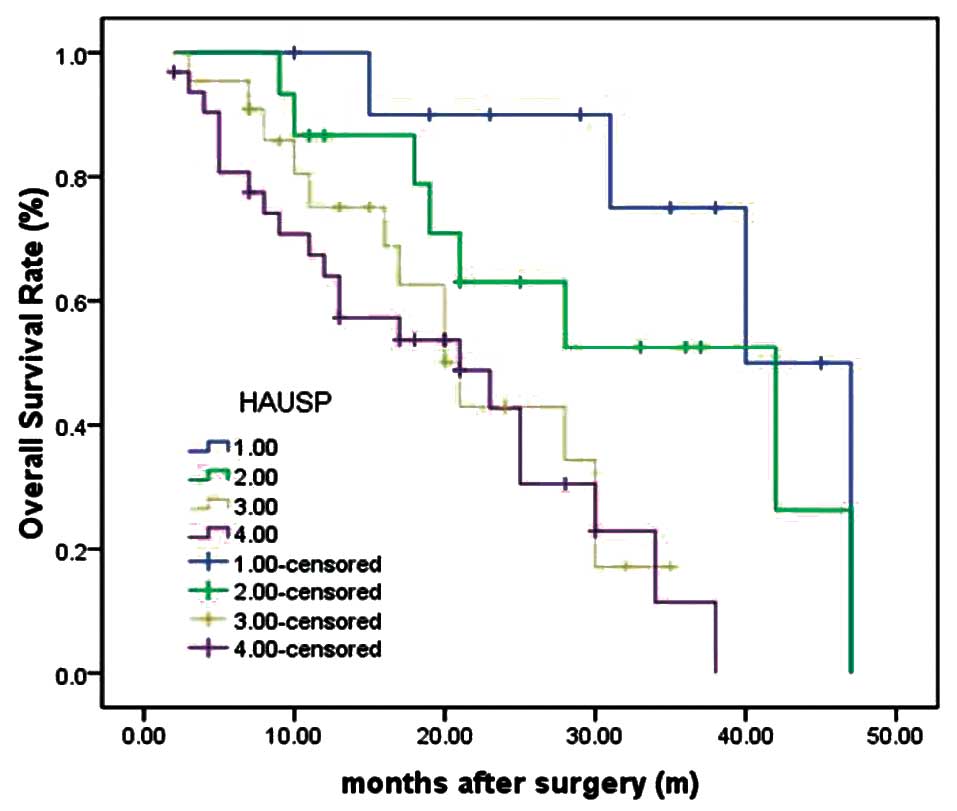
HAUSP-negative group). As determined by the log-rank test, the
survival rate of patients with tumors lacking HAUSP staining was
longer than those with HAUSP-positive staining tumors (P<0.001)
(Fig. 2). The median survival time
of patients with negative expression of HAUSP could not be
estimated by statistical analysis since all patients survived
longer than the overall median level, and median survival time of
patients with tumors with strong positive (+++), moderate positive
(++) and weak positive (+) of HAUSP were 7.9±0.6, 12.8±1.2 and
21.5±2.1 months (log-rank test, P=0.003).
Furthermore, the post-operative survival curve of
patients with glioma and HAUSP expression after adjusting for age,
gender, WHO grade and KPS was plotted. By multivariate analysis,
overexpression of HAUSP was a significant and independent
prognostic indicator for patients with glioma besides age, WHO
grade and KPS. The Cox proportional hazards model showed that
higher HAUSP expression was associated with poor overall
survival.
