Introduction
Gastric cancer remains one of the most aggressive
types of cancer in the world. The highest incidence is in eastern
Asia, in China, Japan and Korea and South America and Eastern
Europe (1–2). When presented, most cases are in the
advanced stage. Despite the advancements in various treatment
modalities, prognosis remains poor (3). Enhancing our understanding of the
carcinogenetic process (4) and
exploring new treatment options are important to improve the
outlook of gastric cancer patients. Gastric carcinogenesis is a
multistep process involving genetic and epigenetic alterations of
protein-coding proto-oncogenes (5)
and tumor-suppressor genes (6–7). In
addition, recent studies have demonstrated the involvement of
microRNAs (8) and emerging evidence
underscores the importance of signaling pathways such as NF-κB,
Wnt/β-catenin signaling and Notch signaling involved in the gastric
cancer development (9).
Human ribonucleotide reductase (RR) is an
enzyme that catalyzes the conversion of ribonucleotide
5′-diphosphates into 2′-deoxyribonucleotides which is essential for
DNA synthesis and cell proliferation (10,11).
RR consists of two subunits, RRM1 and RRM2. These two subunits are
encoded by two distinctive genes on different chromosomes, and
their mRNAs are differentially expressed throughout the cell cycle.
The cellular RRM1 protein level is stabilized throughout the entire
cell cycle, whereas RRM2 is only expressed during the late G1/early
S phase when DNA replication occurs (12). Therefore the catalytic activity of
RR is tightly controlled during the cell cycle by the level of
RRM2.
Given the critical role of RR in cell growth, it has
been considered as a potential therapeutic target for cancer
therapy. RRM2 contributes to malignant cellular phenotype in a
range of human cancers and its overexpression plays a critical role
in tumor invasion (13,14). A previous study showed that
sequence-specific small interfering RNA (siRNA)- mediated
inhibition of RRM2 effectively blocked the cell proliferation and
induced G1/S phase cell cycle arrest in melanoma cell lines
(15). However, the biological
functions of RRM2 in gastric cancer remain unclear. The present
study investigated the tumorigenic role of RRM2 in gastric
cancer.
Materials and methods
Cell lines and clinical gastric
adenocarcinoma samples
The ten gastric cancer cell lines (MKN28, KATO-III,
MKN45, SNU16, SNU1, MKN7, MKN1, NCI-N87, AGS and MGC-803) (16), were cultured in RPMI-1640 (Gibco)
supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml
penicillin and 10 μg/ml streptomycin in a humidified atmosphere of
5% CO2 at 37°C.
Archival tissue blocks from 270 patients with
gastric adenocarcinoma were retrieved from the Department of
Anatomical and Cellular Pathology, Prince of Wales Hospital (PWH),
Hong Kong, and arranged in tissue array blocks. Hematoxylin and
eosin-stained sections were used to define tumor areas, and 3
representative 1-mm cores were obtained from each case and inserted
to a recipient paraffin block using a tissue arrayer (Beecher
Instruments, Silver Spring, MD, USA). Frozen tissues from another
ten pairs of primary gastric cancer and non-tumorous gastric mucosa
were collected from patients who underwent gastrectomy prior to any
therapeutic intervention. In addition, 27 paired gastric cancer and
non-tumorous gastric frozen tissue cDNAs were kindly provided by
the Institute of Digestive Disease (IDD) of Prince Wales Hospital
for RRM2 qRT-PCR analysis.
RNA extraction and qRT-PCR
Total RNA was extracted using RNeasy Mini Kit
according to the manufacturer’s instructions (Qiagen) and the
concentration was measured by NanoDrop 1000 (Thermo Fisher
Scientific). High-Capacity cDNA Reverse Transcription Kits (Applied
Biosystems) were applied to achieve cDNAs. For qRT-PCR, SYBR-Green
PCR Master Mix (Applied Biosystems) was applied for RRM2 expression
(sense, GAAGGCAGAGGCTTCCTTTT and antisense, TACTATGCCATCGCTTGCTG).
Commercially available normal gastric mRNA was obtained from Ambion
(AM 7996). The relative expression level was normalized by
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated
using the 2−ΔΔCt method. Quantitative PCR was performed
in triplicates.
Immunohistochemical staining
Immunohistochemical staining of RRM2 was performed
in 4 μm sections. After de-waxing in xylene, rehydrating and
rinsing in distilled water, sections were incubated at 3%
H2O2 solution for 25 min to block endogenous
peroxidase activities. Antigen retrieval was carried out by using a
pressure cooker with 10 nM citrate buffer (pH 6.0) for a total of
30 min (high power 10 min followed by low power 20 min). The
primary antibody RRM2 (Santa Cruz, 10846; 1:50 dilution in primary
dilution buffer) was incubated at 4°C overnight and chromogen
development was performed using the standard avidin-biotin method
(Dako). The cytoplasmic expression of RRM2 was assessed by
combining the proportional score and the intensity score as
previously described (16). The
proportion score was calculated according to the proportion of
tumor cells with positive cytoplasmic staining (0, none; 1, ≤10%;
2, ≤25%; 3, >25%; 4, >50%). The intensity score was assigned
for the average intensity of positive tumor cells (0, none; 1,
weak; 2, intermediate; 3, strong). The cytoplasmic score of RRM2
was the product of the proportion and intensity scores, ranging
from 0 to 12. The cytoplasmic expression was categorized into
negative (score 0), 1+ (score 1–3), 2+ (score 4–6) and 3+ (score
7–12).
