Introduction
Despite its decreasing trend, gastric cancer (GC)
still remains one of the most troublesome malignant diseases
worldwide. It is the fourth most common cancer and the second
leading cause of cancer-related death worldwide (1–3). In
Asian countries such as Japan and Korea, it has the highest
incidence among all malignancies although its overall survival rate
has improved during the last few decades due to extensive screening
programs (3,4). Due to the poor prognosis and limited
treatment options of advanced GC, it remains a challenging task to
determine effective molecular-pathological markers for early
diagnosis with appropriate histological classification (5).
Lauren classification, which subgroups GCs mainly as
two types, intestinal and diffuse, was first introduced in 1965 and
has been generally accepted as a distinct and simple histological
classification of GCs by most pathologists (6). Since the two major types show
distinctively different phenotypic, epidemiologic and biologic
characteristics, it is believed that different genetic alterations
may be implicated in each histologic type of the classification
(7). However, a considerable number
of GCs accounting for 10–15% of cases, share the histologic
features of these two types and are designated as mixed-type
(8,9). Although this type of tumor shows
poorer prognosis than the common intestinal-type GCs, it is
difficult to define and to diagnose when these tumors are
encountered in a small biopsy. Furthermore, it is even more
difficult to place this type of tumor into a specific category
according to the WHO Classification of Gastric Tumours (2000) that
was mainly founded on the Japanese Classification of Gastric
Carcinoma (1981) and Lauren classification (9). For this reason, a new WHO
classification has recently proposed (2010) this type of tumor as a
mixed adenocarcinoma, while intestinal-type and diffuse-type are
defined as tubular adenocarcinoma and poorly cohesive carcinoma,
respectively (10).
Considering that the tumor cells in diffuse-type
cancers are loosely attached to each other from the early stage
while intestinal-type cancers are derived from intestinal
metaplasia and subsequent dysplasia, different key molecular
changes may occur during the early steps of carcinogenesis in each
type. For this reason, aberrant DNA methylation is being
highlighted as the main change differentiating these subtypes of
GCs from the very first stage (1,11,12).
Accumulating evidence from over 550 studies including the authors’
previous study has demonstrated that aberrant epigenetic changes of
more than 100 genes play a crucial role in the early steps of
gastric carcinogenesis (2,13). However, the results of these studies
were based only on a small portion of GCs and at times were
conflicting in regards to a certain candidate gene. That may be
because most studies reported to date have focused on the presence
of epigenetic alterations and their prognostic value as a screening
tool without accurate consideration of their possible linkage with
histologic or phenotypic features, and the stage of carcinogenesis
that is mainly involved (2).
Furthermore, large scale screenings of epigenetic changes in GCs
have been rarely performed. Since various additional molecular
changes as well as epigenetic alterations are involved throughout
each stage of carcinogenesis, a careful research design is
essential (11).
The present study aimed to identify key differential
epigenetic changes that differentiate the histologic subtypes of
GCs in the early steps of carcinogenesis. We performed a large
scale screening of up to 485,000 CpG sites with Illumina Infinium
methylation assay (IIMA) and selected the most promising sites out
of all the differentially methylated regions (DMRs) between the
groups using one of the most reliable database, KEGG pathway
database (14). We subsequently
verified the results of the screening by pyrosequencing assay
(PA).
Materials and methods
Patient samples
The present study was approved by the Institutional
Review Board of Yonsei University, Wonju College of Medicine
(GR-11–009). Written informed consent to participate in the present
study was properly obtained for each subject. Surgically resected
early gastric cancers (EGCs) from patients treated at the Wonju
Severance Christian Hospital from 2011 to 2012 were prepared using
the following process. First, fresh tissue from the tumors and
adjacent corresponding non-tumor mucosa (CNM) (0.5×0.5 cm-sized)
were collected from each case for the methylation study. Following
routine formalin-fixed paraffin-embedded preparations and
hematoxylin and eosin staining, full pathological evaluation with
confirmatory histological classification according to Lauren were
carried out for each case by three pathologists (Y. Chong, K.
Mia-Jan and M.Y. Cho).
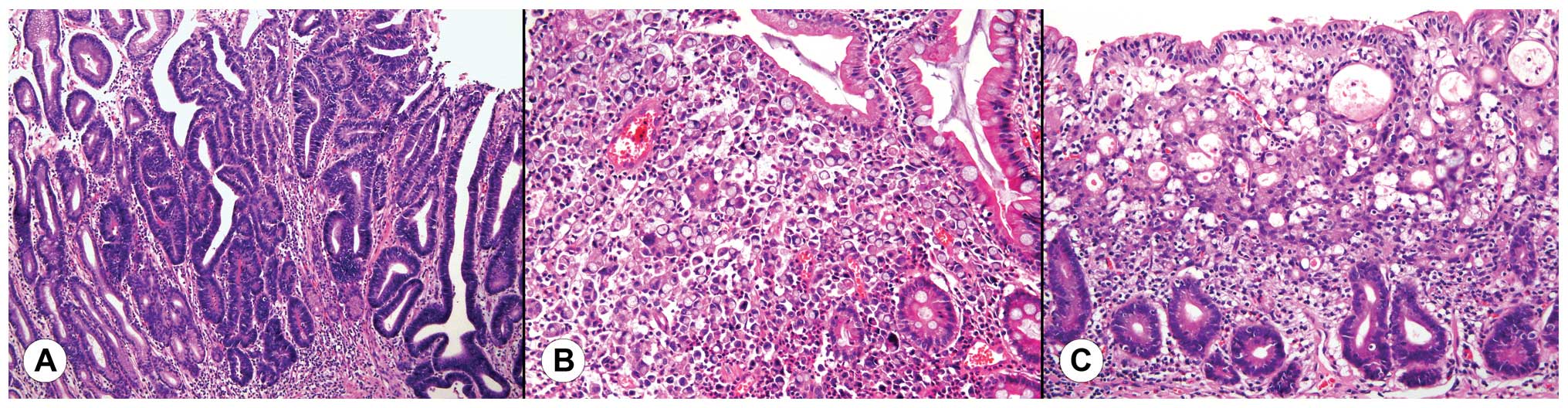
Diffuse-type GCs were defined when the tumor
exclusively consisted of non-cohesive tumor cells diffusely
infiltrating the stroma and exhibiting relatively deep infiltration
with little or no gland formation (Fig.
1A). Intestinal-type GCs were defined when most part of the
tumor consisted of recognizable glands similar to colonic or
gastric mucosa (Fig. 1B). Poorly
differentiated tumors that were difficult to categorize as either
diffuse-type or mixed-type GCs were carefully excluded. Mixed-type
GCs were defined when the tumor exhibited features of both typical
intestinal-type and diffuse-type at the same time. In such cases,
superficial parts of the tumor showed typical gland-like features
as observed in intestinal-type GCs while the neck portion of the
tumor consisted of irregular glands with non-cohesive singly
scattered tumor cells with diffuse infiltration to the deeper part
(Fig. 1C). Due to the histological
similarity that mixed-type and diffuse-type GCs share and the
general proportion of each subtype among overall GCs,
intestinal-type GCs were designated as group A, while mixed and
diffuse-type GC were designated as group B. Presence of
Helicobacter pylori (H. pylori) infection and
intestinal metaplasia in adjacent CNM were also evaluated. Next, 38
cases of EGC and CNM including the previous 12 cases were also
enrolled for validation of DMRs using PA.
DNA extraction and bisulfite
conversion
DNAs for the following studies were extracted from
each sample using DNeasy Blood and Tissue kit® (cat. no.