Quantitative analysis of HAUSP mRNA
expression in glioma by RT-PCR
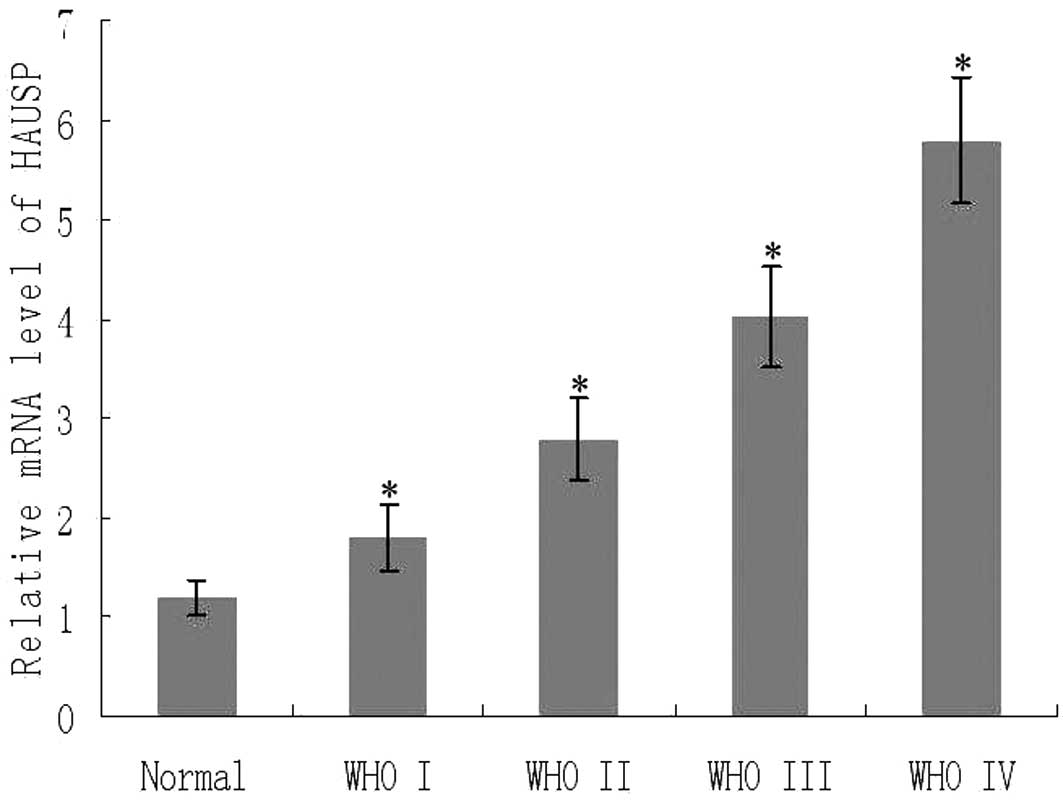
We determined the mRNA expression of HAUSP
normalized to β-actin by real-time PCR. As shown in Fig. 3, there was an obvious increase in
the expression of HAUSP mRNA from the control normal brain tissues
to glioma tissues (P<0.05). We further analyzed the expression
of HAUSP mRNA based on KPS and WHO grade. Notably, HAUSP mRNA
expression increased in patients whose KPS was <80 (P<0.001)
and also increased with advancement of WHO grade I to IV
(P<0.01). There was a significant positive correlation between
the expression of HAUSP mRNA and protein expression levels from the
same glioma tissues (rs=0.878, P<0.001).