Western blot analysis
Western blot analysis was performed as previously
described (17). RRM2 protein was
detected with a polyclonal anti-RRM2 antibody (Santa Cruz, 10846,
1:500). Other antibodies were from Cell Signaling, cleaved-caspase
3 (Asp175) (no. 9541, 1:1,000), cleaved PARP (Asp214) (no. 9541,
1:1000), p-P44/42(T202/Y204) (no. 9154, 1:1,000),
p-P38MAPK(T180/Y182) (no. 4511, 1:1,000), phospho-AKT(S473) (no.
9271, 1:1,000), phospho-Stat3 (T705) (no. 9145, 1:2,000),
anti-Mouse IgG-HRP (Dako, 00049039, 1:30,000) and anti-Rabbit
IgG-HRP (Dako, 00028856, 1:40,000).
In vitro functional assays
Transfection was performed using Lipofectamine 2000
transfection reagent (Invitrogen) and the anti-RRM2 siRNA (verified
siRNA, SI02653441; Qiagen) at the concentration of 25 nM. Mock
transfection (Lipofectamine 2000 only) and scramble siRNA were
included as controls. Cell proliferation was assessed using
CellTiter 96® Non-Radioactive Cell Proliferation Assay
(MTT; Promega, Madison, WI, USA) according to the manufacturer’s
instructions. Monolayer colony formation assay was performed as
described in our previous report (17). Ten days after transfection of siRRM2
or scramble siRNA, the colonies were fixed and stained with 2%
crystal violet. Colonies with >50 cells were counted. The
experiments were performed in triplicate wells and repeated 3
times. Cell invasion assay was performed in a 24-well invasion
chamber (BD Biocoat Matrigel Invasion Chamber, BD Biosciences).
siRRM2 or scramble siRNA transfectants were seeded at
5×104 cells/well and allowed to invade for 24 h through
BD Biocoat Matrigel Invasion Chambers. Cells that invaded through
the Matrigel membrane were counted from 3 microscopic fields
(original magnification, ×400).
Flow cytometry analysis
For cell cycle analysis, the cells were harvested 24
h after transfection, stained with propidium iodide (PI), and
sorted by FACS Calibur Flow Cytometer (Becton, Dickinson, San
Diego, CA, USA). The cell cycle profiles were determined using the
ModFitLT software (Becton-Dickinson). For early apoptosis analysis,
Annexin V-FITC Apoptosis Detection Kit (BioVision, Milpitas, CA,
USA) and 7-Amino-actinomycin (7-AAD, KeyGEN BioTECH) were used
according to the manufacturer’s instructions.
In vivo animal study
MGC-803 cells transfected with si-scramble or siRRM2
were injected subcutaneously into the dorsal flank of five
4-week-old male Balb/c nude mice (si-scramble on the left and
siRRM2 on the right). When the tumors were palpable after 8 days,
the synthetic siRNA complex (25 nM) with siPORT amine transfection
reagent (Ambion) in 50 μl PBS was delivered intratumorally at 3-day
intervals. The mice were sacrificed after 3 weeks and the
xenografts were collected.
Statistical analysis
In the functional assays such as MTT proliferation,