69506; Qiagen, Valencia, CA, USA), according to the manufacturer’s
instructions. The procedure included a proteinase K digestion at
55°C overnight. DNAs were quantified by NanoDrop® UV-Vis
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and
diluted to ~50 ng/l for IIMA. Eight hundred nanograms to 1 μg of
genomic DNAs were treated with sodium bisulfite using the EZ-96 DNA
methylation kit® (cat. no. D5004; Zymo Research, Orange,
CA, USA) according to the manufacturer’s procedure.
DNA methylation profiling by the Infinium
Human Methylation-450K BeadChip kit®
The methylation assay was performed on 4 μl
bisulfite-converted genomic DNA (50 ng/μl) with Illumina HiScanSQ
scanner (Illumina Inc., San Diego, CA, USA) according to the
Infinium methylation assay protocol. Data quality was checked with
the GenomeStudio™ for Methylation Module software (2010.3) and all
samples passed this quality control. Uncorrected β-values were
extracted with the same software. Raw data were submitted to the
Gene Expression Omnibus (GPL8490) dataset.
To determine DMRs, the average β-values were
computed with β-values when they had a signal intensity of
>3,000. β-value is defined as the intensity of the methylated
region over the sum of the intensities of the methylated and
unmethylated regions plus 100, and a value of 0 designates a
totally unmethylated status while the value 1 designates full
methylation. Δβ-value was calculated between two comparison groups
by subtracting one β-value from the other (tumor vs. CNM in group A
and B; CNM of group A vs. B and tumor of group A vs. B). When this
value was >0.2 or <−0.2, it was considered to be meaningful.
The results were considered to be significant when the p-value was
<0.05. A differential score was used for evaluating the
significance of the DMRs and when it was >13 or <−13, it was
considered to have the same significance as a p-value of 0.05, and
when it was >22 or <−22, a p-value of 0.01 was indicated.
Validation by PA and determination of the
eligible candidate genes
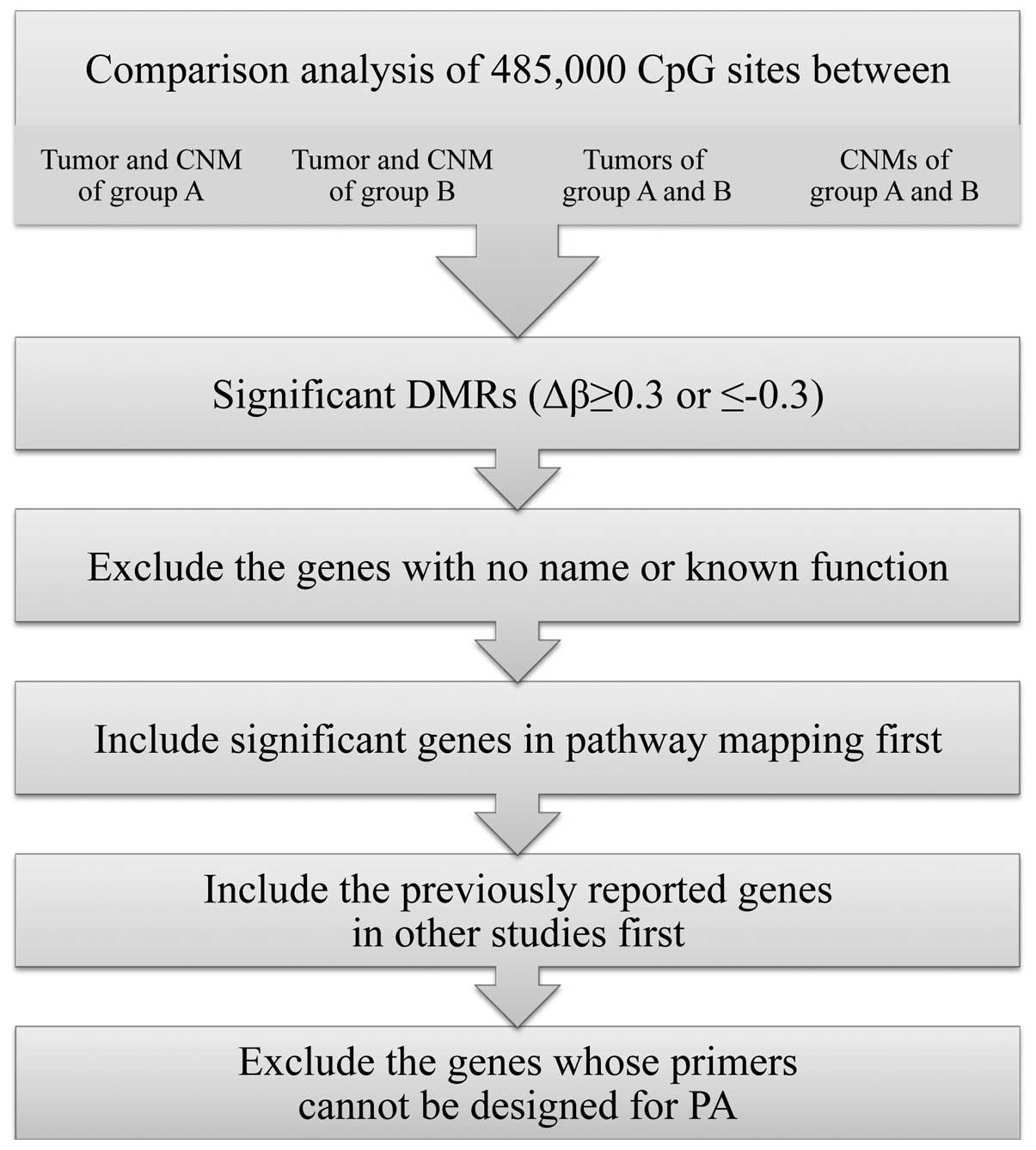
The steps that were used to select the most
significant candidate genes eligible for PA are summarized in
Fig. 2. Among the CpG sites that
showed significant differential methylation between the compared
groups in the hierarchical clustering of IIMA, the sites with high
Δβ-values (>0.3 or <−0.3) were only selected. Next, the DMRs
with no designation or known function were excluded. We then
evaluated the potential functional role of selected DMRs by pathway
mapping with DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) using Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway database
(http://www.kegg.jp/kegg/pathway.html). The pathways
with most DMRs were considered to be significantly implicated with
the DMRs discovered by IIMA, and the DMRs in those pathways were
enrolled to the list of candidate genes first. Next, we reviewed
the literature and included the genes that were previously reported
in other studies. Subsequently, we excluded the genes from the list
whose primers cannot be designed for PA due to the inappropriate CG
content in the targeted sequence.
Eligibility for designing appropriate primers for PA
was checked by PyroMark® Assay Design 2.0 software
(Qiagen, Hilden, Germany). The designed primers were carefully
validated for proper amplification after preliminary polymerase
chain reaction (PCR) tests. One microliter of the converted DNA was
used as a template in each subsequent PCR. PCRs were performed with
HotStarTaq® Plus Master Mix kit (Qiagen, Hilden,
Germany) under the following conditions: 95°C for 15 min; 40 cycles
of 95°C for 1 min; 50°C for 1 min; 72°C for 1 min and 72°C for 10
min. The success of the amplification was assessed with 2% agarose
gel electrophoresis. Pyrosequencing of the PCR products was
performed with the PyroMark™ Q24 system (Qiagen, Uppsala, Sweden)
according to the manufacturer’s instructions. Blue values (perfect
calls) and yellow values (needed to be checked manually) were
considered for subsequent analyses.