Quantitative analysis of HAUSP protein
expression based on WHO grade in gliomas by western blotting
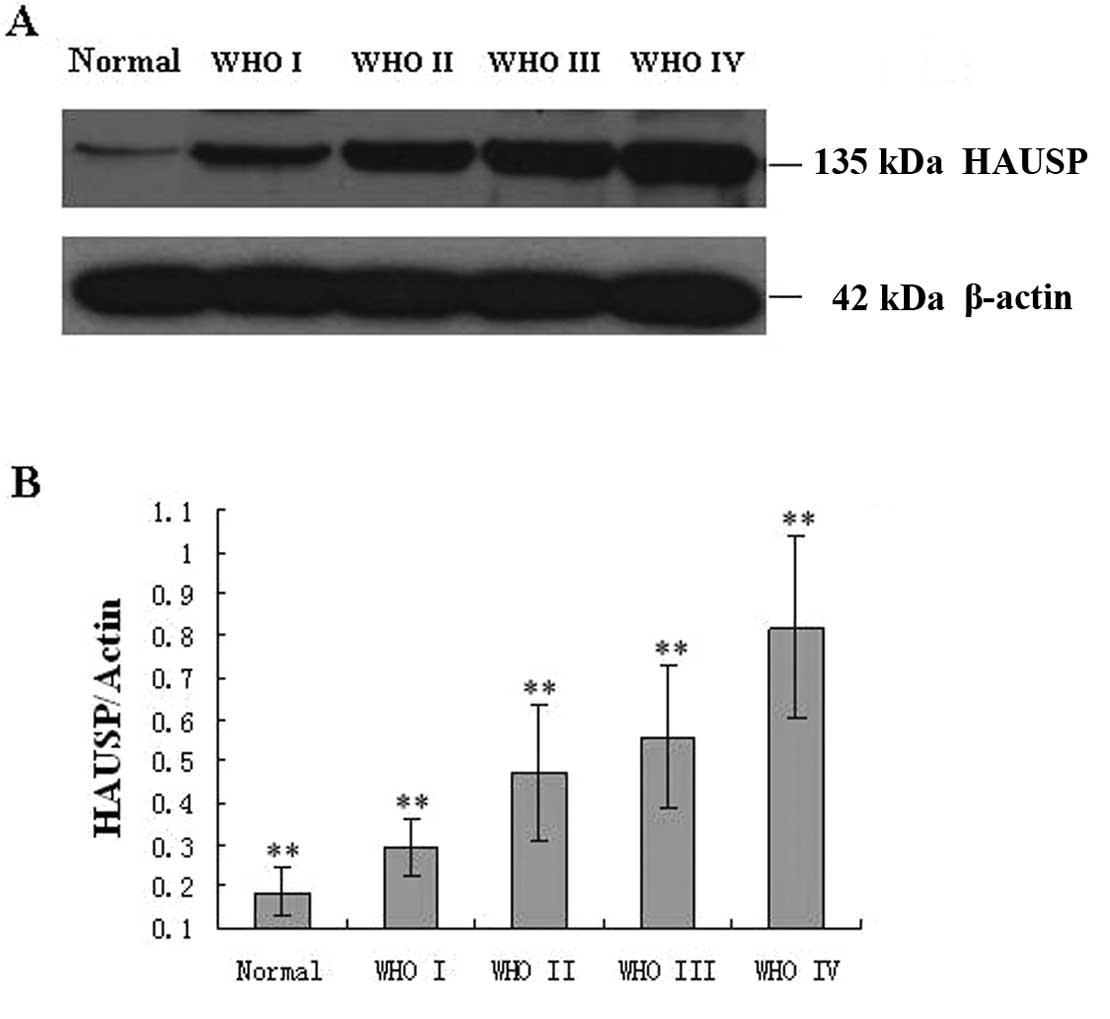
Based on the results of western blot analysis, we
found that HAUSP protein expression tended to increase from the
normal brain tissue to the glioma. We also investigated whether the
expression of HAUSP correlated with WHO grade. HAUSP expression was
lowest in grade I and highest in grade IV (Fig. 4). This result agreed with the
findings of the immunohistochemical analysis and indicated a close
correlation of HAUSP protein expression with WHO grade.
Discussion
In the present study, we investigated the expression
of HAUSP in 80 cases of human glioma and 10 cases of human normal
brain tissues, and compared the expression with tumor grade and the
survival rates of patients. Our data demonstrated that HAUSP
protein was overexpressed in glioma compared to that in the normal
brain tissue. HAUSP mRNA expression was also increased in glioma
compared with the control normal brain tissue. We found an
increasing trend of both HAUSP protein and mRNA levels from WHO
grade I to WHO grade IV glioma. These results suggest that the
transcriptional activation of human HAUSP may participate in the
carcinogenesis and progression of glioma. HAUSP may have an
important role during the genesis or progression of glioma.
