Introduction
Breast cancer is currently the most frequently
diagnosed cancer and the leading cause of cancer-related death in
females worldwide (1). Tumor cell
invasion and metastasis are regarded as a multistep process,
including the proteolytic degradation of the basement membrane and
the extracellular matrix (ECM), altered cell adhesion and the
physical movement of tumor cells. Among the many steps involved in
tumor invasion and metastasis, excessive degradation of ECM is a
crucial step (2).
Matrix metalloproteinases (MMPs) and plasminogen
activator (PA) systems, a family of proteinases in vivo, are
capable of degrading all major components of the ECM (3). These proteinases have been most
closely linked with the invasive and metastatic phenotype of cancer
cells. Both MMP-9 and MMP-3 have been implicated to play a critical
role in breast cancer invasion and metastasis in animal models and
human patients (4). Urokinase-type
plasminogen activator (uPA) is a 55-kDa serine protease that
activates plasminogen to plasmin leading to cell matrix
degradation, which is involved in tumor cell adhesion, migration,
invasion and intravasation (5).
αvβ6 is one member of the αv integrin subfamily
(αvβ1, αvβ3, αvβ5, αvβ6 and αvβ8) defined by the β and αv subunits
(6). Integrin αvβ6 is not expressed
in normal epithelial cells, but is highly expressed during
morphogenetic events, epithelial repair and tumorigenesis (7). Integrin αvβ6 is restrictedly
distributed to the epithelium and is typically focally localized at
the infiltrating edge of tumor cell islands (8). As a specific epithelial-restricted
integrin, αvβ6 has been observed to be upregulated in various
invasive epithelial carcinomas of the colon, pancreas, prostate,
uterus, lung and breast (9–13). More recent evidence suggests that
αvβ6 may be used to identify patients who have a highly significant
increased risk of developing metastatic disease and may serve for
risk stratification of patients with breast ductal carcinoma in
situ (DCIS) (14). Therefore,
targeting integrin αvβ6 can have unexpected consequences which may
represent an opportunity for molecular targeted therapy for
aggressive breast carcinoma.
To date, relatively little is known concerning the
underlying molecular mechanisms between expression of αvβ6 and
degradation of ECM in human breast cancer. Thus, we aimed to
explore whether shRNAs targeting αvβ6 can induce gene silencing
in vitro. In the present study, we investigated the direct
effect and transcriptional modulation mechanism of the
downregulation of αvβ6 expression on the degradation of ECM.
Materials and methods
Reagents
Since the β6 subunit only combines with the αv
subunit, the expression of the β6 subunit represents the expression
of integrin αvβ6. The anti-αvβ6 mouse anti-human monoclonal
antibodies, R6G9 against β6 and 10D5 against αvβ6 (IgG2a), were
purchased from Chemicon International (Harrow, UK). The monoclonal
antibody against ERK1/2 and phospho-ERK1/2 (Thr202/Tyr204,
Thr185/Tyr187) were obtained from Cell Signaling Technology
(Boston, MA, USA).
N-[(2R)-2-(hydroxamidocarbonylmethyl)-4-methylpentanoyl]-L-tryptophan
methylamide (GM6001), amiloride and
1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio] butadiene
(U0126) were from Calbiochem (Darmstadt, Germany). Phenol red free
(PRF)-DMEM was from Gibco.
Cell line and cell culture
Human breast adenocarcinoma cell line MCF-7 was
kindly provided by Dr Xiaolei Wang, and was obtained from the
American Type Culture Collection (Manassas, VA, USA). Cells were
maintained in PRF-DMEM supplemented with 10% heat inactivated fetal
bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin,
and 2 mmol/l L-glutamine in a humidified atmosphere with 5%
CO2 at 37°C. Stably transduced cells were simultaneously
cultured in the absence or presence of the indicated concentrations
of reagents tested for 72 h in PRF-DMEM containing 10% FBS.
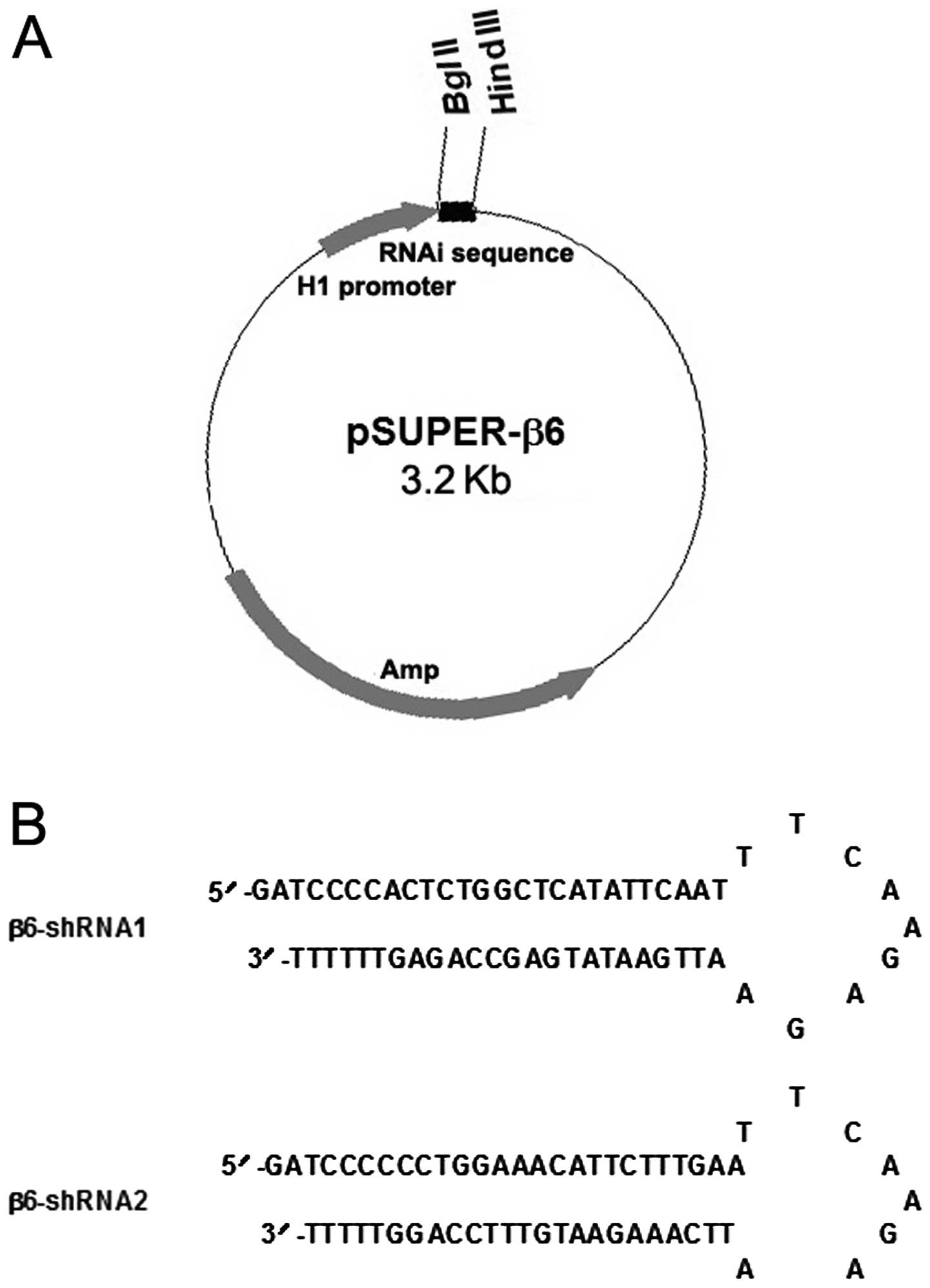
Design of shRNAs and construction of
recombinant vectors
Because not all small interfering RNA (siRNA) target
sequences are equally potent, we designed and selected two
different shRNAs targeting different coding regions within the
human β6 gene (GenBank accession no. A266609) according to the
recommendations (http://www.oligoengine.com and http://sirna.qiagen.com) in our laboratory, which were
synthesized by Integrated DNA Technologies (Coralville, IA, USA).