Statistical analyses
After initial processing of the raw data of IIMA,
general statistical computing was carried out by R project. For
hierarchical clustering analysis, correlation, Euclidean and
Manhattan methods were applied. To search any possible correlations
within the data, Pearson’s correlation method was utilized.
For the PA, univariate analysis using paired t-test,
correlation test, two-sample t-test, one-way ANOVA on the
differences of methylation status were used for comparing the
results according to Lauren classification (intestinal, diffuse and
mixed, and group A and B), location (proximal and distal), age and
presence of H. pylori. For hierarchical clustering analysis,
correlation, Euclidean or Manhattan methods were applied as in the
IIMA. To search for any possible correlations within the data,
Pearson’s correlation method was utilized. Scheffe’s method for
multiple comparisons was used to validate the genes that were found
to have significant differences between any of the two types of the
Lauren classification.
Results
Samples used for analysis
The 12 cases of EGC analyzed using IIMA consisted of
6 cases of intestinal-type, 4 cases of diffuse-type and 2 cases of
mixed-type (M:F, 3:1; age range 44–77 years; median age, 60).
Thirty-eight cases of EGC used for the PA consisted
of 19 cases of intestinal-type, 8 cases of diffuse-type and 11
cases of mixed-type (M:F, 3.75:1; age range 31–82 years; median
age, 67).
DNA methylation profiling by IIMA
Hierarchical clustering analysis
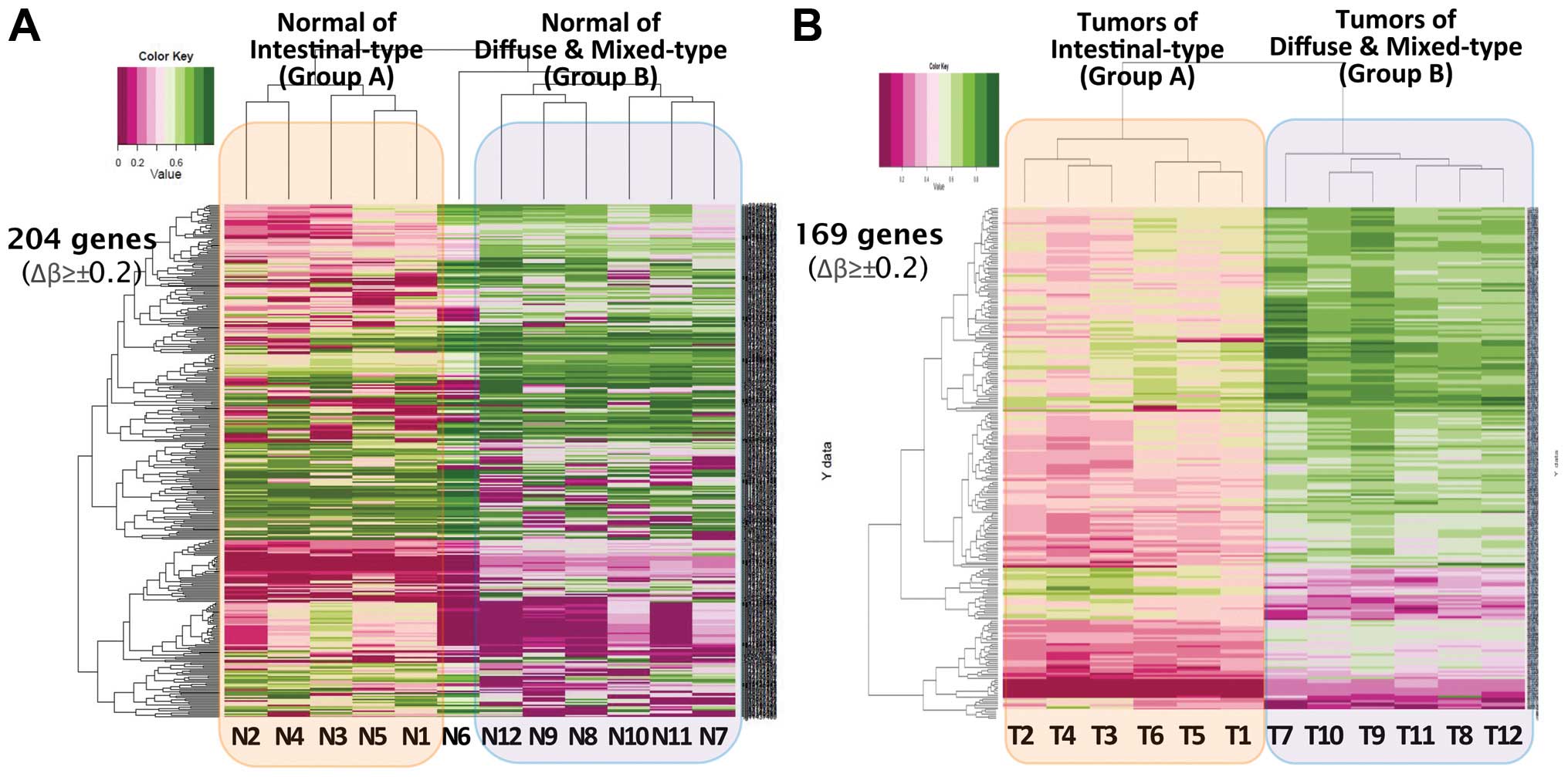
Upon hierarchical clustering analysis of 6 cases of
group A tumors and matched CNM, no distinct grouping was found with
DMRs by either correlation, Euclidean or Manhattan methods. Between
the tumors and CNM of group B, on the other hand, a clear grouping
was made by 208 genes in 3 cases, 2 cases being mixed-type, by both
correlation and Euclidean methods (T7, T11 and T12 vs. N7, N11 and
N12). However, it seemed inappropriate to evaluate the significance
of this grouping since half of the cases were not concordantly
grouped with the other 3 cases.
Hierarchical clustering analysis of CNM showed a
distinct separation between group A and B (except N6) (Fig. 3A). Although it was not statistically
significant, 204 genes showed a differential methylation status
between the two groups with Δβ-value of >0.2. The hierarchical
clustering analysis of the tumors of group A and B showed a
distinct grouping with 169 statistically significant DMRs (Δβ-value
≥±0.2, differential score ≥13, p<0.05) (Fig. 3B). In total, >800 DMRs were found
to be significantly different with a Δβ-value of >0.2 or
<−0.2 between the subgroups. After removal of the DMRs without a
specific gene name or with an unspecified function, 581 genes were
pooled as candidates (208 genes between tumors and CNM in group B,
204 genes found between CNM of group A and B, and 169 genes between
tumors of group A and B). Among these, 197 genes were included in
the list of the candidate genes with Δβ-value >0.3 or
<−0.3.
Validation of ascertained CpGs by PA
Determination of the genes eligible
for PA
As a result of pathway mapping by KEGG pathway
database, 18 genes out of 197 genes were found to be significantly
related to the archived pathways (pathways in cancers,
Notch-signaling pathway, dorso-ventral axis formation, p<0.05).