Protein degradation in eukaryotic cells is mediated
by either the lysosome or ubiquitin-proteasome pathway (13). Ubiquitination and deubiquitination
have a precise role for selective protein degradation in eukaryotic
cells, and are important for the regulation of a number of
intracellular processes including cell cycle, apoptosis,
transcriptional activation, signal transduction, antigen
presentation, oncogenesis, preimplantation, and DNA repair
(3,14,15).
Deregulation of the ubiquitin-proteasome system has been implicated
in the pathogenesis of many human diseases, including cancer.
Ubiquitylation, the covalent attachment of one or
more units of the 76-amino acid peptide ubiquitin, is an important
post-translational modification that regulates the levels, activity
and localization of many cellular proteins (16). Ubiquitylation is a reversible
modification; ubiquitin can also be removed from substrates by DUBs
(17). By specifically removing
ubiquitin chains, DUBs modulate the ubiquitylation status of a
variety of proteins, and thereby contribute to the regulation of
important cellular processes, such as the regulation of the
substrate protein nucleocytoplasmic distribution (18), gene expression (19), proliferation (20), repair of DNA damage (21) and apoptosis (22). Of the DUBs, USP7, also known as
herpes virus-associated ubiquitin-specific protease HAUSP, has been
well characterized.
HAUSP is a 135-kDa protein in the USP family of the
DUB enzymes. It is primarily a nuclear protein and localizes to a
subset of PML bodies (23). In
addition to containing a DUB domain, it also has a C-terminal
domain that contains at least five ubiquitin-like domains (24) and an N-terminal TRAF-like MATH
domain (25). It is produced
ubiquitously and is an evolutionarily highly conserved protein in
eukaryotes, which was originally isolated as a binding partner of
the herpes simplex virus protein Vmw110/ICP0 (4).
A number of studies have indicated, at the molecular
level, by virtue of its deubiquitinating activity, that HAUSP
regulates the steady-state level of several polyubiquitinated
substrates. For example, HAUSP modulates the level of the
p16INK4a and p53 tumor suppressors through changes in
the stabilization of Bmi1/Mel18 and Mdm2, respectively (10,11,26).
HAUSP binding to p53 was recently shown to be regulated by TSPYL5,
a protein potentially involved in breast oncogenesis through its
competition with p53 for binding to the same region of HAUSP
(27). Additional proteins involved
in genomic integrity and regulation, such as the DNMT1 DNA
methylase and the claspin adaptor, are also stabilized by HAUSP
through deubiquitination (28,29).
HAUSP was shown to regulate the cellular compartmentalization of
several monoubiquitinated substrates by deubiquitination.
Concerning this point, in response to oxidative stress, HAUSP
inhibits nuclear localization and transcriptional activity of the
forkhead box O transcription factor (FOXO4) by interacting with and
deubiquitylating FOXO protein (9).
Through a similar mechanism, HAUSP deubiquitylates tumor suppressor
PTEN, and thus regulates its nuclear exclusion (8). HAUSP overexpression in human prostate
cancer was directly associated with tumor aggressiveness, most
likely through PTEN mislocalization (8). Previous in vivo data also
underlined the involvement of HAUSP in cancer cell proliferation
(30). Therefore, HAUSP exerts both
p53-dependent and p53-independent effects on controlling cell
proliferation and apoptosis. Collectively, the connections between
HAUSP and several pathways involving oncogenes and tumor
suppressors, strongly suggest that it may play a role in the
carcinogenesis of different types of tumor.