BLAST search of genome sequence databases (NCBI Unigene and EST
libraries) was performed to ensure that no other human gene was
targeted. The double-stranded DNA (dsDNA) was designed as follows:
a 19-nucleotide target sequence in both sense and antisense
orientations, separated by an 8-nucleotide spacer sequence to form
a hairpin dsRNA and flanked at either end by BglII and
HindIII restriction enzyme sites and the five repeats of T
as transcriptional termination signal. The detailed information of
these shRNAi used in our study is listed in Table I. The oligonucleotides were then
directionally cloned into pSUPER.retro.puro vector at the
BglII and HindIII sites to generate the β6-shRNA
expression vectors (Fig. 1), which
were defined as pSUPER-β6shRNA1 and pSUPER-β6shRNA2, respectively.
In addition, all constructs were further verified by DNA
sequencing.
 | Table IOligonucleotide sequences of the
shRNAi constructs. |
Table I
Oligonucleotide sequences of the
shRNAi constructs.
| β6-shRNA1 |
| Sense |
5′-gatccccACTCTGGCTCATATTCAATTCAAGAGAATTGAATATGAGCCAGAGTtttttgaa-3′ |
| Antisense |
5′-agctttcaaaaaACTCTGGCTCATATTCAATTCAAGAGAATTGAATATGAGCCAGAGTggg-3′ |
| β6-shRNA2 |
| Sense |
5′-gatccccCCTGGAAACATTCTTTGAATCAAGAGATTCAAAGAATGTTTCCAGGtttttgaa-3′ |
| Antisense |
5′-agctttcaaaaaCCTGGAAACATTCTTTGAATCAAGAGATTCAAAGAATGTTTCCAGGggg-3′ |
Transfection of the siRNA constructs
Stable transfection of the recombinant plasmid
pSUPER-β6shRNAs was carried out using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
protocols. Two hundred and fifty milliliters of DMEM without serum
and 4 μg pSUPER-β6shRNAs or control pSUPER per well were
pre-incubated for 5 min at room temperature. During the period of
preincubation, 250 μl of DMEM without serum was mixed with 10 μl
Lipofectamine 2000. The two media were mixed and incubated for 20
min at room temperature for complex formation and then the cells
were transfected. Moreover, stable transformants were subsequently
selected by growth in medium supplemented with 400 μg/ml geneticin
(G418; Gibco) for 3 weeks. After selection with G418, resistant
cell clones were then randomly isolated for cell expansion and
further analysis.
RNA extraction and semi-quantitative
reverse transcription-PCR analysis
Total cellular RNA was isolated from the
untransfected or stably transfected MCF-7 cells using TRIzol (Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions, respectively. A 141-bp fragment of β6 cDNA was
amplified with the following primers: forward,
5′-AGGATAGTTCTGTTTCCTGC-3′ and reverse, 5′-ATCATAGGAATATTTGGAGG-3′.
As an internal control, amplification of the housekeeping gene
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was also
carried out using the following primers: sense, 5′-GTCGGTGT
CAACGGATTTG-3′ and antisense, 5′-ACAAACATGGGGG CATCAG-3′.
Western blot analysis
Cellular protein extracts from the treated or
untreated cells were prepared according to the manufacturer’s
protocols (Upstate Biotechnology, Lake Placid, NY, USA). Fifty
micrograms of cellular protein per lane was electrophoresed and
separated by 4% stacking and 12% resolving SDS-polyacrylamide gel
electrophoresis (SDS-PAGE). Separated proteins were
electrophoretically transferred and blotted onto a polyvinylidene
difluoride filter (PVDF). To avoid unspecific binding, the filters
were incubated in 5% skim milk and 0.05% Tween-20 in TBS for 2 h at
room temperature. Subsequently, the filters were incubated with
mouse monoclonal antibody against human αvβ6 diluted in the same
solution (1:1,000) overnight at 4°C and, afterwards, with
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
secondary antibody diluted at 1:4,000. As control for equivalent
protein loading, the filters were simultaneously incubated with
mouse anti-GAPDH monoclonal antibody (1:5,000). The level of
ERK1/2, phospho-ERK1/2, or uPA expression was also analyzed by
western blotting with the monoclonal antibody against ERK1/2,
phospho-ERK1/2 or uPA as described above. The protein-antibody
complexes were visualized with an enhanced chemiluminescence (ECL)
kit (Amersham Pharmacia Biotech) according to the manufacturer’s
instructions. The intensity of each band was quantified using an
image processing and analysis program.