Five genes, APC, DVL2, ETS1, ITGA3 and
MAML3, among them were eligible for designing primers for PA
and showed a robust amplification in a pilot study. An additional 7
genes from the genes with top Δβ-values, C19orf35,
XKR6, CLIP4, SDC2, CNRIP1,
GAL3ST2 and CCDC57, were selected after checking for
primer eligibility and showed a robust amplification in the pilot
study. As a result, finally 12 genes from the screening result were
determined as eligible for producing tailored primers for the
following PA. Basic information of these 12 genes is listed in
Table I. Each gene includes one or
more (up to 8) CpG sites.
 | Table IPrimer sequences used in the
pyrosequencing assay. |
Table I
Primer sequences used in the
pyrosequencing assay.
| Gene | Ch. | Sequences | Amplicon size
(bp) | CpGs |
|---|
|
C19orf35 | 19 | F- |
AGATGGGGTATTGGGGATATTTTA | 146 | 8 |
| | R- |
CCTCCTAAACAACTCACCC | | |
| XKR6 | 8 | F- |
GGGGTTTTTAGTTTAGTTGG | 158 | 8 |
| | R- |
TCCCTACCCCCTTCCACTA | | |
|
CLIP4a | 2 | F- |
GGGGTGTYGTAGTGTTATTTT | 146 | 7 |
| | R- |
CTAAATCCCCYACCCCAACAACT | | |
| SDC2a | 8 | F- |
GGAGGYGGTAGTGTGATTTTTAGAT | 148 | 6 |
| | R- |
AACYCAAAAACCCTAACTTACC | | |
|
CNRIP1a | 2 | F- |
GGAGGAGTYGGTGAGTAGTT | 153 | 6 |
| | R- |
CCAAACCCTCYCCCAAACATA | | |
| GAL3ST2 | 2 | F- |
GTGTGTTAGAGGTTTGTGATG | 196 | 6 |
| | R- |
CCAAAAACCCTACAAAAATCCCC | | |
| CCDC57 | 17 | F- |
TAGATATTGTTGTTGGGAGTAGTT | 227 | 3 |
| | R- |
TCATTTAATCATTCTCTCCTTCAT | | |
| APC | 5 | F- |
ATATGTGGTTGTATTGGTG | 194 | 3 |
| | R- |
ACATACTACAATAAACCCTATACCA | | |
| DVL2 | | F- |
GTTTAGGGTAGAGATAGTTTGATTG | 180 | 2 |
| | R- |
ACTATACACTAAAACTCCCCTATC | | |
| ETS1 | 11 | F- |
ATTTTTTAATGTTAAGGGGAGATGG | 205 | 4 |
| | R- |
ATCTCCTTACCCTCAACC | | |
| ITGA3 | 17 | F- |
GATTTTGTGGGTTTATGGGGATAT | 201 | 1 |
| | R- |
CTTAAAAACTACCAAACACCTACTT | | |
| MAML3 | 4 | F- |
GGAGTTTTTTGAGGATTTGTAAGTA | 219 | 7 |
| | R- |
AATTCCAAAACCCCAACTACA | | |
Genes with significantly differential
methylation statuses between tumors and CNMs
Univariate analysis showed significant differences
in the methylation status between all tumors and their CNM
regardless of type for 5 genes: C19orf35, XKR6,
SDC2, CNRIP1 and GAL3ST2 (p=0.0262, 0.0021,
0.0081, 0.0467 and 0.0050, respectively) (Fig. 4). Four of the genes showed
hypermethylation in the tumor tissue when compared to the CNM
(XKR6, SDC2, CNRIP1 and GAL3ST2) with
an average of 10.4, 3.6, 5.2 and 7.0, respectively, while
C19orf35 was significantly hypomethylated in the tumor
tissue when compared to the CNM (average −6.5).
Significantly differential methylation
statuses according to Lauren classification
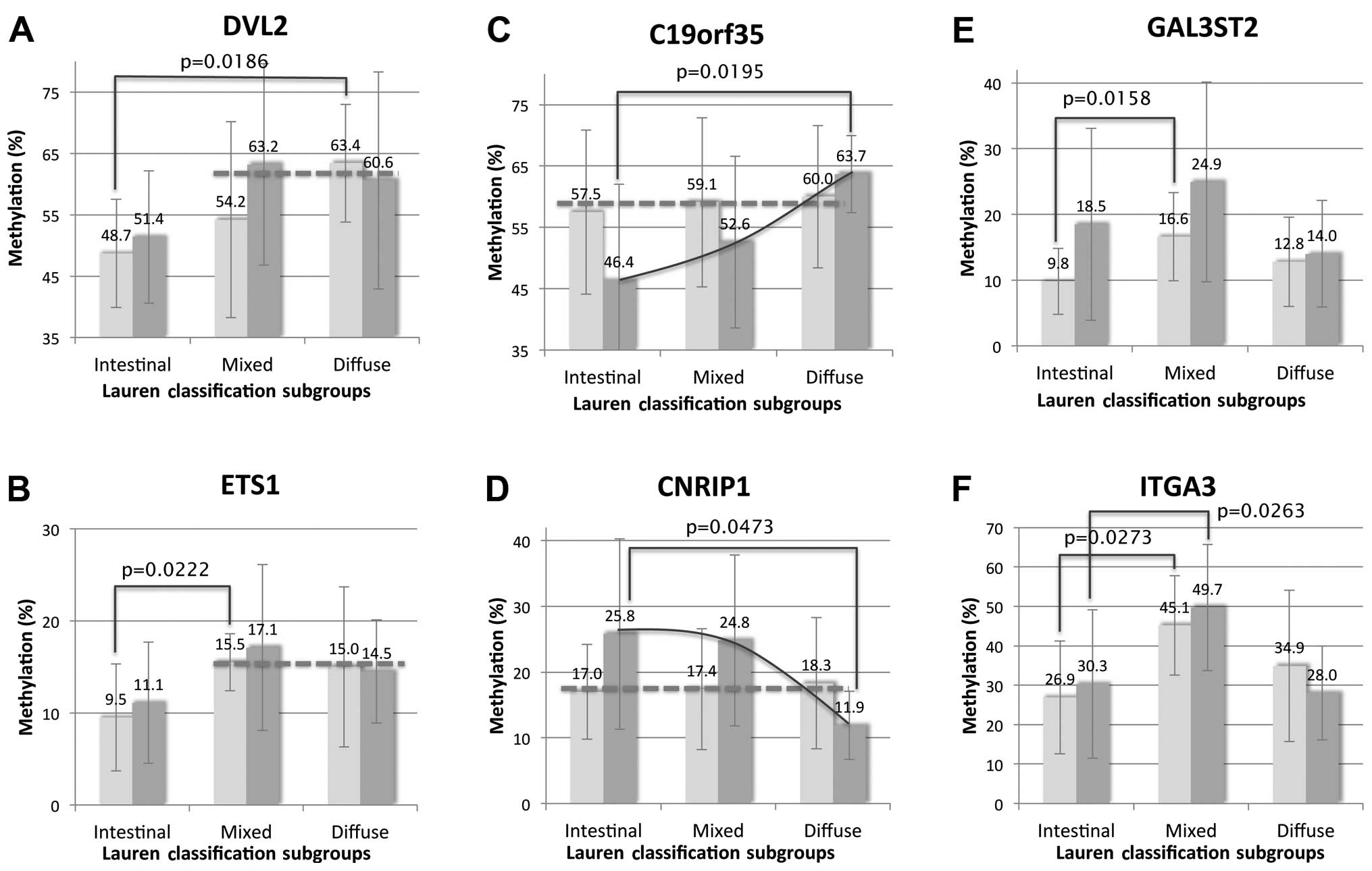
ANOVA revealed a statistical difference in
methylation status according to the Lauren classification in tumor
tissues (C19orf35, CNRIP1 and ITGA3; p=0.0192,
0.0373 and 0.0146, respectively) and matched CNM (GAL3ST2,
DVL2, ETS1 and ITGA3; p=0.0157, 0.0182, 0.0099
and 0.0259, respectively) (Fig. 5).