monolayer colony formation, cell invasion and in vivo tumor
growth, Student’s t-test was used to compare the phenotype
difference of siRRM2 knockdown cells and si-scramble transfected
cells. RRM2 mRNA expression in gastric cancer and paired
non-cancerous tissues were compared by the Mann-Whitney U test.
Correlations between RRM2 expression with other clinicopathological
parameters were evaluated by non-parametric Spearman’s rho rank
test. The Kaplan-Meier method was used to estimate the survival
rates according to the score of RRM2 cytoplasmic staining. For
those statistically significant variables found in the univariate
survival analysis (P<0.05), the multivariate survival was
achieved by the Cox regression analysis. All statistical analysis
was performed by SPSS software (version 16.0; SPSS Inc). A
two-tailed P-value <0.05 was considered to indicate
statistically significant differences.
Results
Upregulation of RRM2 in gastric cell
lines and primary gastric tumors
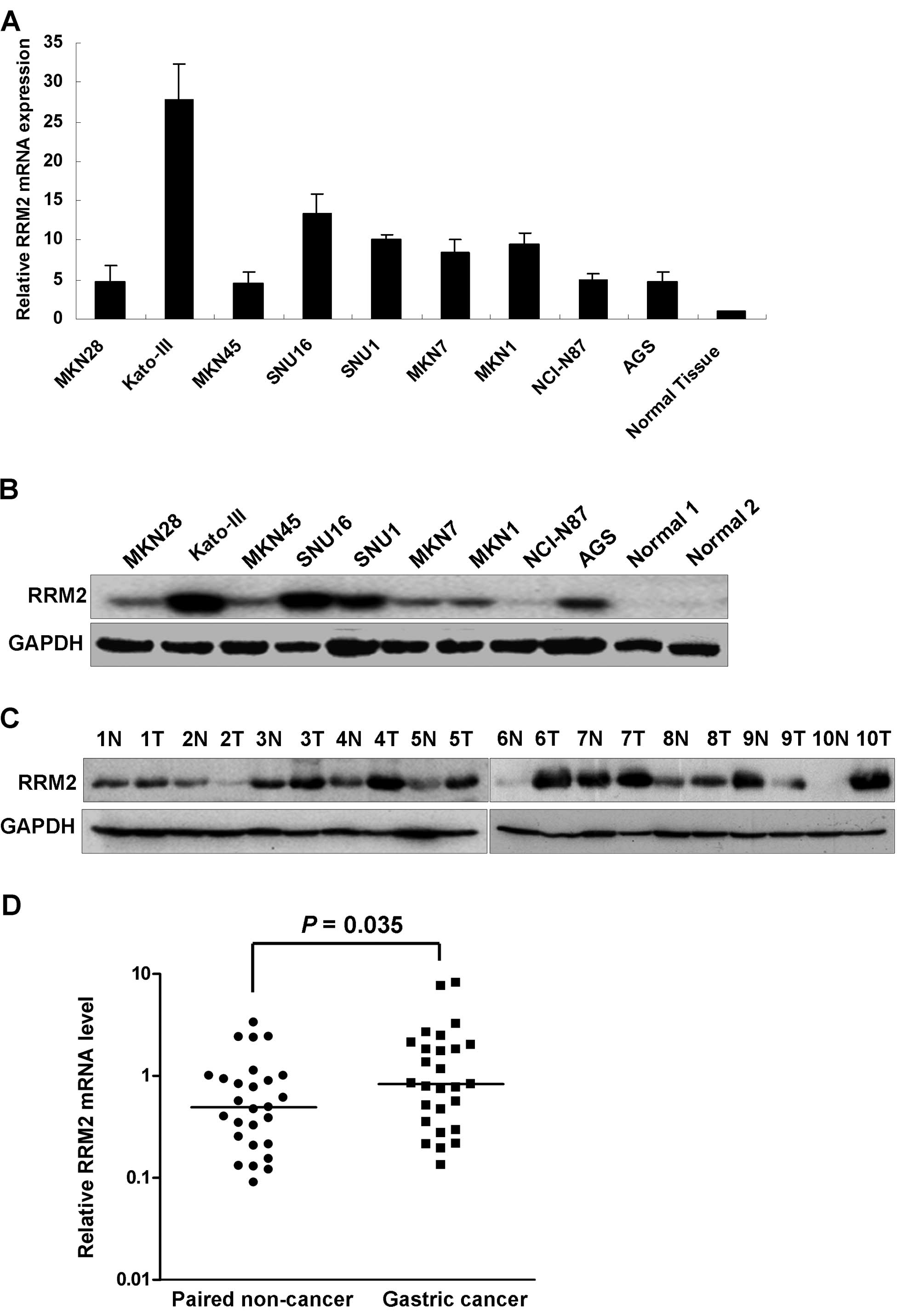
The RRM2 mRNA expression levels were higher in all
nine gastric cancer cell lines examined compared to the normal
gastric tissue as shown in Fig. 1A.
The upregulation of RRM2 in the gastric cancer cells was further
confirmed by western blot analysis. High RRM2 protein expression
was observed in all gastric cancer cell lines compared with the 2
normal gastric mucosal samples from patients who underwent weight
reduction gastric surgery (Fig.
1B). In the panel of gastric cancer cell lines, RRM2 was
strongly expressed in Kato-III, SNU16, SNU1 and AGS cell lines.
When comparing tumor with the corresponding non-tumorous mucosa,
upregulation of RRM2 protein was observed in 8/10 cases by western
blot analysis (Fig. 1C). We further
evaluated the RRM2 mRNA expression in a cohort of 27 pairs of
gastric cancer and corresponding non-tumorous mucosa. Expression of
RRM2 in tumor cells was significantly higher than in non-tumorous
mucosal tissues (P=0.035, Fig. 1D).
Immunohistochemical analysis of another 5 pairs of gastric cancer
samples showed positive staining of RRM2 in all tumor tissues but
not in the non-tumorous gastric glandular epithelium (data not
shown).
RRM2 overexpression correlates with poor
prognosis in gastric cancer patients
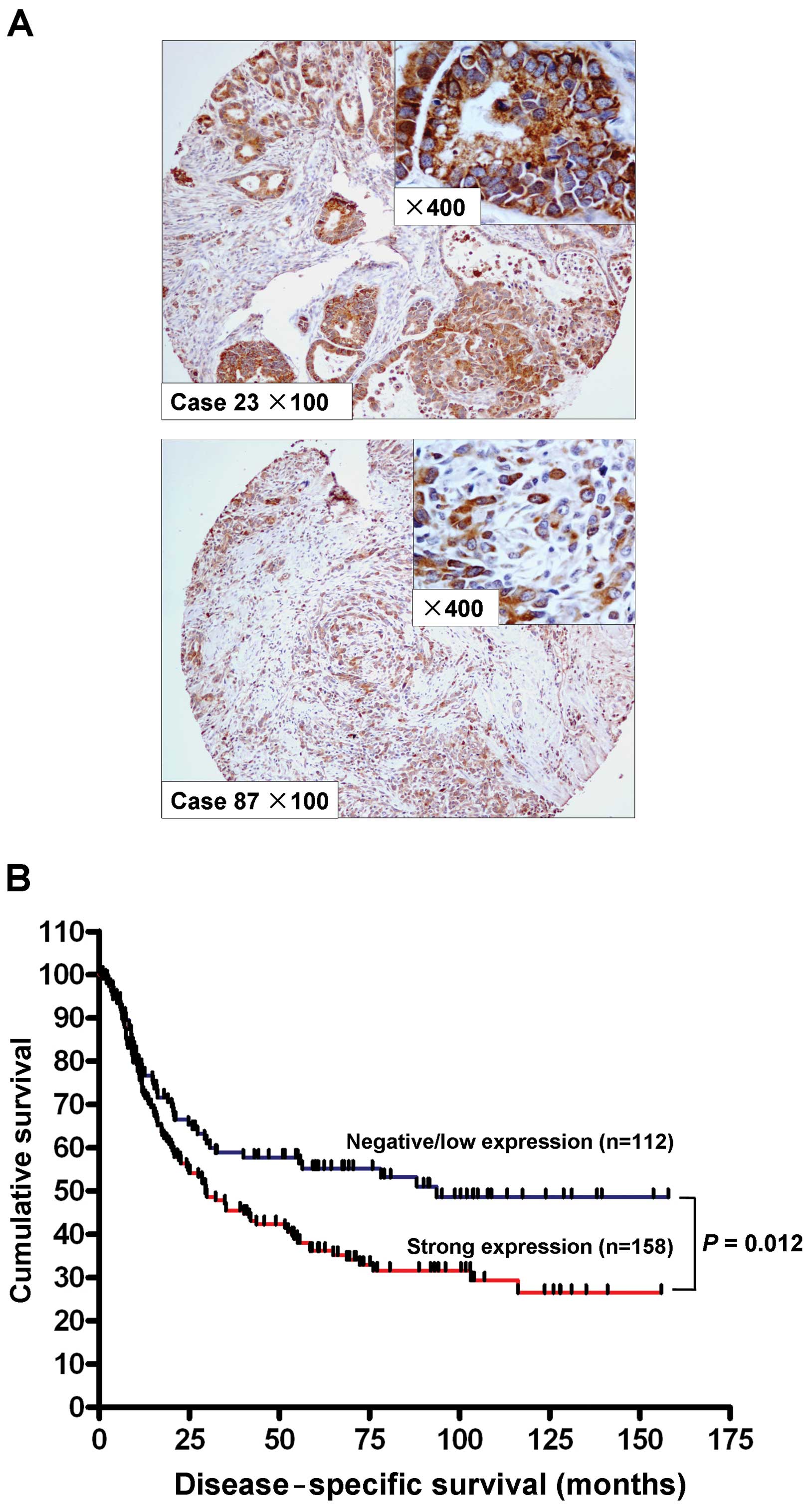
We studied the expression of RRM2 by
immunohistochemistry in a cohort of 270 primary gastric
adenocarcinoma samples. The cytoplasmic expression of RRM2 was
assessed by assigning a proportional score and an intensity score
as described in Materials and methods. No RRM2 expression (score 0)
was observed in 14 cases (5.2%), weak cytoplasmic expression (score
1+) was observed in 98 cases (36.3%), and 116 cases (42.9%) and 42
cases (15.6%) demonstrated moderate (score 2+) and strong (score
3+) cytoplasmic staining, respectively. A total of 112 cases