Gelatin and casein zymography assays
The expression and activity of MMP-9 and MMP-3 were
analyzed by zymography. For assay of MMP-9 (gelatinase B) activity,
gelatin zymography and for assay of MMP-3 activity, casein
zymography were performed from the samples as described previously
(15,16). Briefly, gelatin or casein was added
to the 10% acrylamide separating gel at a final concentration of 1
mg/ml for SDS-PAGE, respectively. Tumor conditioned medium (TCM)
collected from untreated, pSUPER-controlor
pSUPER-β6shRNA-transfected cells under serum-free conditions was
mixed with substrate gel sample buffer [10% SDS, 50% glycerol, 25
mM Tris-HCl (pH 6.8) and 0.1% bromophenol blue], and 70 μl was
loaded onto the gel without prior boiling. Following
electrophoresis, gels were washed twice in 2.5% (v/v) Triton X-100
for 30 min at room temperature to remove the SDS. Gels were then
incubated at 37°C overnight in substrate buffer containing 50 mM
Tris-HCl and 5 mM CaCl2 (pH 8.0). The gels were
subsequently stained with 0.15% (w/v) Coomassie brilliant blue
R-250 in 50% methanol and 10% glacial acetic acid at room
temperature for 20 min, and then destained in the same solution
without Coomassie brilliant blue. Gelatin- or casein-degrading
enzymes were identified as clear zones in a dark blue background of
the stained gel. Zymogram bands were analyzed, and MMP activity was
quantified by densitometry (Personal Densitometer SI) and
ImageQuant software (both from Molecular Dynamics, Sunnyvale, CA,
USA).
[3H]-labeled collagen type IV
degradation assay
Collagen type IV degradation assay was performed as
previously described (17). In
brief, tritium-labeled basement membrane type IV collagen was
denatured to form gelatin by heating at 60°C for 30 min.
Ninety-six-well plates were coated with 65 μl of
[3H]-labeled gelatin (15,000 cpm/well) and allowed to
dry overnight in a laminar flow hood at room temperature. Plates
were then washed 3 times with phosphate-buffered saline (PBS) until
free cpm reached the basal level. For cell-mediated collagen
degradation assays, untreated, pSUPER-control- or
pSUPER-β6shRNA-transfected cells (105 cells/well) were
incubated with the gelatin substrates in 300 μl of serum-free DMEM
at 37°C for 24 h in the absence or presence of various
concentrations of plasminogen. Collagen type IV degradation was
determined by subtracting the cpm released from the buffer only
wells, and was assessed by measuring the cpm released in 300 μl of
medium from triplicate wells for each condition. In studies
concerning inhibition of plasminogen activation, uPA and MMP
activity, untreated cells and pSUPER-control-treated cells
incubated with plasminogen were exposed either to monoclonal
antibody against ανβ6 (10D5), uPA inhibitor amiloride (2 mM), MMP
inhibitor GM6001 (2 mM), or MEK1/2 inhibitor U0126 for 30 min prior
