Introduction
Breast cancer is the most prevalent tumor in women,
and its incidence accounts for 7-10% of all malignant tumors.
Huntington’s disease (HD) is an autosomal dominant hereditary
neurodegenerative disorder characterized by late onset, progressive
psychiatric disruption, cognitive deficits and loss of motor
coordination (1). According to a
previous study, the overall incidence of cancer is significantly
lower among patients with HD, and this seems to be related to
intrinsic biologic factors (2). HD
is caused by a CAG triplet repeat expansion in exon 1 of the
huntingtin (Htt) gene, encoding an abnormal expanded polyglutamine
(polyQ) tract that confers toxicity to the mutant Htt protein. When
the mutant Htt gradually accumulates in the cell, it can affect the
function of several proteins and the conduction of some signaling
pathways. Huntingtin-interacting protein 1 (HIP1) and
huntingtin-associated protein 1 (HAP1) are two known ligands of Htt
(3,4). Research on the relevance between HIP1
and cancer showed that HIP1 had a high expression level in breast,
colon, prostate and other types of cancer (5,6).
However, there are currently no studies regarding the association
between HAP1 and cancer. In this study, we first examined the level
of HAP1 in human breast tumor and normal breast tissues, and
investigated the roles and possible mechanisms of HAP1 in human
breast cancer cells. In addition, we selected ER positive MCF-7 and
triple negative MDA-MB-231 cells, representing two main types of
breast cancer, for research.
Materials and methods
Tumor samples
A total of 43 breast carcinoma specimens with
non-neoplastic adjacent tissues from patients with primary breast
tumor were collected during the surgical procedures at the
Department of General Surgery, Jiangsu Cancer Hospital, Nanjing
Medical University (Nanjing, China). The sample collection was
carried out in accordance with the National Regulation of Clinical
Sampling in China.
Cell culture
The human breast cancer cell lines MCF-7 and
MDA-MB-231 were maintained in DMEM with high glucose (Gibco, Grand
Island, NY, USA) and 10% FBS (Gibco).
Viral production and infection of target
cells
Retrovirus was generated by cotransfection of
pBabe-puro empty vector (as control) or pBabe-puro-Hap1
plasmid along with pVSP-G (envelope) and pVSV-GP (packaging)
plasmids in 293FT cells, growing at 85-90% confluency in 10-cm
petri dishes. After 48 h, the culture medium containing the viral
particles was harvested. Target cells were infected with virus
containing medium in the presence of 10 μg/ml polybrene (Sigma, St.
Louis, MO, USA). Then, changing back to fresh medium, cells were
selected with 2 μg/ml puromycin (Fischer, USA) for 7-10 days,
re-cultured in a larger dish containing puromycin and used for
further assay. All plasmids and cells were kindly provided by
Professor Jinrong Zhou, at Harvard University (Cambridge, MA,
USA).
Cell viability assay
Cells (3,000/well) were plated in 96-well plates.
Each group had five replicates. The growth was monitored 72 h later
and cell viability was measured by CCK-8 (Dojindo, Kumamoto
Prefecture, Kyushu, Japan).
Colony formation assay
Cells were seeded in six-well culture plates at 200
cells/well. After 10 days of incubation, the cells were stained
with 0.1% crystal violet (Enox, Shanghai, China). The colonies with
>50 cells were counted. The colony formation rate was acquired
through colony numbers/total seeding cells.
Semiquantitative RT-PCR
Total RNA was extracted with the TRIzol Reagent
(Invitrogen, Carlsbad, CA, USA) and was then reverse transcribed
using PrimeScript RT Master mix (Takara, Kumamoto Prefecture,
Kyushu, Japan). Real-time PCR was performed using an ABI 7300
Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) with
the SYBR Premix Ex Taq (Takara). All samples were analyzed in
triplicate and in optically clear 96-well plates (Corning Inc.,
Corning, NY, USA). The cycling parameters were: 95°C for 30 sec
followed by 40 cycles of 95°C for 5 sec, 60°C for 31 sec and 95°C
for 15 sec, 60°C for 60 sec, 95°C for 15 sec. The human β-actin
transcript was used as an internal reference to control for
variations in the total mRNA quantity of each sample. Each RNA
sample was analyzed in triplicate. Primer sequences are listed in
Table I.
 | Table IPrimers used for Q-PCR
amplification. |
Table I
Primers used for Q-PCR
amplification.