(41.5%) were considered negative/low expression of RRM2 (score 0
and 1+), whereas 158 cases (58.5%) with scores 2+ and 3+ were
considered strongly positive for RRM2. Expression of RRM2 was
primarily localized in the cytoplasm of the tumor cells (Fig. 2A). The association of RRM2
expression with clinicopathological parameters is shown in Table I. Strong RRM2 expression correlated
with higher tumor grade (P=0.010) and advanced T stage (P=0.025).
Univariate analysis indicated that expression of RRM2 in gastric
adenocarcinoma associated with poorer disease-specific survival
(P=0.012, Fig. 2B). Old age (age,
>60 years, P=0.043), diffuse type histology (P<0.001), higher
tumor grade (P=0.035) and advanced stage (P<0.001) were also
correlated with poor disease-specific survival by Univariate
analysis (Table II). Using
multivariate Cox proportional hazards regression analysis, only age
(P<0.001) and stage (P<0.001) were independently associated
with disease-specific survival (Table
II).
 | Table ICorrelation between RRM2 expression
and other clinicopathological parameters. |
Table I
Correlation between RRM2 expression
and other clinicopathological parameters.
| RRM2 expression
(n=270) | |
|---|
|
| |
|---|
| Characteristics | Negative and low | Strong | P-value |
|---|
| Gender |
| Male | 75 | 106 | 0.983 |
| Female | 37 | 52 | |
| Age, years |
| ≤60 | 48 | 53 | 0.127 |
| >60 | 64 | 105 | |
| Type |
| Intestinal | 63 | 84 | 0.710 |
| Diffuse | 49 | 73 | |
| Grade |
| 1 | 8 | 1 | 0.010 |
| 2 | 43 | 57 | |
| 3 | 61 | 99 | |
| Stage |
| I | 29 | 29 | 0.241 |
| II | 16 | 16 | |
| III | 30 | 55 | |
| IV | 37 | 57 | |
| Stage (T) |
| 1 | 20 | 16 | 0.025 |
| 2 | 31 | 42 | |
| 3 | 52 | 95 | |
| 4 | 9 | 4 | |
| Stage (N) |
| 0 | 32 | 27 | 0.070 |
| 1 | 31 | 39 | |
| 2 | 30 | 50 | |
| 3 | 19 | 41 | |
| Stage (M) |
| 0 | 93 | 134 | 0.613 |
| 1 | 19 | 23 | |
| H.
pylori |
| Absence | 45 | 75 | 0.254 |
| Presence | 62 | 75 | |
 | Table IIUnivariate and multivariate Cox
regression analysis of clinicopathological factors in patients with
gastric adenocarcinoma. |
Table II
Univariate and multivariate Cox
regression analysis of clinicopathological factors in patients with
gastric adenocarcinoma.
|
Characteristics | Univariate analysis
RR (95% CI) | P-value | Multivariate
analysis |
|---|
| Gender |
| Male | 0.832
(0.593–1.167) | 0.286 | |
| Female | 1.000 | | |
| Age, years |
| ≤60 | 0.700
(0.496–0.988) | 0.043 |
<0.001 |
| >60 | 1.000 | | |
| Type |
| Intestinal | 0.543
(0.390–0.755) |
<0.001 | 0.958 |
| Diffuse | 1.000 | | |
| Grade |
| 1 | 0.221
(0.054–0.897) | 0.035 | 0.740 |
| 2 | 0.720
(0.508–1.021) | 0.065 | |
| 3 | 1.000 | | |
| Stage |
| I | 0.098
(0.054–0.178) |
<0.001 |
<0.001 |
| II | 0.127
(0.061–0.264) |
<0.001 | |
| III | 0.385
(0.265–0.560) |
<0.001 | |
| IV | 1.000 | | |
| H.