In Scheffe’s method of multiple comparisons, C19orf35,
CNRIP1 and DVL2 were significantly different between
the diffuse-type and intestinal-type (p=0.0195, 0.0473 and 0.0186,
respectively) while GAL3ST2, ETS1 and ITGA3
were different between the mixed-type and intestinal-type GCs
(p=0.0158, 0.0222 and 0.0273, respectively). Integrated
interpretation of the methylation pattern of these 6 genes
according to histologic subtype allowed us insight that three
groups, DVL2 and ETS1, C19orf35 and
CNRIP, GAL3ST2 and ITGA3, present different
transition between tumors and matched CNM. DVL2 and
ETS1 were hypomethylated both in the tumor tissue and the
CNM of intestinal-type EGCs while they were hypermethylated in the
mixed-type and diffuse-type EGCs (Fig.
5A and B). On the other hand, the methylation statuses of
C19orf35 and CNRIP in the three histologic subtypes
exhibited a similar level in the CNM while those varied in the
tumors according to histologic subtype (Fig. 5C and D). Notably, GAL3ST2 and
ITGA3 were significantly hypermethylated in the tumors and
the CNM of the mixed-type rather than in the intestinal-type and
diffuse-type subgroups (Fig. 5E and
F).
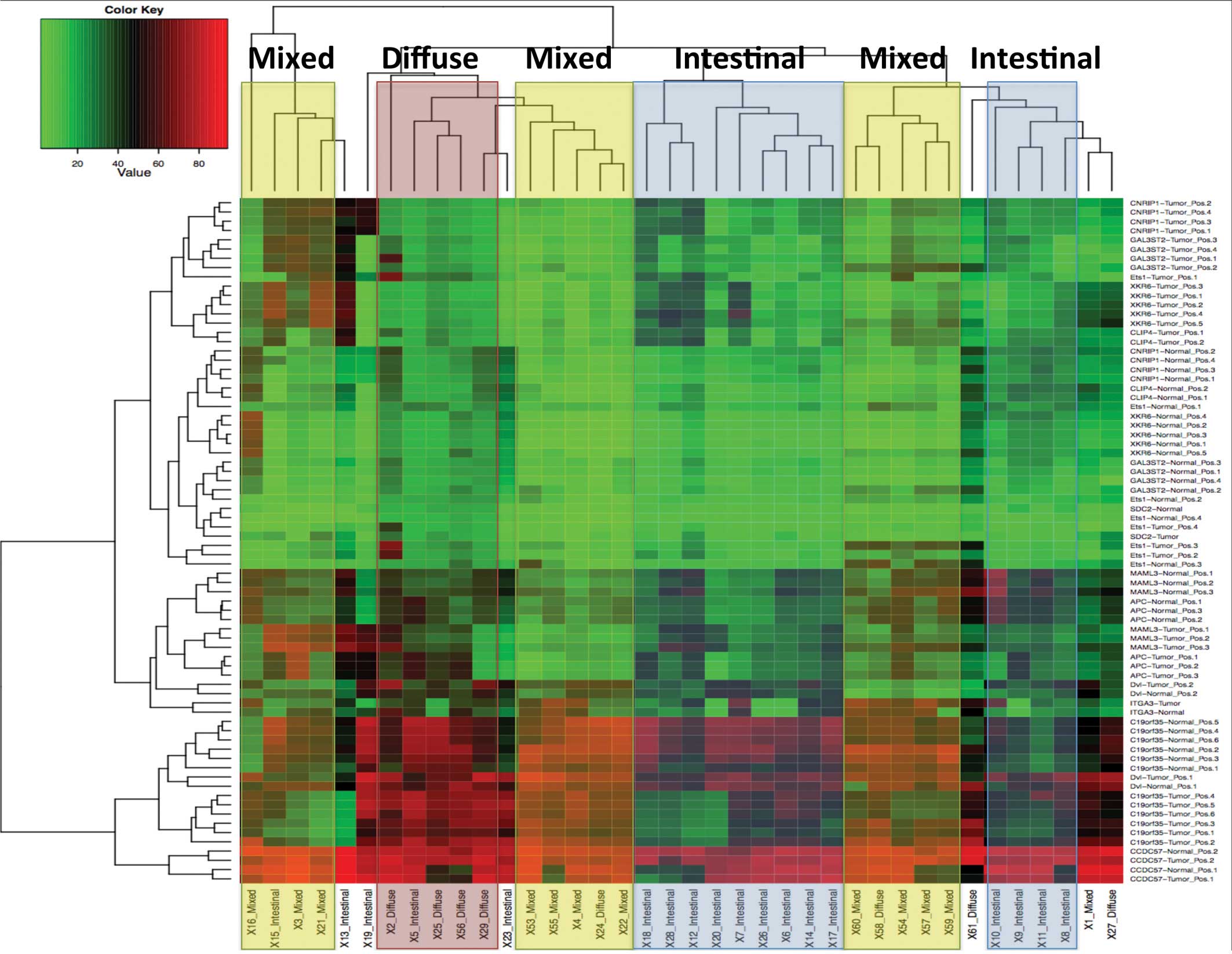
The hierarchical clustering analysis using the
methylation status of these 6 genes showed a relatively distinct
grouping between each type as the results of the hierarchical
clustering analysis using IIMA (Fig.
6).
Relationship between genes with
significantly differential methylation status and patient age,
tissue location and H. pylori infection
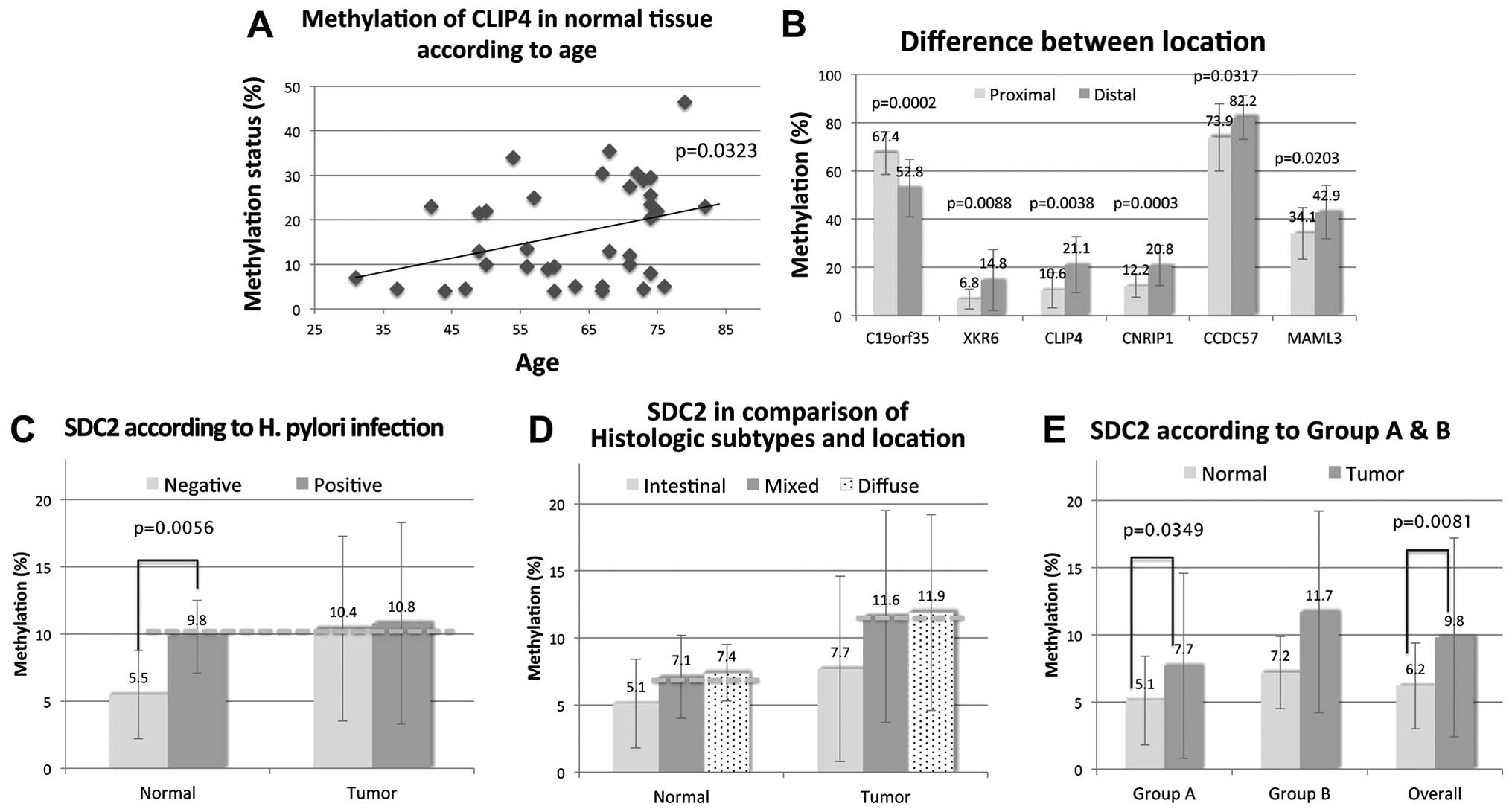
To determine whether changes in methylation status
are related with non-tumor factors, we statistically analyzed the
correlation between methylation status in the non-neoplastic mucosa
with patient age, location and H. pylori infection (15). There was a significant correlation
between patient age and the methylation status of CLIP4
(Fig. 7A). Paired t-test revealed a
significant difference in the methylation in C19orf35,
XKR6, CLIP4, CNRIP1, CCDC57 and
MAML3 between proximal and distal location (p=0.0002,
0.0088, 0.0038, 0.0003, 0.0317 and 0.0203, respectively; average
difference, −14.6, 8.0, 10.5, 8.6 and 8.3, respectively) (Fig. 7B). The methylation level of all
genes except that of C19orf35 was significantly higher in
the distal compared to proximal stomach. However, no significant
difference was found in the tumor tissues according to their
location.
In the one-way ANOVA, SDC2 was significantly
hypermethylated in CNM with H. pylori infection (p=0.0056,
average difference, 4.4) (Fig. 7C).
Aberrant methylation of SDC2 was found to correlate with
diffuse-type and mixed-type GCs (Fig.
7D and E).
Discussion
Based on methylation analysis using IIMA validated
by PA, we indirectly verified that certain methylation changes are
closely associated with the carcinogenesis of GC. Moreover,
different subsets of aberrantly methylated genes are involved in
the carcinogenesis of a specific histologic subtype of GC,
logically suggesting that mixed-type GC may be a distinct subtype.
Hierarchical clustering analysis of IIMA was broadly concordant
with the results of PA. Tumors of group A and B in IIMA showed a
sharply demarcated distinction in the methylation pattern for 169
genes (Fig. 3B). A rather clear
grouping of 208 genes in 3 cases of group B indirectly denoted the
molecular uniqueness of mixed-type GCs, although the data should be
interpreted with extreme caution due to the limited number of cases
and incomplete grouping.
Based on an integrated interpretation of the
statistical analysis of the results of PA, we classified the genes
with their methylation patterns according to the histologic
subtypes into three groups: i) DVL2 and ETS1, ii)
C19orf35 and CNRIP1, and iii) GAL3ST2 and
ITGA3 (Fig. 5).
DVL2 and ETS1 were specific to
diffuse-type and mixed-type compared to the intestinal-type
subgroup. Both genes showed a similar methylation pattern both in
the CNM and in the tumors of the intestinal-type while they showed