to plating onto the [3H]-labeled extracellular matrix.
All of the experiments were performed independently and repeated at
least three times.
Statistical analysis
All statistical analyses were performed using SPSS
13.0 software package. The two-sided unpaired Student’s t-test and
one-way ANOVA were used to evaluate the statistical significance of
differences in two groups and multiple groups, respectively. All
data were presented as mean ± standard deviation (SD). P<0.05
was considered to indicate a statistically significant
difference.
Results
Stable expression of β6shRNAs in the
MCF-7 cells
The two shRNAs selected targeting the β6 gene were
cloned into the pSUPER.retro vectors (Fig. 1A). The predicted forms and sequences
of these shRNAs are shown in Fig.
1B. In addition, the recombinant vectors were validated by
restriction enzyme digestion, and the inserted sequences were
verified by DNA sequencing. After transfection and selection, the
stably transfected cells were named as MCF-7/ανβ6-1 (transfected
with pSUPER-β6shRNA1), MCF-7/ανβ6-2 (transfected with
pSUPER-β6shRNA2) and MCF-7/CON (transfected with parental vector
pSUPER.retro), respectively.
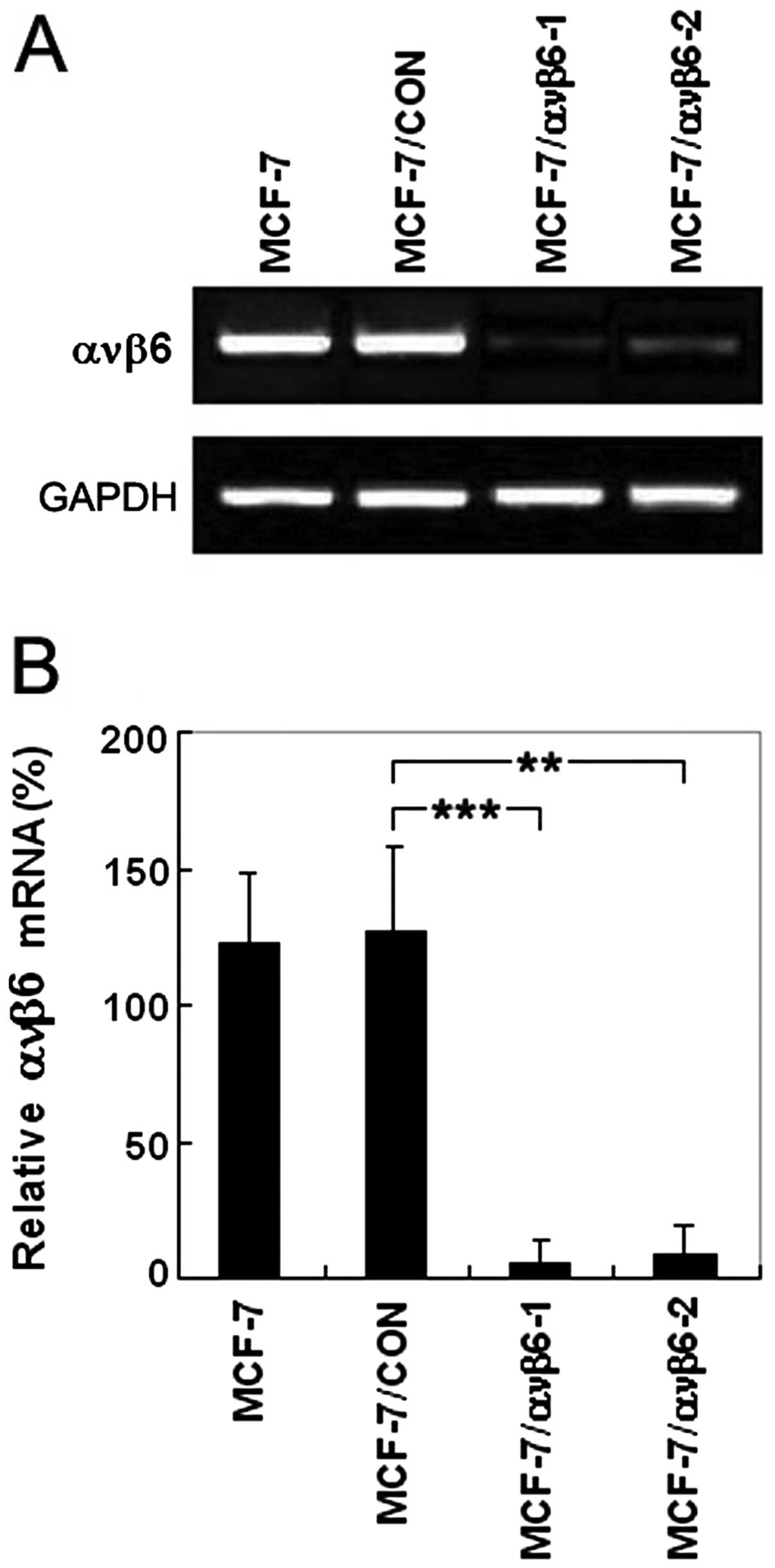
ανβ6 mRNA expression is efficiently
suppressed by pSUPER-β6shRNAs in the MCF-7 cells
After β6shRNA stably transfected cells (MCF-7/ανβ6-1
and MCF-7/ανβ6-2) were generated, ανβ6 expression at the mRNA level
was investigated by semi-quantitative RT-PCR. In comparison with
the control cells, the levels of ανβ6 mRNA were markedly decreased
by 95.2 and 91.7% in the MCF-7/ανβ6-1 and MCF-7/ανβ6-2 cells,
respectively (Fig. 2A and B). In
addition, no effects of RNAi were observed on the expression of
GAPDH used as an internal control. These results suggest that
β6shRNA can efficiently downregulate ανβ6 mRNA. In summary,
shRNA-mediated silencing was markedly pronounced, and no effect
could be observed by any other of the controls.
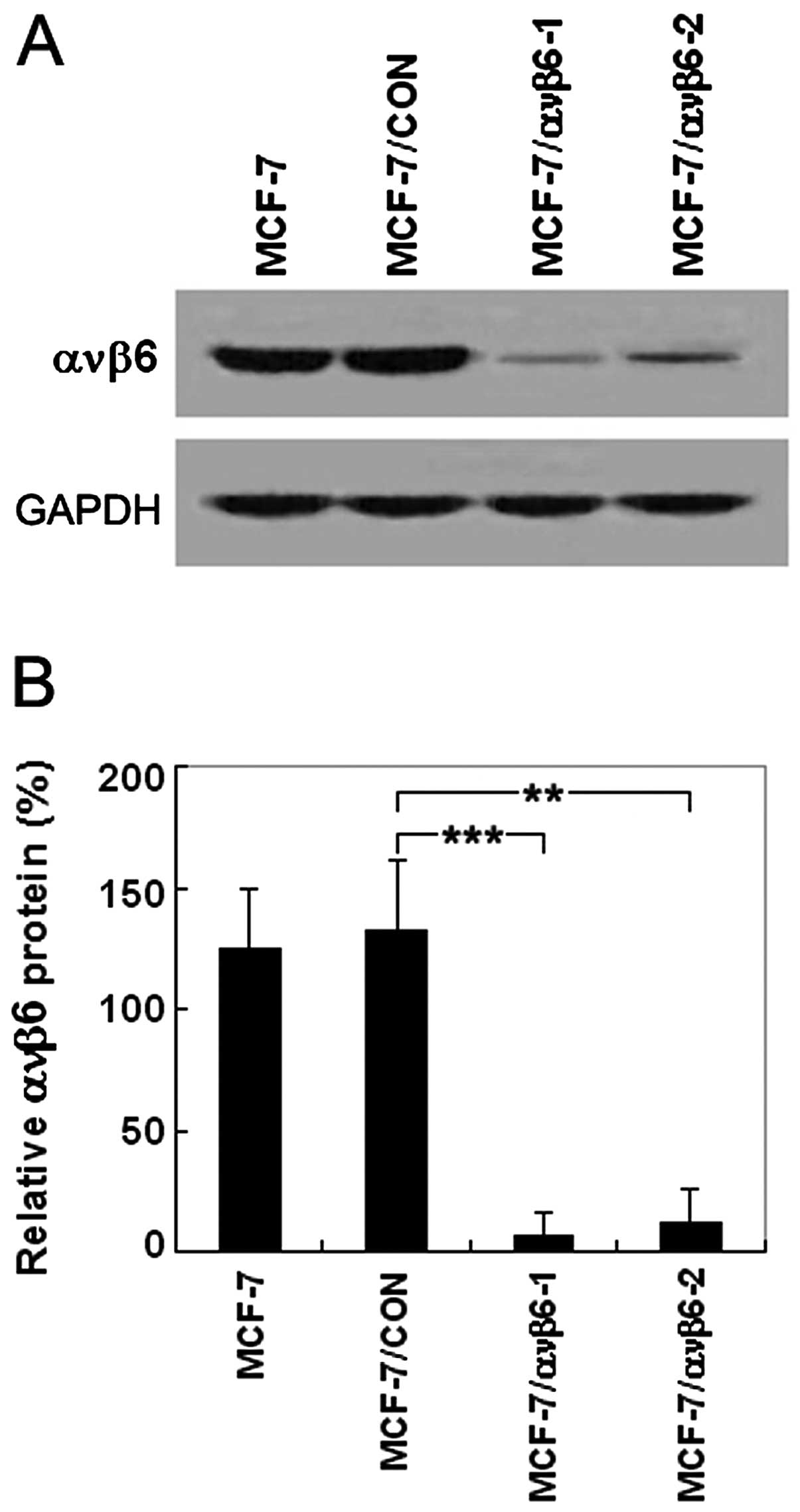
Knockdown of ανβ6 protein level in the
MCF-7 cells by shRNA recombinant plasmids
The pSUPER system used in this study was able to
markedly suppress the expression of the β6 gene. The shRNA
expression plasmids and a control vector were transfected into
MCF-7 cells, and ανβ6 protein expression was monitored by western
blotting. As shown in Fig. 3A and
B, pSUPER-β6shRNA1 and pSUPER-β6shRNA2 significantly decreased
the ανβ6 protein level in the MCF-7 cells to 6.9 and 9.7%,
respectively, whereas the control vector did not inhibit ανβ6
protein expression. In other words, the ανβ6 protein expression
inhibition rates were 93.1 and 90.3% in the MCF-7/ανβ6-1 and
MCF-7/ανβ6-2 cells, respectively. However, GAPDH expression was not
affected by the same experimental conditions. This change in ανβ6
protein expression was consistent with that of the ανβ6 mRNA level.