| Gene | Forward | Reverse |
|---|
| HAP1 |
5′-ATGCGCCCGAAGAGGTTGG-3′ |
5′-CTGCAGATCGTCGTGCCGATGA-3′ |
| Caspase-3 |
5′-GATGGAAGCGAATCAATGGACT-3′ |
5′-CTGTACCAGACCGAGATGTCA-3′ |
| Caspase-8 |
5′-TCATGGACCACAGTAACATGGA-3′ |
5′-AGTGAACTGAGATGTCAGCTCAT-3′ |
| Caspase-9 |
5′-CACTCCCCCTGAAGACGAGTC-3′ |
5′-GTGGGCAAACTAGATATGGCG-3′ |
| Bcl-2 |
5′-GGTGGGGTCATGTGTGTGG-3′ |
5′-CGGTTCAGGTACTCAGTCATCC-3′ |
| Bax |
5′-CCCGAGAGGTCTTTTTCCGAG-3′ |
5′-CCAGCCCATGATGGTTCTGAT-3′ |
| Survivin |
5′-TGCGGGAATCCAAAGGATAATTCA-3′ |
5′-CTTCATCTTTGTCATACTTCATGGCT-3′ |
| EGFR |
5′-GAAGGAGCTGCCCATGAGAA-3′ |
5′-GACTATGTCCCGCCACTGGAT-3′ |
| β-actin |
5′-TTCTACAATGAGCTGCGTGTG-3′ |
5′-CAGCCTGGATAGCAACGTACA-3′ |
Western blot analysis
Cells were collected and lysed with lysis buffer
(Beyotime, Jiangsu, China), and concentrated to obtain the
proteins. The amount of total proteins was estimated by Onedrop
OD-1000+ Spectrophotometer (Onedrop, Shanghai, China), and then
proteins were mixed with SDS-PAGE buffer (Beyotime) and boiled for
5 min. The proteins were loaded onto 10% SDS-PAGE gel and,
following electrophoresis, the proteins were transferred onto a
PVDF membrane (Bio-Rad, Hercules, CA, USA). Then, the membrane was
blocked with nonfat-dried milk, incubated with primary and
secondary antibody (Dako, Japan), and protein bands were visualized
by ECL detection reagent (Millipore, Billerica, MA, USA).
In vitro migration and invasion
assays
For the migration assays, the cells were detached
and aliquots of 2×105 cells/ml were plated onto the
inserts of the 8-μm pore-sized Transwell chambers (Corning Inc.).
For the Transwell invasion assays, 1×106 cells were
plated onto inserts containing a polycarbonate membrane with a thin
layer of BD Matrigel Matrix (BD Biosciences, Franklin Lakes, NJ,
USA). Both migration and invasion were assayed 24 h later by
counting the cell numbers across the membrane.
Flow cytometric assay
For cell apoptosis, a total of 1×106
cells were transferred to a tube in which 5 μl Annexin V and 1 μl
propidium iodide (PI) were added after being resuspended with 100
μl binding buffer (Invitrogen). The cells were then allowed to
incubate at room temperature for 15 min and were analyzed using
flow cytometry. For cell cycle analysis, cells were collected,
washed twice with PBS, supplemented with 1 ml 75% ethanol, and kept
at -20°C overnight. The cells were then resuspended with PBS,
supplemented with PI and RNaseA, incubated for 30 min at 37°C and
analyzed using flow cytometry.
Statistical analyses
Data are expressed as the means ± SD, and
statistical significance was assessed by Student’s t-test and
one-way ANOVA. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of HAP1 in breast tumor
samples
HAP1 expression was assessed by qRT-PCR in
surgical specimens from patients with breast cancer. As shown in
Fig. 1, HAP1 expression was
markedly reduced in most cancer tissues compared with normal breast
tissues. Of 43 cases, ~70% showed HAP1 expression levels in
normal breast tissues >3-fold higher than those of matched
tumors (Fig. 1).
Expression of HAP1 in transfected and
control cells
The MCF-7 and MDA-MB-231 cells transfected with
pBabe-puro-HAP1 and pBabe-puro were labeled MCF-7/HAP1, MCF-7/pBabe
and MDA-MB-231/HAP1, MDA-MB-231/pBabe, respectively. Western
blotting and qRT-PCR results of different cells showed that
MCF-7/HAP1 (P<0.006) and MDA-MB-231/HAP1 (P=0.001) had a
significantly higher expression of HAP1 than their control groups.
Moreover, there was no obvious difference between pBabe and
negative control groups (P>0.05) (Fig. 2A and B).