pylori |
| Absence | 1.244
(0.884–1.750) | 0.211 | |
| Presence | 1.000 | | |
| RRM2 |
| Negative/low | 0.637
(0.448–0.905) | 0.012 | 0.402 |
| Strong | 1.000 | | |
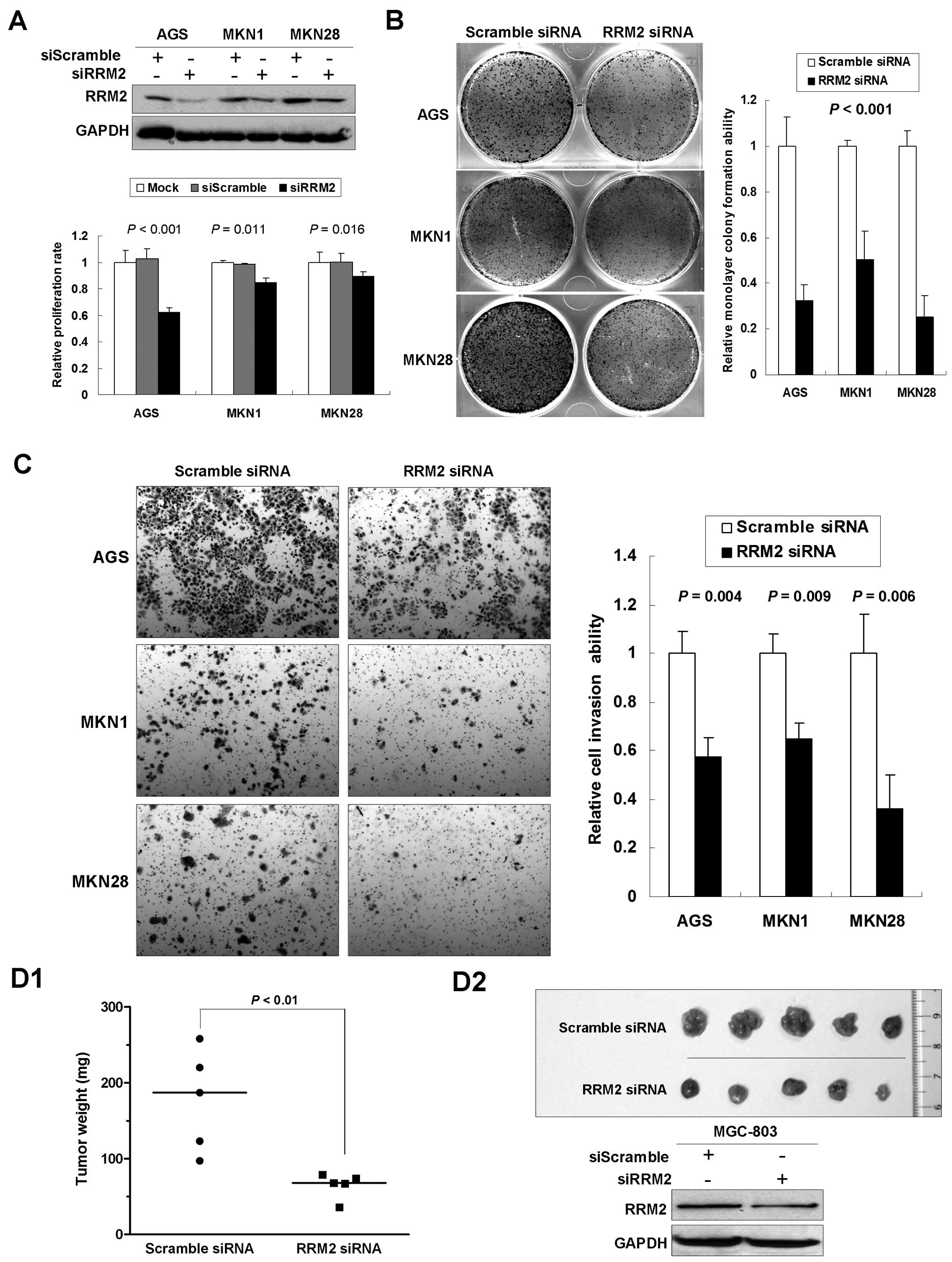
RRM2 knockdown reduces proliferation and
invasiveness in gastric cancer cells
Upregulation of RRM2 in gastric cancer suggested a
potential tumorigenic role of RRM2 in cancer development. To
elucidate the functional role of RRM2 in gastric tumorigenesis, we
first investigated the effect of RRM2 knockdown by siRNA in
vitro. To avoid cell-context dependent effects, the experiments
were performed in multiple gastric cancer cell lines. The 5-day MTT
assay indicated that knocking down RRM2 significantly reduced cell
proliferation (AGS, 62.2%, P<0.001; MKN1, 84.4%, P=0.011; MKN28,
89.1%, P=0.016; Fig. 3A) compared
with the mock and the scramble siRNA control groups. In
anchorage-dependent monolayer colony formation assays, we observed
a significant reduction of colony number in cell lines transfected
with anti-RRM2 siRNA compared with the scramble siRNA control group
(reduced to 32.4, 50.2 and 25.1% in AGS, MKN1 and MKN28,
respectively, P<0.001; Fig. 3B).
Matrigel invasion assays were carried out to monitor the invasion
potential of gastric cancer cells transfected with siRRM2. By using
BD Biocoat chamber, we found that the number of invaded cells was
significantly lower in siRRM2 transfectants than in the control
groups (reduced to 57.3% in AGS, P=0.004; 64.9% in MKN1, P=0.009;
and 36.2% in MKN28, P=0.006; Fig.
3C).