The present results confirm that HAUSP is
overexpressed during glioma progression. We further analyzed the
correlation of HAUSP expression and survival rates of patients. Our
data revealed that only 13.75% (11/80) of glioma cases showed
negative staining for HAUSP. The survival rate of patients with
strong positive or moderate positive staining tumors for HAUSP was
lower than the survival rates of patients with tumors showing
HAUSP-negative or weak positive staining. Kaplan-Meier analysis of
the survival curves showed a significantly worse overall survival
for patients whose tumors had high HAUSP levels, indicating that
high HAUSP protein level is a marker of poor prognosis for patients
with glioma in our study. Moreover, multivariate analysis showed
that overexpression of HAUSP may be a marker of worse outcome
independent of known clinical prognostic indicators such as age,
KPS and grade. These data indicate that high expression of HAUSP is
correlated with a worse outcome of patients with glioma in China.
Thus, HAUSP may be an independent predictor of survival for glioma
patients. In our study, which consisted of 90 cases of glioma and
normal brain samples, HAUSP expression was analyzed by
immunohistochemistry, real-time PCR and western blot analysis.
Thus, using a comprehensive methodology and a detailed clinical
follow-up in our study the results were reliable.
A recent study revealed that HAUSP counterbalances
REST (also known as neuron restrictive silencer factor, NRSF)
ubiquitination and prevents nasopharyngeal carcinoma (NPC)
differentiation. HAUSP expression was found to decline concordantly
with REST upon neuronal differentiation and reciprocally with
β-TrCP levels. HAUSP knockdown in NPCs decreased REST and induced
differentiation. In contrast, HAUSP overexpression upregulated REST
by overriding β-TrCP-mediated ubiquitination. Furthermore, REST
overexpression in NPCs rescued the differentiation phenotype
induced by HAUSP knockdown. It demonstrated that HAUSP stabilized
REST through deubiquitination and antagonized β-TrCP in regulating
REST at the post-translational level. Thus, the HAUSP-mediated
deubiquitination represents a critical regulatory mechanism
involved in the maintenance of NPCs. Expression of functional HAUSP
is critical for preventing REST ubiquitination and suppressing NPC
differentiation. All-trans retinoic acid (RA) induced NPCs
to undergo cellular differentiation, HAUSP and REST protein levels
gradually decreased during NPC differentiation. HAUSP may play
critical roles in the stabilization of stem cell transcription
factors and may promote maintenance of ‘stemness’ (31).
Embryonic stem cells (ESCs) and cancer cells share
many key biological properties, such as self-renewal, an
undifferentiated state, extensive proliferative potency,
pluripotency and differentiation capacity. These parallel features
suggest that similar mechanisms may be involved in regulating ESCs
and cancer cells. The cancer stem cell (CSC) theory of
tumorigenesis assumes the possibility of the identification of a
small group of tumor cells responsible for the occurrence, growth,
and recurrence of tumors in different types of cancers including
gliomas (32–34). Transcriptional regulators of stem
cell maintenance and differentiation require exquisite control to
direct cell fate determination. Uncontrolled activation of core
stem cell pathways drives transformation while loss of function in
these cellular mechanisms leads to degenerative conditions
(35). A number of transcriptional
factors (Nanog, c-Myc, Sox2 and Oct3/4) serving as core regulators
of self-renewal and maintenance of ESCs and tissue stem cells have
been found to be ubiquitylated by different E3 ubiquitin ligases.
It is likely that each stem cell transcription factor can be
deubiquitylated by a specific deubiquitylase (35), and different types of
transcriptional factors play a key role in the pathogenesis of
gliomas and maintain the undifferentiated state of glioma cells.
For example, our previous study indicated that the ESC
transcriptional factor, NANOG, was overexpression in glioma tissues
when compared with that in normal brain tissues at the mRNA and
protein levels. An association between higher NANOG expression and
aggressive grades of gliomas was also demonstrated. It may
contribute to the existence of BTSCs and may be related to
tumorigenesis of the cerebrum by maintaining the undifferentiated
state of glioma cells (36). We
speculate that overexpression of HAUSP in gliomas may be involved
in a network that counterbalances transcriptional factor
ubiquitination, stabilizes stem cell transcription factors,
prevents glioma cell differentiation and promotes maintenance of
‘stemness’.