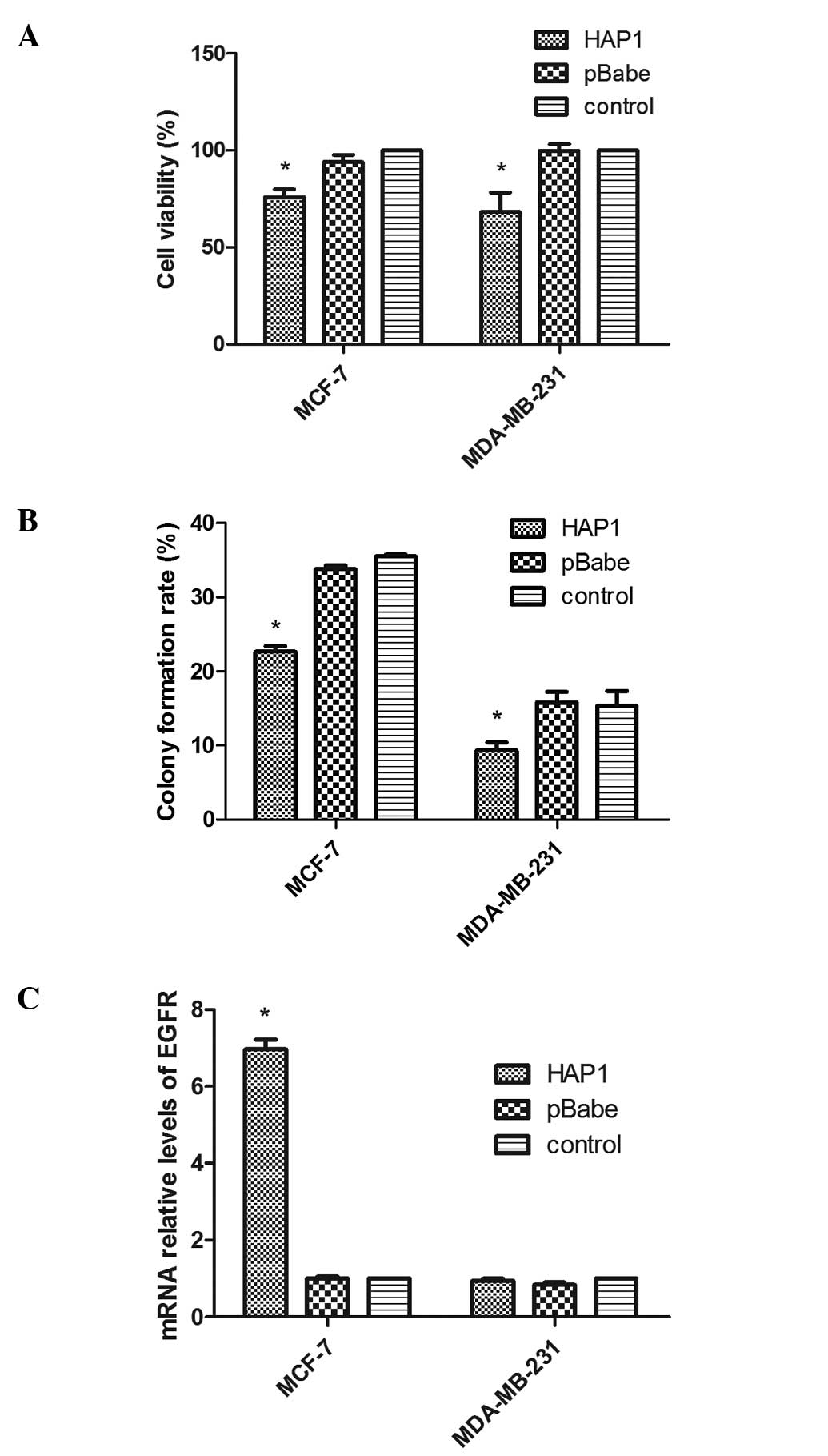
Effects of HAP1 on breast cancer cell
growth and expression of EGFR in cells
At 72 h after cells were plated in 96-well plates,
upregulation of HAP1 significantly increased the cell
viability of both HAP1 overexpression cells in MCF-7 [HAP1
mean, 76.0±6.8 vs. pBabe mean, 94.0±6.6 (P=0.03) vs. control mean,
100±0.0 (P=0.004)] and MDA-MB-231 [HAP1 mean, 68.2±17.4 vs. pBabe
mean, 99.8±6.0 (P=0.041) vs. control mean, 100±0.0 (P=0.034)]
compared with empty vector plasmid transfected cells and negative
control cells (Fig. 3A). In
addition, colony formation rates were significantly decreased in
MCF-7 cells with HAP1 overexpression [HAP1 mean, 22.7±1.3% vs.
pBabe mean, 33.8±0.8% (P=0.000) vs. control mean, 35±0.5%
(P=0.000)] and MDA-MB-231 [HAP1 mean, 9.3±1.9% vs. pBabe mean,
15.8±2.5% (P=0.022) vs. control mean, 16±2.6% (P=0.024)] when
compared with those of pBabe and control cells (Fig. 3B). We also compared the expression
of EGFR in three groups of the cell lines. We observed that in the
MCF-7 cell lines, EGFR expression of MCF-7/HAP1 [HAP1 mean, 7.0±0.4
vs. pBabe mean, 1.0±0.1 (P=0.000) vs. control mean, 1.0±0.0
(P=0.000)] was ~7-fold higher than that of the other two groups.