To further investigate the effects of anti-RRM2
siRNA on tumorigenicity of gastric cancer, in vivo tumor
formation assay was carried out. A gastric cancer cell line
MGC-803, which is able to form xenograft tumor in nude mice, was
transfected with siRRM2. Three weeks later, mice were sacrificed
and the transplanted tumors were removed and weighted. A
significant decrease in xenograft weight was observed upon RRM2
knockdown (P<0.01; Fig. 3D1).
Successful knockdown of RRM2 was confirmed by western blot analysis
in xenograft tumors (Fig. 3D2). The
findings support that RRM2 plays an important role in promoting
malignant growth of gastric cancer cells in vivo.
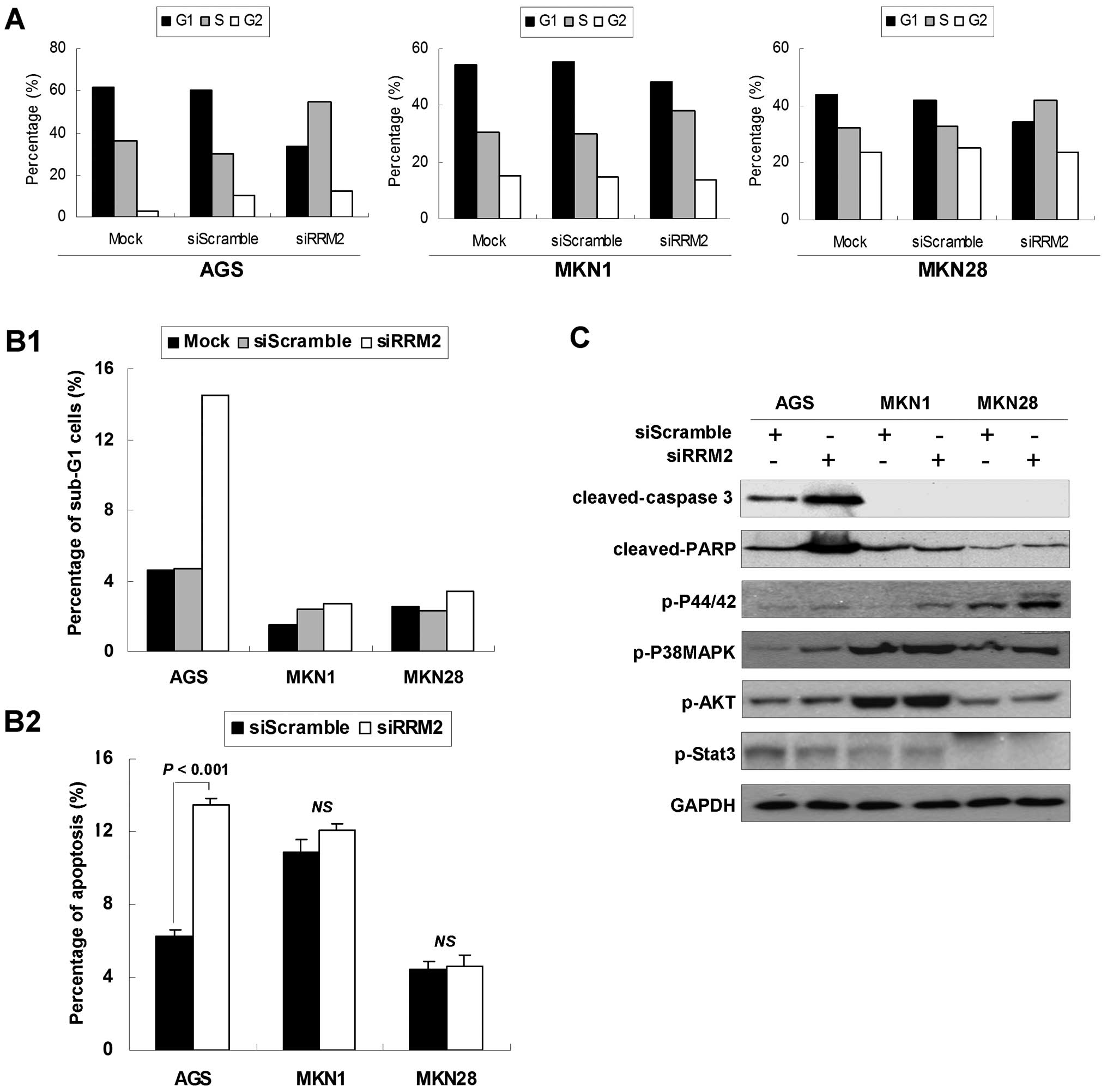
RRM2 knockdown changes the cell cycle
distribution in gastric cancer cells
To investigate the mechanisms underlying the growth
suppressive role of anti-RRM2 siRNA, the effects of anti-RRM2 siRNA
on cell cycle distribution were analyzed. Twenty-four hours after
transfection, accumulation of S-phase cells was observed in the
anti-RRM2-transfected group. As shown in Fig. 4A, the anti-RRM2-transfected cells
showed increased percentages of S-phase cells (AGS, from 35.9 to
54.6%; MKN1, from 30.4 to 38.1%; MKN28, from 32.4 to 42.1%) and
decreased percentages of G1 phase cells (AGS, from 61.6 to 33.4%;
MKN1, from 54.2 to 48.4%; MKN28, from 44.0 to 34.4%) compared with
the si-scramble control group cells.
Furthermore, the RRM2 knockdown was shown to induce
both early and late apoptosis in AGS cells. The sub-G1 AGS cell
population increased to 14.5% in siRRM2 transfectants compared with
si-scramble transfectants (4.7%; Fig.
4B1). This was further validated by Annexin V-FITC apoptosis
analysis, as shown in Fig. 4B2.
There was an increase in early apoptotic cell population in AGS
cells after anti-RRM2 siRNA transfection (from 6.3 to 13.5%,
P<0.01). Late apoptosis, represented by a significant increase
of cleaved-caspase 3 and cleaved-PARP (Fig. 4C), was also observed in AGS after
anti-RRM2 siRNA transfection compared with the control groups.