Therefore, these results indicate that β6shRNA strongly suppresses
ανβ6 protein as well as the mRNA level and can be used to target
the β6 gene for breast cancer therapy.
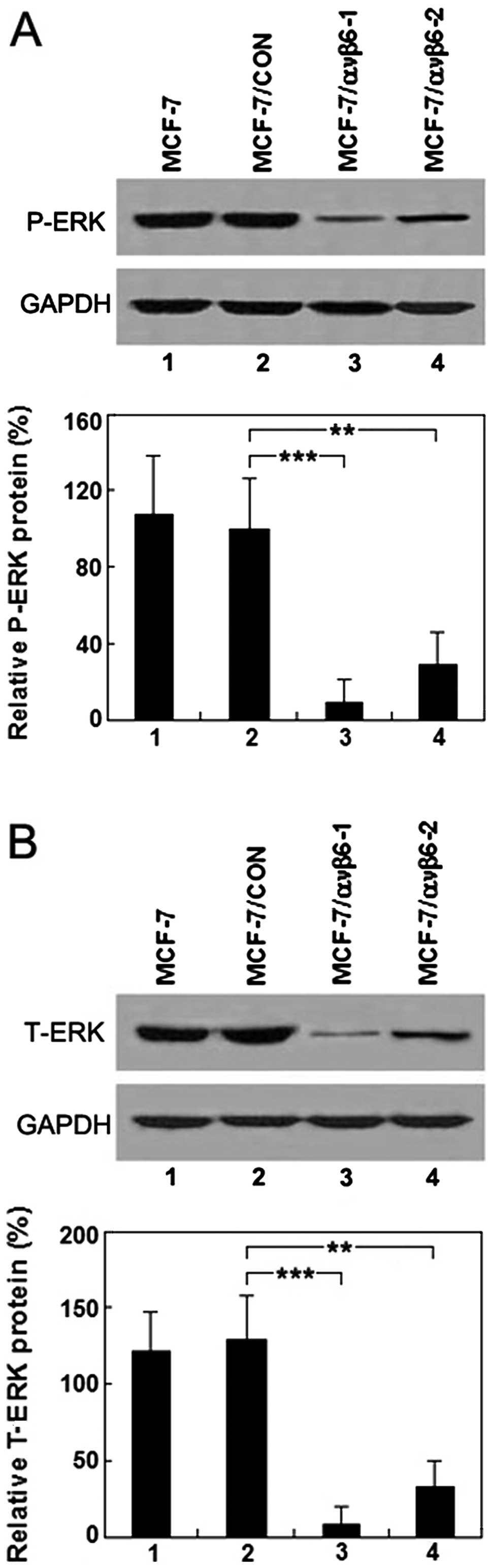
Suppression of integrin ανβ6 expression
downregulates ERK1/2 levels
To investigate the possible causal link between ανβ6
and ERK1/2, untreated and treated MCF-7 cells for 72 h after stable
transfection with pSUPER-β6shRNAs or pSUPER-control were harvested.
The levels of ERK1/2 and phospho-ERK1/2 were assessed by western
blot analysis. The inhibitory effects of the downregulation of ανβ6
expression on the phosphorylation and nonphosphorylation levels of
ERK1/2 in the MCF-7/ανβ6-1 and MCF-7/ανβ6-2 cells are shown in
Fig. 4A and B. These findings are
in agreement with the change in ανβ6 mRNA and protein levels and
further support our hypothesis that the suppression of ανβ6
expression by β6shRNAs leads to the inactivation of the MAP kinase
pathway.
Inhibition of integrin ανβ6 suppresses
the secretion of pro-MMP-9, pro-MMP-3 and uPA in tumor conditioned
medium from the human breast cancer MCF-7 cell line
The effects of reduced ανβ6 on MMP-9, MMP-3 and uPA
expression in vitro were evaluated by gelatin zymography,
casein zymography and western blot analysis, respectively.
Untreated MCF-7 cells and cells after stable transfection with