In conclusion, our comprehensive analysis indicated
that overexpression of HAUSP appears to be intimately involved in
the pathogenesis of gliomas, and the activity of HAUSP may
contribute to maintaining an undifferentiated state of glioma
cells. On the basis of these findings, we assume that inhibition of
HAUSP, important in the progression of glioma cell transformation,
may block the tumorigenesis of gliomas, and targeting HAUSP may be
an approach to improve the therapeutic intervention for poorly
differentiated gliomas. However, further study is required to
determine the precise role of HAUSP and the mechanism of HAUSP
transcriptional regulation in gliomas, in particular, the
relationship between the transcriptional factors of ESCs that are
involved in the pathogenesis of gliomas.
Acknowledgements
This study was supported by a grant (no. 81172407)
from the National Natural Science Foundation of China (NSFC). We
would like to thank all surgeons from the Department of
Neurosurgery of Anhui Provincial Hospital for aiding in the
collection of tumor samples.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: the avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010.PubMed/NCBI
|
|
3
|
Hershko A and Ciechanover A: The ubiquitin
system. Annu Rev Biochem. 67:425–479. 1998. View Article : Google Scholar
|
|
4
|
Everett RD, Meredith M, Orr A, Cross A,
Kathoria M and Parkinson J: A novel ubiquitin-specific protease is
dynamically associated with the PML nuclear domain and binds to a
herpesvirus regulatory protein. EMBO J. 16:1519–1530. 1997.
View Article : Google Scholar
|
|
5
|
Sowa ME, Bennett EJ, Gygi SP and Harper
JW: Defining the human deubiquitinating enzyme interaction
landscape. Cell. 138:389–403. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kessler BM, Fortunati E, Melis M, Pals CE,
Clevers H and Maurice MM: Proteome changes induced by knockdown of
the deubiquitylating enzyme HAUSP/USP7. J Proteome Res.
6:4163–4172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicholson B and Suresh Kumar KG: The
multifaceted roles of USP7: new therapeutic opportunities. Cell
Biochem Biophys. 60:61–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song MS, Salmena L, Carracedo A, Egia A,
Lo-Coco F, Teruya-Feldstein J and Pandolfi PP: The
deubiquitinylation and localization of PTEN are regulated by a
HAUSP-PML network. Nature. 455:813–817. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Horst A, de Vries-Smits AM,
Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM
and Burgering BM: FOXO4 transcriptional activity is regulated by
monoubiquitination and USP7/HAUSP. Nat Cell Biol. 8:1064–1073.
2006.PubMed/NCBI
|
|
10
|
Cummins JM, Rago C, Kohli M, Kinzler KW,
Lengauer C and Vogelstein B: Tumour suppression: disruption of
HAUSP gene stabilizes p53. Nature. 428:1 p following 486. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Brooks CL, Kon N and Gu W: A dynamic
role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 13:879–886. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Masuya D, Huang C, Liu D, Nakashima T,
Yokomise H, Ueno M, Nakashima N and Sumitomo S: The HAUSP gene
plays an important role in non-small cell lung carcinogenesis
through p53-dependent pathways. J Pathol. 208:724–732. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Glickman MH and Ciechanover A: The
ubiquitin-proteasome proteolytic pathway: destruction for the sake
of construction. Physiol Rev. 82:373–428. 2002.PubMed/NCBI
|
|
14
|
Ciechanover A: The ubiquitin-proteasome
pathway: on protein death and cell life. EMBO J. 17:7151–7160.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baek KH: Conjugation and deconjugation of
ubiquitin regulating the destiny of proteins. Exp Mol Med. 35:1–7.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grabbe C, Husnjak K and Dikic I: The
spatial and temporal organization of ubiquitin networks. Nat Rev
Mol Cell Biol. 12:295–307. 2011. View
Article : Google Scholar
|
|
17
|
Komander D, Clague MJ and Urbé S: Breaking
the chains: structure and function of the deubiquitinases. Nat Rev
Mol Cell Biol. 10:550–563. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
García-Santisteban I, Banuelos S and
Rodríguez JA: A global survey of CRM1-dependent nuclear export
sequences in the human deubiquitinase family. Biochem J.