However, there was no difference in EGFR expression between the
three groups in the MDA-MB-231 cell lines (F=3.2; P=0.1) (Fig. 3C).
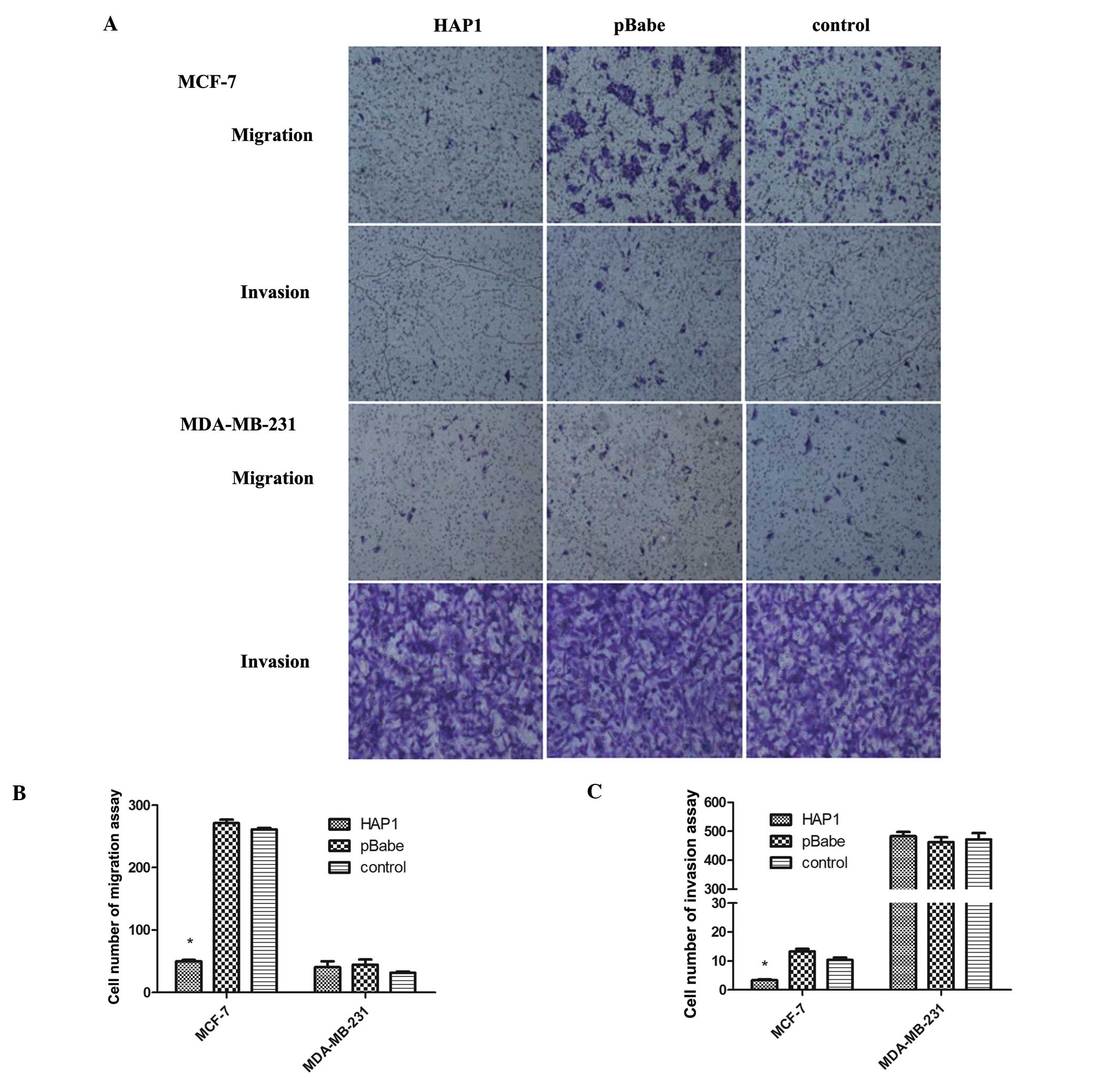
Overexpression of HAP1 impairs migration
and invasion
We observed that MCF-7/HAP1 had fewer cells across
the membrane than MCF-7/pBabe and MCF-7 in migration assays [HAP1
mean, 50.0±3.6 vs. pBabe mean, 271±9.5 (P=0.000) vs. control mean,
261±4.6 (P=0.000)] (Fig. 4A and B).