pSUPER-β6shRNA1, pSUPER-β6shRNA2 or pSUPER-control for 72 h were
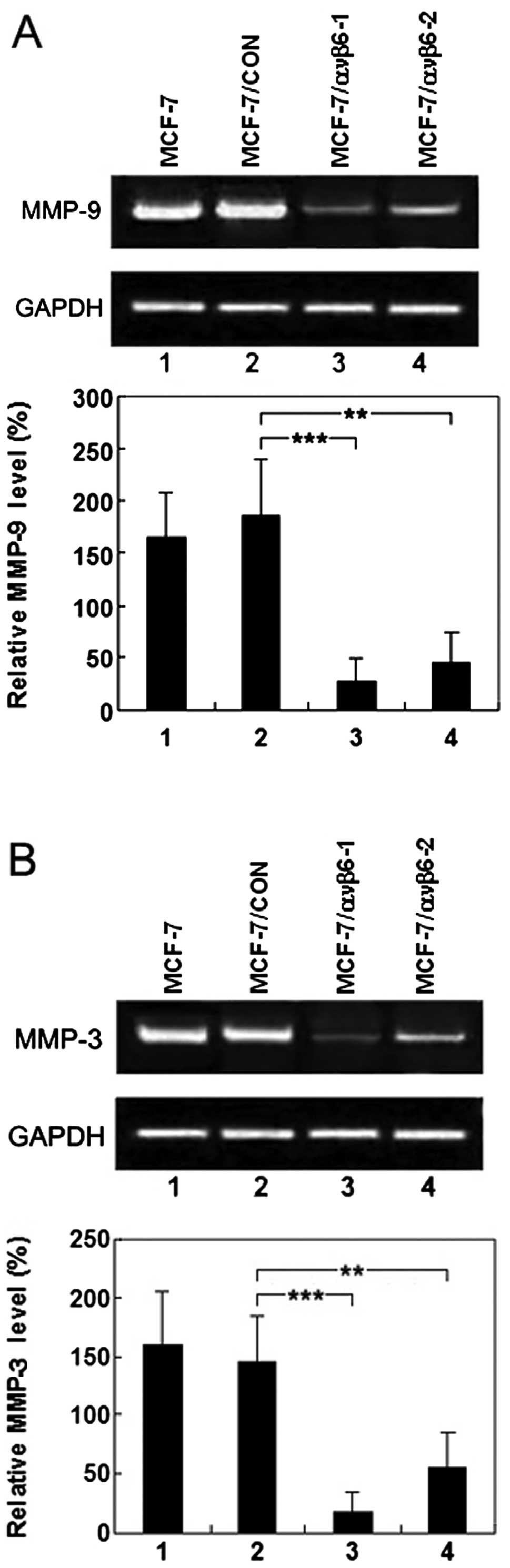
harvested and TCM was prepared. As shown in Fig. 5A and B, compared with the control
cells, MMP-9 and MMP-3 production was decreased by 90.7 and 93.8%
in the MCF-7/ανβ6-1 cells, respectively. Next, we aimed to
ascertain whether a similar trend would be observed in the
MCF-7/ανβ6-2 cells, stably transfected with pSUPER-β6shRNA2. MMP-9
and MMP-3 production was reduced by 70.4 and 75.6%, respectively
(Fig. 5A and B). Furthermore,
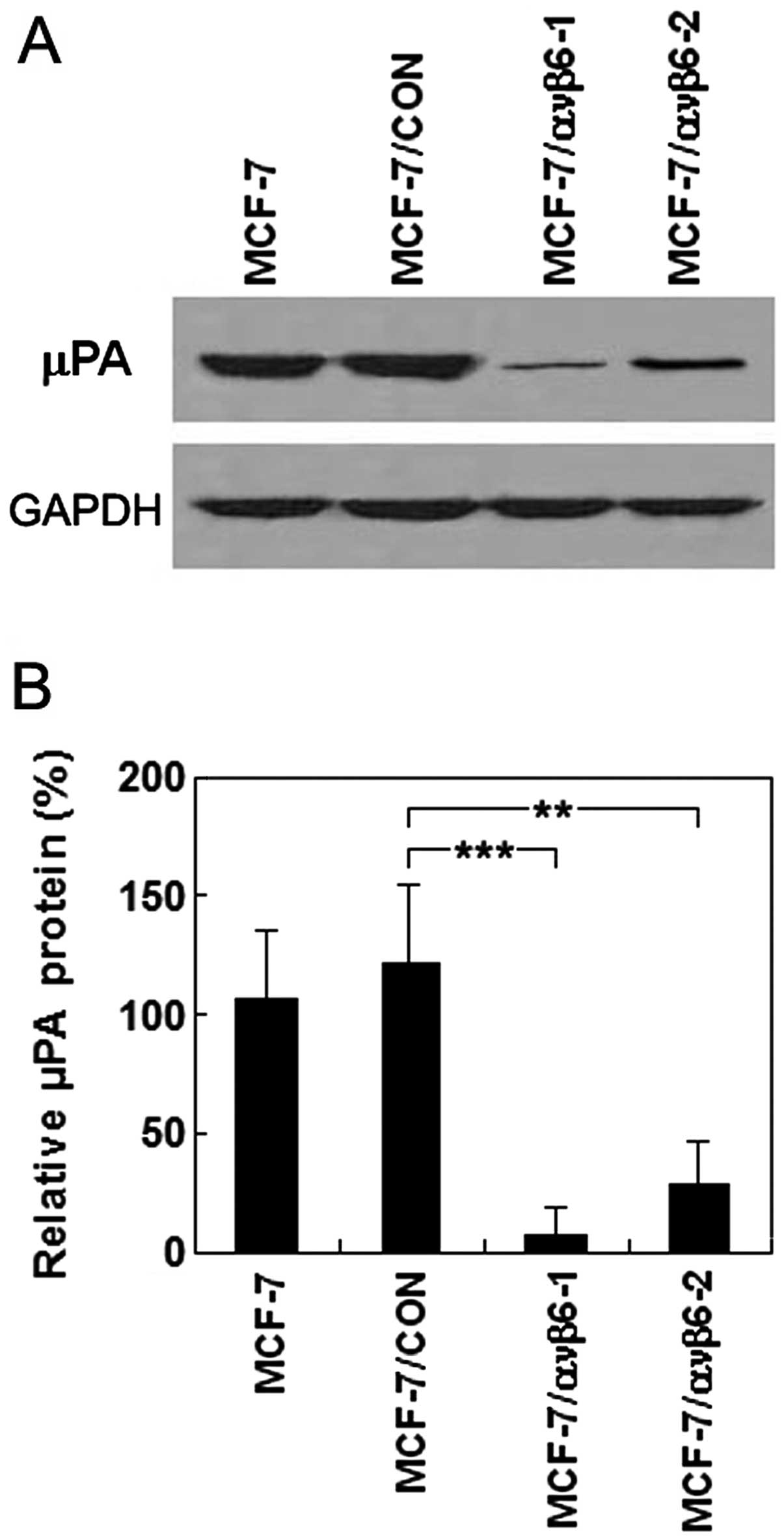
western blot analysis demonstrated that relative uPA protein levels
were 7.1±0.6 and 28.3±1.2% in the MCF-7/ανβ6-1 and MCF-7/ανβ6-2
cells, respectively, significantly lower than that of the control
cells (121.4±3.5%; P<0.05) (Fig. 6A
and B). In other words, the uPA protein expression was
decreased by 94.2 and 76.7% in the MCF-7/ανβ6-1 (transfected with
pSUPER-β6shRNA1) and MCF-7/ανβ6-2 cells (transfected with
pSUPER-β6shRNA2), respectively, compared with that of the MCF-7/CON
cells (transfected with parental vector pSUPER.retro). No effects
of RNAi were observed in regards to the expression of GAPDH, which
was used as an internal control. Therefore, these results suggest
that inhibition of integrin ανβ6 by RNAi could efficiently suppress
the secretion of pro-MMP-9, pro-MMP-3 and uPA in the human breast
cancer MCF-7 cell line.