441:209–217. 2012.PubMed/NCBI
|
|
19
|
Frappier L and Verrijzer CP: Gene
expression control by protein deubiquitinases. Curr Opin Genet Dev.
21:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song L and Rape M: Reverse the curse - the
role of deubiquitination in cell cycle control. Curr Opin Cell
Biol. 20:156–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bergink S and Jentsch S: Principles of
ubiquitin and SUMO modifications in DNA repair. Nature.
458:461–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramakrishna S, Suresh B and Baek KH: The
role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci.
68:15–26. 2011. View Article : Google Scholar
|
|
23
|
Muratani M, Gerlich D, Janicki SM, Gebhard
M, Eils R and Spector DL: Metabolic-energy-dependent movement of
PML bodies within the mammalian cell nucleus. Nat Cell Biol.
4:106–110. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Faesen AC, Dirac AM, Shanmugham A, Ovaa H,
Perrakis A and Sixma TK: Mechanism of USP7/HAUSP activation by its
C-terminal ubiquitin-like domain and allosteric regulation by
GMP-synthetase. Mol Cell. 44:147–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zapata JM, Pawlowski K, Haas E, Ware CF,
Godzik A and Reed JC: A diverse family of proteins containing tumor
necrosis factor receptor-associated factor domains. J Biol Chem.
276:24242–24252. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Maertens GN, El Messaoudi-Aubert S,
Elderkin S, Hiom K and Peters G: Ubiquitin-specific proteases 7 and
11 modulate Polycomb regulation of the INK4a tumour suppressor.
EMBO J. 29:2553–2565. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Epping MT, Meijer LA, Krijgsman O, Bos JL,
Pandolfi PP and Bernards R: TSPYL5 suppresses p53 levels and
function by physical interaction with USP7. Nat Cell Biol.
13:102–108. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du Z, Song J, Wang Y, et al: DNMT1
stability is regulated by proteins coordinating deubiquitination
and acetylation-driven ubiquitination. Sci Signal.
3:ra802010.PubMed/NCBI
|
|
29
|
Faustrup H, Bekker-Jensen S, Bartek J,
Lukas J and Mailand N: USP7 counteracts SCFbetaTrCP- but not
APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 184:13–19.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Becker K, Marchenko ND, Palacios G and
Moll UM: A role of HAUSP in tumor suppression in a human colon
carcinoma xenograft model. Cell Cycle. 7:1205–1213. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Z, Wu Q, Guryanova OA, Cheng L, Shou
W, Rich JN and Bao S: Deubiquitylase HAUSP stabilizes REST and
promotes maintenance of neural progenitor cells. Nat Cell Biol.
13:142–152. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
33
|
Bertrand J, Begaud-Grimaud G, Bessette B,
Verdier M, Battu S and Jauberteau MO: Cancer stem cells from human
glioma cell line are resistant to Fas-induced apoptosis. Int J
Oncol. 34:717–727. 2009.PubMed/NCBI
|
|
34
|
Li G, Chen Z, Hu YD, Wei H, Li D, Ji H and
Wang DL: Autocrine factors sustain glioblastoma stem cell
self-renewal. Oncol Rep. 21:419–424. 2009.PubMed/NCBI
|
|
35
|
Huang Z, Zhou W and Bao S: Role of
deubiquitylase HAUSP in stem cell maintenance. Cell Cycle.
10:1182–1183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu CS, Li DX, Liu YH, Fu XM, Tang SF and
Li J: Expression of NANOG in human gliomas and its relationship
with undifferentiated glioma cells. Oncol Rep. 26:593–601.
2011.PubMed/NCBI
|