In addition, compared to the number of the other two cell groups,
the number of MCF-7/HAP1 cells that invaded the Matrigel-coated
filter was significantly lower [HAP1 mean, 3.3±0.6 vs. pBabe mean,
13.3±1.5 (P=0.000) vs. control mean, 10.3±1.5 (P=0.000)] (Fig. 4A and C). Thus, the overexpression of
HAP1 in MCF-7 can curb tumor cell migration and invasion.
However, in MDA-MB-231 cells, there was no significant difference
in the number of cells across the membrane in both cell migration
(F=0.8; P=0.5) and invasion (F=0.3; P=0.7) (Fig. 4A-C).
Effects of HAP1 on cell cycle, apoptosis
and expression of relative genes
According to cell cycle distribution analysis by
flow cytometry, HAP1 increased the percentage of cells in the
G2M phase. In MCF-7 cells, the percentage in the
G2M phase of the HAP1 group was 30.84%, higher than the
13.17% and the 10.61% in the pBabe and control groups. In
MDA-MB-231 cells, the percentage in the G2M phase of the
HAP1 group was 14.84%, also higher than the other two groups
(Table II). The results indicated
that HAP1 induced G2M arrest in both cell lines.
Furthermore, HAP1 overexpression significantly induced
apoptosis of MCF-7 cells compared with pBabe and control groups,
and there was also no significant difference in MDA-MB-231 cells
(Fig. 5A). Therefore, we detected
the expression of genes, including caspase-3, -8, -9, Bcl-2, Bax
and survivin, involved in cell apoptosis and survival, by RT-PCR
(Fig. 5B and C). The results showed
that the expression of caspase-3, -9, Bcl-2, Bax and survivin in
MCF-7/HAP1 was significantly higher than in the other two groups.
Moreover, the expression of caspase-3 was >800 times higher in
MCF-7/HAP1 compared with the other two groups [HAP1 mean,
857.7±278.9 vs. pBabe mean, 1.0±0.1 (P=0.000) vs. control mean,
1.0±0.0 (P=0.000)], and it was confirmed by western blotting
(Fig. 5D).
 | Table IICell cycle distribution in MCF-7 and
MDA-MB-231 cells. |
Table II
Cell cycle distribution in MCF-7 and
MDA-MB-231 cells.
| MCF-7 | MDA-MB-231 |
|---|
|
|
|
|---|
| HAP1 | pBabe | Control | HAP1 | pBabe | Control |
|---|
|
G0G1 (%) | 36.53±0.96 | 46.04±2.22 | 58.11±2.18 | 56.69±0.83 | 56.79±1.08 | 55.63±1.02 |
| S (%) | 32.63±0.34 | 40.78±0.41 | 31.27±0.66 | 28.46±0.22 | 31.45±0.22 | 33.17±0.17 |
| G2M
(%) | 30.84±1.09a | 13.17±1.83 | 10.61±1.53 | 14.84±0.90a | 11.76±0.93 | 11.19±0.94 |
Discussion
HAP1 is a type of cytoplasmic protein distributed
mainly in the microtubules and membranous organelles, including the
mitochondria, endoplasmic reticulum, tubulovesicles, endosomal and
lysosomal organelles, and synaptic vesicles (7,8). HAP1
affects the synthesis of certain proteins and the conduction of
signals (9-11). As a ligand of the production of HD,
it was initially studied in the nervous system, whereas later
research showed that it was involved in the other systems.
Herein, we detected the expression of HAP1 in
breast tumor and normal breast tissues. In contrast to normal
breast tissues from breast cancer patients, we observed that
HAP1 expression was reduced in the majority of breast tumor
samples. To the best of our knowledge, this is the first published
study to show that HAP1 is downregulated in breast tumor
tissues and, based on these findings, it is suggested that
HAP1 plays a role in the pathogenesis of breast cancer.
HAP1 is involved in numerous cellular functions,
including cell proliferation and apoptosis (12,13).