significantly higher methylation in the CNM as well as in the
tumors of the mixed-type and diffuse-type rather than the
intestinal-type (Fig. 5A and B).
These results are consistent with previous findings that some
aberrantly methylated genes in tumors of certain subtypes are also
aberrantly methylated in CNM (16).
This suggests that the aberrant methylation of these two genes may
start from the CNM exclusively in mixed-type and diffuse-type
subgroups. Similarly to these results, these genes showed
significant difference in methylation in the CNM between group A
and B in the hierarchical clustering analysis of IIMA.
Dishevelled, symbolized as DVL2, is a positive
mediator of Wnt/β-catenin signaling and its aberrant overexpression
has been known to result in neoplastic transformation, yet the
direct involvement in human cancer has not been reported to date
(17). Since the function of Wnt
signaling components in gastric carcinogenesis has been recently
described, it seems advisable to evaluate the methylation change of
this gene in relation to other components of Wnt signaling.
Contrary to our screening results and the accumulated evidence on
the aberrant methylation of APC in GC, we could not find any
connection between APC and the parameters examined (16). The ETS1 proto-oncoprotein is
a member of the Ets family of transcription factors and has been
implicated in invasive behavior in relation to angiogenesis in many
types of human cancer such as astrocytomas, breast cancer and some
types of colon cancer (18). It is
not expressed in normal gastric epithelium and is overexpressed in
GC, resulting in its value as a marker of poor prognosis (18–20).
To our knowledge, no information exists to date on the methylation
change of this gene in GC. Further evaluation for comprehensive
understanding is needed.
C19orf35 and CNRIP1 were specific to
diffuse-type compared to the intestinal-type subgroups. Both
revealed a similar level of methylation in CNM regardless of the
histologic subtype of tumor while the methylation of the tumor
tissue varied with the histologic type (Fig. 5C and D). C19orf35 was
hypermethylated but CNRIP1 was hypomethylated in
diffuse-type tumors compared to the CNM while it had an opposite
trend in the tumors of the intestinal-type and mixed-type
subgroups. Importantly, these findings suggest the possibility of
utilizing these genes as specific markers for diffuse-type GCs. On
the other hand, the function of C19orf35 is still unknown
while frequent methylation of CNRIP1 has recently been
reported in colorectal cancers and adenoma (21,22).
GAL3ST2 and ITGA3 were specific to
mixed-type compared to intestinal-type and diffuse-type subgroups.
They were specifically hypermethylated in CNM as well as in the
tumor tissue of mixed-type GC (Fig. 5E
and F). Thus, this analogously represents the molecular
uniqueness of mixed-type GCs contrary to the idea that these are
intermingled tumors of intestinal-type and diffuse-type GCs.
Hierarchical clustering of IIMA also showed a distinctive
methylation pattern of mixed-type compared to diffuse-type GCs,
although the data were not perfect. Little is known about the
functional role of GAL3ST2, a member of the
galactose-3-O-sulfotransferase protein family, in human
types of cancers. However, a recent series of evidence indicates
that downregulation of GAL3ST2 expression is observed in
colon cancers, and GAL3ST2 has also been implicated in the
metastatic potential of breast and laryngeal cancers (23–25).
Considering the mounting evidence on the central role of selectins
such as galectin-3 in thyroid, pancreatic, colon and even GCs in
relation to several relevant genes, the associative function of
Gal:3-O-sulfotransferase in GCs should be emphasized and
evaluated in more detail (26–29).
For ITGA3, it has been established that α3β1 integrin is
involved in the maintenance of basement membrane integrity through
adhesion to laminin 5 and plays an essential role in the
invasiveness and metastatic potential of tumor cells (30–32).