In the meantime, the expression levels of several
phosphorylated proteins which were involved in cell cycle arrest,
cell growth or cell apoptosis were determined. As shown in Fig. 4C, p-P44/42(T202/Y204) and
p-P38MAPK(T180/Y182) were found to be phosphorylated to high level
upon anti-RRM2 siRNA transfection compared with the si-scramble
control, whereas p-AKT (S473) and p-Stat3 (T705) phosphorylation
status showed no change between these two groups.
Discussion
In the present study, we found both mRNA and protein
levels of RRM2 were highly expressed in gastric cancer samples
compared with normal gastric mucosa. The expression of RRM2 in
primary gastric cancer correlated with advanced T stage and poor
prognosis. These results suggested that RRM2 might play a role in
gastric tumorigenesis. In keeping with this finding, a previous
analysis of RRM2 genomic sequence and promoter region, including
consensus TATA box, three CCAAT boxes, and several GC rich sites,
provided the hypothesis of its upregulation in favor of cancer
transformation and drug resistance (18). According to a previous study
(19), RRM2 overexpression that was
significantly associated with Epstein-Barr virus, expression of
survivin and DNA methyltransferase 1, predicted poor prognosis in
patients with gastric cancer. Collectively, these results provided
evidence that RRM2 served as a potential biomarker and therapeutic
target for gastric cancer. In addition, RRM2 has been proposed to
be a biomarker for a several types of human cancer. These previous
studies suggested that RRM2 may serve as a potential diagnostic
biomarker in breast cancer (20),
bladder cancer (21) and ovarian
cancer (10). RRM2 may also be a
potential prognostic marker in non-small cell lung cancer receiving
chemotherapeutic agents (22) and a
potential therapeutic target for hepatocellular carcinoma (HCC)
(23), bladder cancer (21) and pancreatic carcinoma (24,25).
The functional studies demonstrated for the first
time that RRM2 knockdown by siRNA suppressed in vitro and
in vivo growth of gastric cancer cells, and reduced cancer
cell invasiveness. These results were concordant with a previous
report (26) that anti-RRM2 siRNA exhibited
anti-proliferative activity in various types of cancer cells. In
addition to the apoptotic aspect, we found that anti-RRM2 siRNA
induced both early and late apoptosis in AGS cells but not in MKN1
and MKN28 cells. Hence, the apoptotic response to siRRM2 appeared
to be cell context-dependent. It is uncertain whether the different
TP53 status might contribute to the different apoptotic responses
as MKN1 and MKN28 cells harbor the TP53 mutation, whereas AGS cells
carry the wild type TP53 (17). It
has been reported that antisense RRM2 downregulated RRM2 expression
and sensitized a prostate cancer cell line PC3 to UV radiation
(27). In pancreatic
adenocarcinoma, synergism between RRM2 siRNA and gemcitabine
resulted in increased tumor apoptosis (28). RRM2 is a downstream target of the
ATM-p53 pathway that mediates radiation-induced DNA repair
(29) and silencing of RRM2 was
found to enhance DNA damage as measured by histone γ-H2AX. Under
DNA damage and Chk1 activation mediated by ATM and ATR,
upregulation of RRM2 expression level coupled with its nuclear
recruitment suggests an active role of RRM2 in the cellular process
in response to DNA damage (30).
RRM2 knockdown by siRNA-reduced gastric cancer cell
invasion ability was also observed in this study. RRM2
overexpression in pancreatic adenocarcinoma increases cellular
invasiveness and MMP-9 expression in an NF-κB-dependent manner,
whereas RNA interference-mediated silencing of RRM2 expression
attenuates cellular invasiveness (13). In KB cells, RRM2 plays a positive
role in angiogenesis and growth through regulation of the
expression of antiangiogenic thrombospondin-1 (TSP-1) and
proangiogenic vascular endothelial growth factor (VEGF) (14). These results suggested that RRM2 has
a significant role in driving tumor cell invasion and
metastasis.
In conclusion, we demonstrated the upregulation of
RRM2 in gastric adenocarcinoma and its overexpression was
correlated with poor prognosis in patients with gastric cancer,
suggesting that RRM2 plays a crucial role in gastric tumorigenesis
and could be used as a potential prognostic biomarker in gastric
cancer. Functional studies demonstrated that downregulation of RRM2
expression by RRM2-specific siRNA quenched its oncogenic properties
by inhibiting cell growth in vitro and in vivo,
decreasing cell invasiveness. These findings suggested that RRM2
might be involved in gastric tumorigenesis and might potentially
serve as a prognostic marker and a therapeutic target in gastric
cancer.
Acknowledgements
This study was financially supported by the National
Natural Science Foundation of China (81201591).
References
|
1
|
Kelley JR and Duggan JM: Gastric cancer
epidemiology and risk factors. J Clin Epidemiol. 56:1–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith MG, Hold GL, Tahara E and El-Omar
EM: Cellular and molecular aspects of gastric cancer. World J
Gastroenterol. 12:2979–2990. 2006.
|
|
3
|
Alberts SR, Cervantes A and van de Velde
CJ: Gastric cancer: epidemiology, pathology and treatment. Ann
Oncol. 14(Suppl 2): ii31–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fox JG and Wang TC: Inflammation, atrophy,
and gastric cancer. J Clin Invest. 117:60–69. 2007. View Article : Google Scholar
|
|
5
|
Li J, Ng EK, Ng YP, et al: Identification
of retinoic acid-regulated nuclear matrix-associated protein as a
novel regulator of gastric cancer. Br J Cancer. 101:691–698. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheung KF, Lam CN, Wu K, et al:
Characterization of the gene structure, functional significance,
and clinical application of RNF180, a novel gene in gastric cancer.