Effect of ανβ6 gene expression silencing
by RNAi on degradation of [3H]-labeled collagen type
IV
To determine whether inhibition of integrin ανβ6 by
RNAi suppresses extracellular matrix degradation,
plasminogen-dependent [3H]-labeled collagen type IV
degradation assay was performed. Collagen type IV, the major
structural component of the basement membrane, was used as the
substrate for both collagenase MMP-9 and MMP-3. Degradation of the
basement membrane was measured by the release of tritium from
[3H]-labeled, heat-denatured radiolabeled type IV
collagen. Exposure of the gelatin substrate to serum-free
nonconditioned culture medium DMEM for 24 h resulted in
spontaneous, non-proteinase-mediated release of tritium into the
fluid phase background cpm, the counts per minute measured
(Fig. 7A). Exposure of the collagen
substrate to TCM obtained from the untreated cells, pSUPER-β6shRNA-
and pSUPER-control transfected cells did not result in tritium
release in either the presence or absence of 8 μg/ml plasminogen
above background levels (Fig. 7A),
indicating that the released collagenases in the culture
supernatants were neither active nor activatable by plasminogen in
the absence of cells. In contrast, exposure of collagen to
untreated and pSUPER-control-treated human breast cancer MCF-7
cells in the presence of exogenous plasminogen significantly
increased the basal level of collagen type IV degradation, compared
to the corresponding control cells in the absence of
plasminogen.
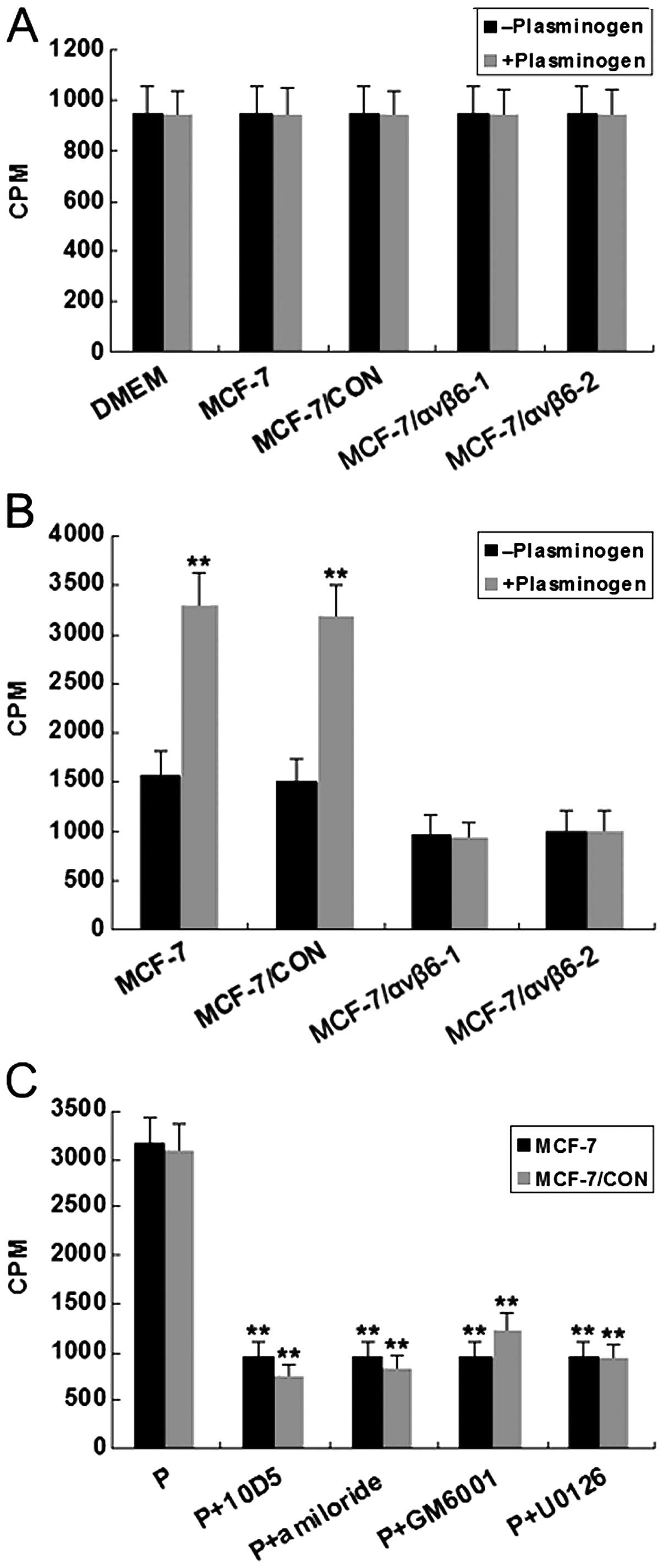 | Figure 7Effect of ανβ6 gene expression
silencing by RNAi on the degradation of [3H]-labeled
collagen type IV. Collagen type IV degradation was measured by the
release of tritium into the fluid phase, and triplicate wells were
used for each experimental condition. (A) Untreated MCF-7 and
treated cells (MCF-7/CON, MCF-7/ανβ6-1 and MCF-7/ανβ6-2 cells) were
harvested and incubated for 24 h in the absence or presence of
plasminogen (8 μg/ml) in 24-well plates coated with
[3H]-labeled heat-denatured collagen type IV. Background
cpm (spontaneous release of tritium in the presence of DMEM) is
shown on the left. **P<0.01, vs. the corresponding
control cells in the absence of plasminogen. (B) Untreated MCF-7
and treated cells (MCF-7/CON, MCF-7/ανβ6-1 and MCF-7/ανβ6-2 cells)
were incubated for 24 h in the absence or presence of plasminogen
(8 μg/ml) in 24-well plates coated with [3H]-labeled,
heat-denatured collagen type IV. **P<0.01, vs. the
corresponding control cells in the absence of plasminogen. (C)
Untreated cells and pSUPER-control treated cells incubated with
plasminogen (P, 8 μg/ml) were exposed either to a monoclonal
antibody against ανβ6 (10D5), uPA inhibitor amiloride (2 mM), MMP
inhibitor GM6001 (2 mM), or MEK1/2 inhibitor U0126 for the duration
of the experiment. Results are shown as mean values ± SEM of three
different experiments performed in triplicate.
**P<0.01, vs. the corresponding control cells in the
absence of plasminogen. |
Not unexpectedly, there was no such effect in the
pSUPER-β6shRNA-transfected cells (Fig.
7B). Furthermore, as shown in Fig.
7C, the increased and more extensive collagen degradation
monitored in the untreated and pSUPER-control-treated cells, was
abolished by the addition of either anti-αvβ6 antibody 10D5, MMP
inhibitor GM6001, uPA inhibitor amiloride or MEK1/2 inhibitor
U0126. These results strongly suggest that silencing of ανβ6 gene
expression by RNAi could effectively suppress mitogen-activated
protein kinase (MAPK)-dependent degradation of the extracellular
matrix, and also demonstrated that MMPs and uPA-mediated
plasminogen dependent-proteolysis are essential to the degradation
of the basement membrane.