However, there are contradictory reports regarding the significance
of ITGA3 expression in various types of cancers (33–36).
Considering the results demonstrated recently that GAL3ST3
is involved in tumor metastasis by regulating the ability of
adhesion to selectins and expression of integrins, more evidence is
needed to clarify the relationship among these molecules (25). A recent study by Park et al
strengthens the results of the present study by showing
significantly higher frequencies of CpG island hypermethylation of
16 genes in mixed-type GCs when compared to diffuse-type and
intestinal-type subgroups (37).
They found no difference in the number of methylated genes between
diffuse-type and intestinal-type-like components of mixed-type
cancers, rejecting the hypothesis that mixed-type GCs may be the
mixture of intestinal-type and diffuse-type GCs.
The heatmap made with the pyrosequencing data of the
aforementioned 6 genes also demonstrated a rather clear distinction
between each subtype (Fig. 6).
Regarding the results of SDC2 methylation in
relation to H. pylori infection, this hypothesis seems to be
more credible since SDC2 in CNM with H. pylori
infection revealed significant hypermethylation compared to CNM
without H. pylori infection up to the level of tumor
regardless of H. pylori infection (Fig. 7C). These results are in line with
the results of previous studies that some genes such as
CDH1, known for having diffuse-type specificity, are
hypermethylated in cases with H. pylori infection (12,16).
These indirectly suggest the possible pathogenic relationship
between the genes and H. pylori infection as well as between
H. pylori infection and certain histologic types of GC. In
our results, aberrant methylation of SDC2 was found to
correlate with diffuse-type and mixed-type GC (Fig. 7D and E). Recently, a series of genes
have been implicated in gastric carcinogenesis in relation to H.
pylori infection (38–43). However, SDC2 was not included
in the list. SDC2 is known as one of a four-member family of
transmembrane heparin sulfate proteoglycans and plays important
roles in many human types of cancers such as colorectal, hepatic
and pancreatic cancers (44,45).
The function of SDC2 in the gastric carcinogenesis of GC is
still unclear. Interestingly, a recent study raised the possibility
that upregulation of SDC2 may play an important role in
peritoneal metastasis of human GC cells (46). Taken together, these results
emphasize the importance of molecular assessment of premalignant or
even non-neoplastic mucosa of stomach in the presence of H.
pylori infection.
Even in the non-tumor tissues, the methylation
status of certain genes was affected by various factors such as
age, location and the status of H. pylori infection similar
to the results of previous studies (16). Similar to CLIP4 in the
present study, other genes have also been documented to show a
general progressive increase in methylation in normal gastric
tissues as the age of the patient increases (16). Six genes showed significantly
differential methylation by PA between the anatomic locations
(Fig. 7B). Considering that the
component of gastric mucosa varies with location, a differential
methylation status of genes according to location in normal tissues
may be justifiable. Similar results in previous studies have
demonstrated differences in the methylation status of genes in
non-neoplastic tissues of different organs or different sites in an
organ (15).
XKR6 showed significant hypermethylation in
the CNM of the distal stomach (p=0.0009) as well as in overall
tumor tissues (p=0.008). This suggests that certain methylation
changes may be involved in making specific locations of the stomach
susceptible for carcinogenesis. Recent studies have reported that
the methylation statuses of certain genes such as CDKN2A,
COX-2 and HMLH1 are significantly different according
to tumor location (38,39). Little is known concerning
XKR6 in human disease except that it is a member of the Kell
blood group complex subunit-related family and its SNP (rs6985109)
may be a potential risk factor in systemic lupus erythematosus
(47).
The evaluation of the methylation status of certain
genes may be helpful to distinguish between the histologic subtype
of EGC. The methylation statuses of DVL2, ETS1,
C19orf35, CNRIP, GAL3ST2 and ITGA3 were
significantly different according to the histologic subtype of the
EGC. We found that the methylation patterns of GAL3ST2 and
ITGA3 were specifically different in mixed-type compared to
the intestinal-type or diffuse-type subgroups, suggesting that the
mixed-type is a distinct histologic subtype. The methylation of
various genes that are associated with non-tumor factors such as
age, location and H. pylori infection should be
considered.
Acknowledgements
This study was supported by a Korea Science and
Engineering Foundation (KOSEF) grant funded by the Korean
government (MEST) (grant code: 2012-8-5113).
References
|
1
|
Dicken BJ, Bigam DL, Cass C, Mackey JR,
Joy AA and Hamilton SM: Gastric adenocarcinoma: review and
considerations for future directions. Ann Surg. 241:27–39.
2005.PubMed/NCBI
|
|
2
|
Sapari NS, Loh M, Vaithilingam A and Soong
R: Clinical potential of DNA methylation in gastric cancer: a
meta-analysis. PLoS One. 7:e362752012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shin A, Kim J and Park S: Gastric cancer
epidemiology in Korea. J Gastric Cancer. 11:135–140. 2011.
View Article : Google Scholar
|
|
4
|
Inoue M and Tsugane S: Epidemiology of
gastric cancer in Japan. Postgrad Med J. 81:419–424. 2005.
View Article : Google Scholar
|
|
5
|
Yasui W, Oue N, Aung PP, Matsumura S,
Shutoh M and Nakayama H: Molecular-pathological prognostic factors
of gastric cancer: a review. Gastric Cancer. 8:86–94. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lauren P: The two histological main types
of gastric carcinoma: diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965.PubMed/NCBI
|
|
7
|
Tan IB, Ivanova T, Lim KH, et al:
Intrinsic subtypes of gastric cancer, based on gene expression
pattern, predict survival and respond differently to chemotherapy.
Gastroenterology. 141:476–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng HC, Li XH, Hara T, et al: Mixed-type
gastric carcinomas exhibit more aggressive features and indicate
the histogenesis of carcinomas. Virchows Arch. 452:525–534. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ackerman LV and Rosai J: Rosai and
Ackerman’s Surgical Pathology. 9th edition. Mosby; St. Louis, MO:
2004
|
|
10
|
Bosman FT; World Health Organization
International Agency for Research on Cancer. WHO Classification of
Tumors of the Digestive System. International Agency for Research
on Cancer. 4th edition. IARC Press; Lyon: 2010
|
|
11
|
Oue N, Mitani Y, Motoshita J, et al:
Accumulation of DNA methylation is associated with tumor stage in
gastric cancer. Cancer. 106:1250–1259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamamoto E, Suzuki H, Takamaru H, Yamamoto
H, Toyota M and Shinomura Y: Role of DNA methylation in the
development of diffuse-type gastric cancer. Digestion. 83:241–249.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choi J, Cho MY, Jung SY, Jan KM and Kim
HS: CpG island methylation according to the histologic patterns of
early gastric adenocarcinoma. Korean J Pathol. 45:469–476. 2011.
View Article : Google Scholar
|
|
14
|
Dedeurwaerder S, Defrance M, Calonne E,
Denis H, Sotiriou C and Fuks F: Evaluation of the Infinium
Methylation 450K technology. Epigenomics. 3:771–784. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hong SJ, Kang MI, Oh JH, et al: DNA
methylation and expression patterns of key tissue-specific genes in
adult stem cells and stomach tissues. J Korean Med Sci. 24:918–929.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao C and Bu X: Promoter methylation of
tumor-related genes in gastric carcinogenesis. Histol Histopathol.