In this study, upregulation of HAP1 resulted in a
significant decrease in the cell growth and colony formation rate
of both breast cancer cells. Furthermore, cell cycle results showed
that the percentage of G2M phase increased in both MCF-7
and MDA-MB-231 cells, indicating that cell growth could be arrested
at the G2M phase by HAP1. Moreover, apoptosis may be
involved in the inhibition of cell growth. It has been reported
that apoptosis plays an important role during the malignant
transformation of normal cells. The above studies, along with ours,
indicate that loss or downregulation of HAP1 may disrupt the
balance between proliferation and apoptosis and may represent a key
pathogenic step in the development of breast cancer. The cell
apoptosis assay confirmed that HAP1 induced cell apoptosis and
increased expression of relative genes in MCF-7 cells, particularly
caspase-3. Caspase-3 is located at the downstream of the apoptosis
signaling pathway, and plays a key role in cell apoptosis (14,15).
Collectively, HAP1-induced apoptosis in MCF-7 cell lines may be
mainly mediated by the caspase-dependent pathway; in the MDA-MB-231
cells, however, the three groups showed no difference in the
expression of those genes.
Previous studies showed that inhibition of
HAP1 expression decreased EGFR signaling and cell viability,
whereas overexpression of HAP1 enhanced this signaling
activity (12). EGFR is expressed
in various types of tissue, including epithelial, mesenchymal and
of neuronal origin, and plays a major role in normal cellular
processes such as proliferation, differentiation and development.
EGFR is also highly expressed in a number of solid tumors and its
expression correlates with tumor progression, resistance to
chemotherapy and a poor prognosis (16,17).
Our detection of EGFR showed that EGFR expression was higher in
MCF-7/HAP1, which negatively correlated with the results of cell
proliferation. Therefore, we considered that EGFR might play a role
in MCF/HAP1, although not a major one; on the contrary, the effect
of EGFR was counteracted by others.
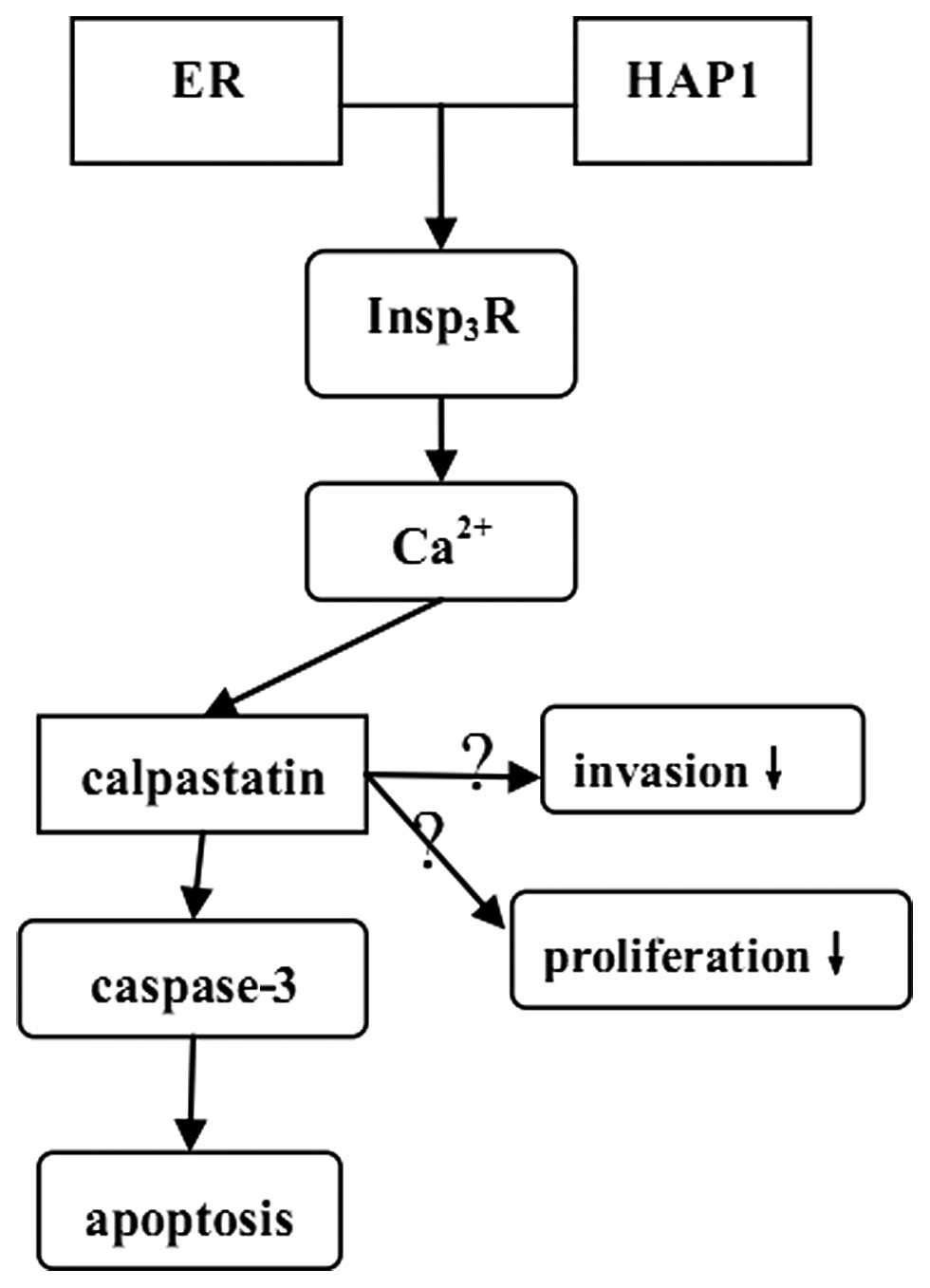
Although it is unclear why the effect of HAP1 is
significant in MCF-7 cells and not in MDA-MB-231 cells, we propose
a hypothesis (Fig. 6). It was
reported that more than 90% sporadically lurking
HAP1-immunoreactive (HAP1-ir) cells expressed nuclear estrogen
receptor (ER) (18). Therefore, we
hypothesized that ER-positive MCF-7 cells gathered more HAP1 than
triple negative MDA-MB-231 cells and interacted with them.