Cancer. 118:947–959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu J, Cheng YY, Tao Q, et al: Methylation
of protocadherin 10, a novel tumor suppressor, is associated with
poor prognosis in patients with gastric cancer. Gastroenterology.
136:640–651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu WK, Lee CW, Cho CH, et al: MicroRNA
dysregulation in gastric cancer: a new player enters the game.
Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu WK, Cho CH, Lee CW, et al:
Dysregulation of cellular signaling in gastric cancer. Cancer Lett.
295:144–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferrandina G, Mey V, Nannizzi S, et al:
Expression of nucleoside transporters, deoxycitidine kinase,
ribonucleotide reductase regulatory subunits, and gemcitabine
catabolic enzymes in primary ovarian cancer. Cancer Chemother
Pharmacol. 65:679–686. 2010. View Article : Google Scholar
|
|
11
|
Goan YG, Zhou B, Hu E, Mi S and Yen Y:
Overexpression of ribonucleotide reductase as a mechanism of
resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell
line. Cancer Res. 59:4204–4207. 1999.PubMed/NCBI
|
|
12
|
Engström Y, Eriksson S, Jildevik I, Skog
S, Thelander L and Tribukait B: Cell cycle-dependent expression of
mammalian ribonucleotide reductase. Differential regulation of the
two subunits. J Biol Chem. 260:9114–9116. 1985.PubMed/NCBI
|
|
13
|
Duxbury MS and Whang EE: RRM2 induces
NF-kappaB -dependent MMP-9 activation and enhances cellular
invasiveness. Biochem Biophys Res Commun. 354:190–196. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang K, Hu S, Wu J, et al: Overexpression
of RRM2 decreases thrombspondin-1 and increases VEGF production in
human cancer cells in vitro and in vivo: implication of RRM2 in
angiogenesis. Mol Cancer. 8:112009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zuckerman JE, Hsueh T, Koya RC, Davis ME
and Ribas A: siRNA knockdown of ribonucleotide reductase inhibits
melanoma cell line proliferation alone or synergistically with
temozolomide. J Invest Dermatol. 131:453–460. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang W, Tong JH, Chan AW, et al: Stathmin1
plays oncogenic role and is a target of microRNA-223 in gastric
cancer. PLoS One. 7:e339192012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang W, Tong JH, Chan AW, et al:
Yes-associated protein 1 exhibits oncogenic property in gastric
cancer and its nuclear accumulation associates with poor prognosis.
Clin Cancer Res. 7:2130–2139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou B and Yen Y: Characterization of the
human ribonucleotide reductase M2 subunit gene; genomic structure
and promoter analyses. Cytogenet Cell Genet. 95:52–59. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morikawa T, Hino R, Uozaki H, et al:
Expression of ribonucleotide reductase M2 subunit in gastric cancer
and effects of RRM2 inhibition in vitro. Hum Pathol. 41:1742–1748.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kretschxmer C, Sterner-Kock A, Siedentopf
F, Schoenegg W, Schlag PM and Kemmner W: Identification of early
molecular markers for breast cancer. Mol Cancer. 10:152011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morikawa T, Maeda D, Kume H, Homma Y and
Fukayama M: Ribonucleotide reductase M2 subunit is a novel
diagnostic marker and a potential therapeutic target in bladder
cancer. Histopathology. 57:885–892. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Souglakos J, Boukovinas I, Taron M, et al:
Ribonucleotide reductase subunits M1 and M2 mRNA expression levels
and clinical outcome of lung adenocarcinoma patients treated with
docetaxel/gemcitabine. Br J Cancer. 98:1710–1715. 2008. View Article : Google Scholar
|
|
23
|
Satow R, Shitashige M, Kanai Y, et al:
Combined functional genome survey of therapeutic targets for
hepatocellular carcinoma. Clin Cancer Res. 16:2518–2528. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Itoi T, Sofuni A, Fukushima N, et al:
Ribonucleotide reductase subunit M2 mRNA expression in pretreatment
biopsies obtained from unresectable pancreatic carcinomas. J
Gastroenterol. 42:389–394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duxbury MS, Ito H, Benoit E, Zinner MJ,
Ashley SW and Whang EE: Retrovirally mediated RNA interference
targeting the M2 subunit of ribonucleotide reductase: a novel
therapeutic strategy in pancreatic cancer. Surgery. 136:261–269.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heidel JD, Liu JY, Yen Y, et al: Potent
siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce
cell proliferation in vitro and in vivo. Clin Cancer Res.
13:2207–2215. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou B, Liu X, Mo X, et al: The human
ribonucleotide reductase subunit hRRM2 complements p53R2 in
response to UV-induced DNA repair in cells with mutant p53. Cancer
Res. 63:6583–6594. 2003.PubMed/NCBI
|
|
28
|
Duxbury MS, Ito H, Zinner MJ, Ashley SW
and Whang EE: RNA interference targeting the M2 subunit of
ribonucleotide reductase enhances pancreatic adenocarcinoma
chemosensitivity to gemcitabine. Oncogene. 23:1539–1548. 2004.
View Article : Google Scholar
|
|
29
|
Hu ZZ, Huang H, Cheema A, Jung M,
Dritschilo A and Wu CH: Integrated bioinformatics for
radiation-induced pathway analysis from proteomics and microarray
data. J Proteomics Bioinform. 1:47–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang YW, Jones TL, Martin SE, Caplen NJ
and Pommier Y: Implication of checkpoint kinase-dependent
up-regulation of ribonucleotide reductase R2 in DNA damage
response. J Biol Chem. 284:18085–18095. 2009. View Article : Google Scholar : PubMed/NCBI
|