27:1271–1282. 2012.PubMed/NCBI
|
|
17
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.
|
|
18
|
Dittmer J: The biology of the Ets1
proto-oncogene. Mol Cancer. 2:292003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsutsumi S, Kuwano H, Nagashima N, Shimura
T, Mochiki E and Asao T: Ets-1 expression in gastric cancer.
Hepatogastroenterology. 52:654–656. 2005.PubMed/NCBI
|
|
20
|
Yu Y, Zhang YC, Zhang WZ, et al: Ets1 as a
marker of malignant potential in gastric carcinoma. World J
Gastroenterol. 9:2154–2159. 2003.PubMed/NCBI
|
|
21
|
Lind GE, Danielsen SA, Ahlquist T, et al:
Identification of an epigenetic biomarker panel with high
sensitivity and specificity for colorectal cancer and adenomas. Mol
Cancer. 10:852011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oster B, Thorsen K, Lamy P, et al:
Identification and validation of highly frequent CpG island
hypermethylation in colorectal adenomas and carcinomas. Int J
Cancer. 129:2855–2866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrasekaran EV, Lakhaman SS, Chawda R,
Piskorz CF, Neelamegham S and Matta KL: Identification of
physiologically relevant substrates for cloned Gal:
3-O-sulfotransferases (Gal3STs): distinct high affinity of
Gal3ST-2 and LS180 sulfotransferase for the globo H backbone,
Gal3ST-3 for N-glycan multiterminal Galβ1, 4GlcNAcβ units
and 6-sulfoGalβ1, 4GlcNAcβ, and Gal3ST-4 for the mucin core-2
trisaccharide. J Biol Chem. 279:10032–10041. 2004.PubMed/NCBI
|
|
24
|
Seko A, Nagata K, Yonezawa S and Yamashita
K: Down-regulation of Gal 3-O-sulfotransferase-2 (Gal3ST-2)
expression in human colonic non-mucinous adenocarcinoma. Jpn J
Cancer Res. 93:507–515. 2002.
|
|
25
|
Shi BZ, Hu P, Geng F, He PJ and Wu XZ:
Gal3ST-2 involved in tumor metastasis process by regulation of
adhesion ability to selectins and expression of integrins. Biochem
Biophys Res Commun. 332:934–940. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheong TC, Shin JY and Chun KH: Silencing
of galectin-3 changes the gene expression and augments the
sensitivity of gastric cancer cells to chemotherapeutic agents.
Cancer Sci. 101:94–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim SJ, Choi IJ, Cheong TC, et al:
Galectin-3 increases gastric cancer cell motility by up-regulating
fascin-1 expression. Gastroenterology. 138:1035–1045. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim SJ, Shin JY, Lee KD, et al: Galectin-3
facilitates cell motility in gastric cancer by up-regulating
protease-activated receptor-1 (PAR-1) and matrix
metalloproteinase-1 (MMP-1). PLoS One. 6:e251032011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kwak JE, Kim HS, Joo M, Chang SH, Shim SH
and Lee HR: The expression of galectin-3 and galectin-7 in
epithelial dysplasia and adenocarcinoma of the stomach. Korean J
Pathol. 42:365–372. 2008.
|
|
30
|
Saito Y, Sekine W, Sano R, et al:
Potentiation of cell invasion and matrix metalloproteinase
production by α3β1 integrin-mediated adhesion of gastric carcinoma
cells to laminin-5. Clin Exp Metastasis. 27:197–205. 2010.
|
|
31
|
Takatsuki H, Komatsu S, Sano R, Takada Y
and Tsuji T: Adhesion of gastric carcinoma cells to peritoneum
mediated by α3β1 integrin (VLA-3). Cancer Res. 64:6065–6070.
2004.
|
|
32
|
Ura H, Denno R, Hirata K, Yamaguchi K and
Yasoshima T: Separate functions of α2β1 and α3β1 integrins in the
metastatic process of human gastric carcinoma. Surg Today.
28:1001–1006. 1998.
|
|
33
|
Adachi M, Taki T, Huang C, et al: Reduced
integrin alpha3 expression as a factor of poor prognosis of
patients with adenocarcinoma of the lung. J Clin Oncol.
16:1060–1067. 1998.PubMed/NCBI
|
|
34
|
Hashida H, Takabayashi A, Tokuhara T, et
al: Integrin α3 expression as a prognostic factor in colon cancer:
association with MRP-1/CD9 and KAI1/CD82. Int J Cancer. 97:518–525.
2002.
|
|
35
|
Kurokawa A, Nagata M, Kitamura N, et al:
Diagnostic value of integrin α3, β4, and β5 gene expression levels
for the clinical outcome of tongue squamous cell carcinoma. Cancer.
112:1272–1281. 2008.
|
|
36
|
Zhu GH, Huang C, Qiu ZJ, et al: Expression
and prognostic significance of CD151, c-Met, and integrin
alpha3/alpha6 in pancreatic ductal adenocarcinoma. Dig Dis Sci.
56:1090–1098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park SY, Kook MC, Kim YW, Cho NY, Kim TY
and Kang GH: Mixed-type gastric cancer and its association with
high-frequency CpG island hypermethylation. Virchows Arch.
456:625–633. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alves MK, Ferrasi AC, Lima VP, Ferreira
MV, de Moura Campos Pardini MI and Rabenhorst SH: Inactivation of
COX-2, HMLH1 and CDKN2A genes by promoter
methylation in gastric cancer: relationship with histological
subtype, tumor location and Helicobacter pylori genotype.
Pathobiology. 78:266–276. 2011.
|
|
39
|
Alves MK, Lima VP, Ferrasi AC, Rodrigues
MA, De Moura Campos Pardini MI and Rabenhorst SH: CDKN2A
promoter methylation is related to the tumor location and
histological subtype and associated with Helicobacter pylori
flaA(+) strains in gastric adenocarcinomas. APMIS. 118:297–307.
2010. View Article : Google Scholar
|
|
40
|
Dong CX, Deng DJ, Pan KF, et al: Promoter
methylation of p16 associated with Helicobacter
pylori infection in precancerous gastric lesions: a
population-based study. Int J Cancer. 124:434–439. 2009.
|
|
41
|
Kang GH, Lee S, Cho NY, et al: DNA
methylation profiles of gastric carcinoma characterized by
quantitative DNA methylation analysis. Lab Invest. 88:161–170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maekita T, Nakazawa K, Mihara M, et al:
High levels of aberrant DNA methylation in Helicobacter
pylori-infected gastric mucosae and its possible association
with gastric cancer risk. Clin Cancer Res. 12:989–995.
2006.PubMed/NCBI
|
|
43
|
Peterson AJ, Menheniott TR, O’Connor L, et
al: Helicobacter pylori infection promotes methylation and
silencing of trefoil factor 2, leading to gastric tumor
development in mice and humans. Gastroenterology. 139:2005–2017.
2010. View Article : Google Scholar
|
|
44
|
Essner JJ, Chen E and Ekker SC:
Syndecan-2. Int J Biochem Cell Biol. 38:152–156. 2006. View Article : Google Scholar
|
|
45
|
Han I, Park H and Oh ES: New insights into
syndecan-2 expression and tumourigenic activity in colon carcinoma
cells. J Mol Histol. 35:319–326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bai F, Guo X, Yang L, et al: Establishment
and characterization of a high metastatic potential in the
peritoneum for human gastric cancer by orthotopic tumor cell
implantation. Dig Dis Sci. 52:1571–1578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Budarf ML, Goyette P, Boucher G, et al: A
targeted association study in systemic lupus erythematosus
identifies multiple susceptibility alleles. Genes Immun. 12:51–58.
2011. View Article : Google Scholar : PubMed/NCBI
|