In various cells, the Ca2+ signaling
process is mediated by the
endoplasmic-reticulum-membrane-associated 1,4,5-trisphosphate
(Insp3) receptor (Insp3R), which has a
critical role in the control of cellular and physiological
processes as diverse as cell division, cell proliferation and
apoptosis (19,21). Tang et al found that
Insp3R could bind with HAP1A in vitro and in
vivo, suggesting that HAP1 may impact the function of
Insp3R (20). When
Insp3R is activated by HAP1, the Ca2+ release
is increased, subsequently inducing the activation of calpains.
The calpains are a family of neutral cysteine
proteases that require calcium for their catalytic activity. The
cellular activity of calpain is, in part, regulated by their
endogenous inhibitor calpastatin (22,23).
The calpains function in the controlled proteolysis of a large
number of specific substrates involved in various cellular
processes such as migration, cell signalling and apoptosis. In
addition, previous studies showed that calpain cleaved caspase-9
and therefore blocked the caspase-3 activation (24). Furthermore, Storr et al
suggested that ER-positive breast cancer had a significantly high
calpastatin expression compared with ER-negative breast cancer, and
the expression of calpastatin in non-triple negative breast cancer
was also significantly higher than in triple-negative breast cancer
(22). The expression of calpain 1,
however, was the opposite of calpastatin. Based on these data, we
deduced that in ER-positive or (and) non-triple negative breast
cancers, more apoptosis was induced than in ER-negative or (and)
triple-negative breast cancer.
Collectively, ER, expressed in MCF-7, induced HAP1
assembly, then sequentially activated Insp3R,
calpastatin and caspase-3, finally increasing apoptosis. Moreover,
the procedure may also impact invasion. This pathway does not exist
in MDA-MB-231. However, further research is required to verify the
hypothesis and to clarify the mechanism.
In summary, our findings demonstrated for the first
time that HAP1 downregulation occurs in breast tumor, and
that HAP1 suppresses breast cancer cell growth. However, there were
also some different effects of HAP1 between ER-positive
MCF-7 and triple negative MDA-MB-231 cells, which we hypothesize
may reflect the different types of breast cancer in the clinic. The
findings of this study may provide a new therapeutic target for the
treatment of breast cancer patients.
Acknowledgements
We thank Professor Jinrong Zhou of Harvard
University and Dr Shuchun Li of Nanjing University of Technology
for their technical assistance and helpful discussion.
References
|
1
|
Schulte J and Littleton JT: The biological
function of the Huntingtin protein and its relevance to
Huntington’s Disease pathology. Curr Trends Neurol. 5:65–78.
2011.
|
|
2
|
Sorensen SA, Fenger K and Olsen JH:
Significantly lower incidence of cancer among patients with
Huntington disease: an apoptotic effect of an expanded
polyglutamine tract? Cancer. 86:1342–1346. 1999. View Article : Google Scholar
|
|
3
|
Kalchman MA, Koide HB, McCutcheon K, et
al: HIP1, a human homologue of S. cerevisiae Sla2p,
interacts with membrane-associated huntingtin in the brain. Nat
Genet. 16:44–53. 1997.PubMed/NCBI
|
|
4
|
Li XJ, Li SH, Sharp AH, et al: A
huntingtin-associated protein enriched in brain with implications
for pathology. Nature. 378:398–402. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rao DS, Hyun TS, Kumar PD, et al:
Huntingtin-interacting protein 1 is overexpressed in prostate and
colon cancer and is critical for cellular survival. J Clin Invest.
110:351–360. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rao DS, Bradley SV, Kumar PD, et al:
Altered receptor trafficking in Huntingtin Interacting Protein
1-transformed cells. Cancer Cell. 3:471–482. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marcora E and Kennedy MB: The Huntington’s
disease mutation impairs Huntingtin’s role in the transport of
NF-kappaB from the synapse to the nucleus. Hum Mol Genet.
19:4373–4384. 2010.
|
|
8
|
Gutekunst CA, Li SH, Yi H, Ferrante RJ, Li
XJ and Hersch SM: The cellular and subcellular localization of
huntingtin-associated protein 1 (HAP1): comparison with huntingtin
in rat and human. J Neurosci. 18:7674–7686. 1998.PubMed/NCBI
|
|
9
|
Cape A, Chen X, Wang CE, et al: Loss of
huntingtin-associated protein 1 impairs insulin secretion from
pancreatic β-cells. Cell Mol Life Sci. 69:1305–1317. 2012.
|
|
10
|
Yang GZ, Yang M, Lim Y, et al: Huntingtin
associated protein 1 regulates trafficking of the amyloid precursor
protein and modulates amyloid beta levels in neurons. J Neurochem.
122:1010–1022. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuen EY, Wei J, Zhong P and Yan Z:
Disrupted GABAAR trafficking and synaptic inhibition in a mouse
model of Huntington’s disease. Neurobiol Dis. 46:497–502.
2012.PubMed/NCBI
|
|
12
|
Li SH, Yu ZX, Li CL, et al: Lack of
huntingtin-associated protein-1 causes neuronal death resembling
hypothalamic degeneration in Huntington’s disease. J Neurosci.
23:6956–6964. 2003.PubMed/NCBI
|
|
13
|
Takeshita Y, Fujinaga R, Zhao C, Yanai A
and Shinoda K: Huntingtin-associated protein 1 (HAP1) interacts
with androgen receptor (AR) and suppresses SBMA-mutant-AR-induced
apoptosis. Hum Mol Genet. 15:2298–2312. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Galluzzi L, Kepp O and Kroemer G:
Caspase-3 and prostaglandins signal for tumor regrowth in cancer
therapy. Oncogene. 31:2805–2808. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang Q, Li F, Liu X, et al: Caspase
3-mediated stimulation of tumor cell repopulation during cancer
radiotherapy. Nat Med. 17:860–866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Pandey V, Kessler T, Lehrach H and
Wierling C: Modeling of miRNA and drug action in the EGFR signaling
pathway. PLoS One. 7:e301402012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yano S, Kondo K, Yamaguchi M, et al:
Distribution and function of EGFR in human tissue and the effect of
EGFR tyrosine kinase inhibition. Anticancer Res. 23:3639–3650.
2003.PubMed/NCBI
|
|
18
|
Islam MN, Fujinaga R, Yanai A, et al:
Characterization of the ‘sporadically lurking HAP1-immunoreactive
(SLH) cells’ in the hippocampus, with special reference to the
expression of steroid receptors, GABA, and progenitor cell markers.
Neuroscience. 210:67–81. 2012.
|
|
19
|
Bosanac I, Alattia JR, Mal TK, et al:
Structure of the inositol 1,4,5-trisphosphate receptor binding core
in complex with its ligand. Nature. 420:696–700. 2002. View Article : Google Scholar
|
|
20
|
Tang TS, Tu H, Chan EY, et al: Huntingtin
and huntingtin associated protein 1 influence neuronal calcium
signaling mediated by inositol-(1,4,5) triphosphate receptor type
1. Neuron. 39:227–239. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kopil CM, Siebert AP, Kevin FJ and Neumar
RW: Calpain-cleaved type 1 inositol 1,4,5-trisphosphate receptor
impairs ER Ca(2+) buffering and causes neurodegeneration in primary
cortical neurons. J Neurochem. 123:147–158. 2012.PubMed/NCBI
|
|
22
|
Storr SJ, Lee KW, Woolston CM, et al:
Calpain system protein expression in basal-like and triple-negative
invasive breast cancer. Ann Oncol. 23:2289–2296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mataga MA, Rosenthal S, Heerboth S, et al:
Anti-breast cancer effects of histone deacetylase inhibitors and
calpain inhibitor. Anticancer Res. 32:2523–2529. 2012.PubMed/NCBI
|
|
24
|
Chua BT, Guo K and Li P: Direct cleavage
by the calcium-activated protease calpain can lead to inactivation
of caspases. J Biol Chem. 275:5131–5135. 2000. View Article : Google Scholar : PubMed/NCBI
|