Discussion
Degradation of extracellular matrix components and
basement membranes is crucial for invasion and metastasis of cancer
cells. During this process, MMPs appear to be primarily responsible
for much of the ECM degradation (18). Recently, integrin ανβ6 has emerged
as a novel potential target for anticancer therapy and plays a
major role in promoting malignant tumor progression. In addition,
it has previously been reported that integrin αvβ6 is restrictedly
distributed to the epithelium and is typically focally localized at
the infiltrating edge of tumor cell islands.
In the present study, we further investigated the
effect and regulatory mechanism of ανβ6 gene knockdown by RNAi on
the degradation of the extracellular matrix in MCF-7 breast cancer
cells. RNAi is a novel and powerful tool for specific inhibition of
a targeted gene by introducing double-stranded RNA into cells
leading to sequence-specific destruction (19–21).
RNAi is a sequence-specific, post-transcriptional gene silencing
process in many organisms, which can be triggered by dsRNA and is
cleaved into 21- to 23-nucleotide RNA fragments, known as siRNAs,
by the ribonuclease III (RNase III)-like enzyme, Dicer.
Subsequently, these siRNAs are incorporated into a protein complex,
which is also called RNA-induced silencing complex (RISC). This
protein complex is able to degrade homologous mRNA and then to
inhibit the targeting gene at the post-transcriptional level
(22,23). RNAi constitutes a promising source
for new therapeutic approaches, including cancer gene therapy. For
years, many research groups have focused on effective tools to
specifically downregulate gene expression, such as antisense
oligonucleotide strategy. However, its success has been limited,
due to the lack of specificity and potency. RNAi-induced knockdown
of target gene expression is an attractive approach for gene
therapy since multiple targets may be manipulated simultaneously
(24–26). Therefore, we explored whether shRNAs
targeting αvβ6 can induce gene silencing in vitro.
The present results demonstrated that siRNA can
efficiently suppress ανβ6 expression with high specificity at the
mRNA and protein levels. Because of the pSUPER vector containing
the H1 RNA polymerase III promoter upstream of the inserted DNA
sequence, the shRNAs can be effectively expressed after being
transfected into tumor cells (27).
In our study, by using a new retroviral pSUPER.retro vector system,
we successfully generated permanent cell lines that constitutively
express specific siRNA. First, two shRNAs targeting the ανβ6 gene
were selected and cloned into the expression vector pSUPER. We
successfully established stably transfected cells: MCF-7/ανβ6-1,
MCF-7/ανβ6-2 and MCF-7/CON. In addition, our data showed that both
shRNAs against αvβ6 expressed by the recombinant plasmids were
markedly effective in suppressing αvβ6 mRNA and protein in MCF-7
cells. RT-PCR detection revealed that pSUPER-β6shRNA1 and
pSUPER-β6shRNA2 decreased ανβ6 mRNA by 95.2 and 91.7%,
respectively. ανβ6 protein expression was reduced by 93.1 and 90.3%
in the MCF-7/ανβ6-1 and MCF-7/ανβ6-2 cells compared to the
MCF-7/CON cells, respectively, by western blot analysis. Relative
ανβ6 mRNA and protein levels were significantly lower than these
levels in the untreated control cells. In this experiment, both
pSUPER-β6shRNA constructs showed a pronounced ανβ6 gene silencing
activity in the MCF-7 cells at the mRNA level as well as the
protein level and were similar to each other. These data thus
indicate that ανβ6 could specifically serve as a biomarker and a
potential and attractive therapeutic target for directed cancer
genetic therapy.
Integrin ανβ6 in cancer cells is of particular
interest for the fact that it is not expressed in normal
epithelium, but is highly expressed during tumorigenesis, leading
to enhanced tumor cell migration and invasion (28). Recently, it has been reported that
the upregulation of the integrin β6 gene could be involved in
oxytocin-induced cell growth in human breast tumor-derived
endothelial cells (29). A more
recent study also showed that ανβ6-positive patients have a
markedly high risk of progression to invasive breast cancer
(14). To date, it remains unclear
whether and how ανβ6 is involved in regulation of breast cancer
cell migration and metastasis, or whether there is any relationship
between ανβ6 expression and ECM degradation in human breast cancer
cells. In the present study, the effect and possible regulatory
mechanism of the downregulation of ανβ6 expression on matrix
degrading enzyme secretion in breast cancer cells were
investigated. Degradation of the extracellular matrix and
components of the basement membrane by proteases facilitates the
detachment of tumor cells, their crossing of tissue boundaries, and
invasion into adjacent tissue compartments. MMPs and the serine
protease uPA, the class of proteolytic enzymes, are thought to play
a central role in this process, because of their ability to degrade
many ECM components (30). In the
present study, MMP-9, MMP-3 and uPA protein expression levels were
decreased by 93.1, 90.3 and 94.2% in the MCF-7/ανβ6-1 cells, and
70.4, 75.6 and 76.7% in the MCF-7/ανβ6-2 cells compared to the
MCF-7/CON cells, respectively. To the best of our knowledge, we
showed for the first time that suppression of integrin ανβ6 in
MCF-7 human breast carcinoma cells dramatically reduced pro-MMP-9,
pro-MMP-3 and uPA secretion in tumor conditioned medium. These
findings are in agreement with the observation by Gu et al
that inhibition of β6 expression, MEK inhibition, or deletion of
the β6-ERK2 binding site suppressed MMP-9 secretion in human colon
cancer cell lines WiDr and HT29 (31). Our results thus indicate a potential
regulatory mechanism whereby integrin ανβ6 contributes to breast
cancer invasion by enhancing matrix degrading enzyme secretion and
activity.
In addition, to assess whether downregulation of
pro-MMP-9, pro-MMP-2, uPA expression in serum-free tumor
conditioned medium affects the degradation of the extracellular
matrix, radiolabeled type IV collagen degradation assay was
performed. Loss of basement membrane type IV collagen has
previously been shown to be associated with breast cancer
tumorigenesis, and the absence of type IV collagen was found to be
involved in the overexpression of MMPs. uPA has been identified as
the critical trigger for active plasmin generation resulting in an
overall increase in catalytic efficiency (32). Moreover, it has been recently
reported that activation of pro-MMPs by plasminogen occurs through
uPA-mediated generation of plasmin (33). In the present study, the
plasminogen-dependent ECM degradation in the untreated and
pSUPER-control cells was completely abrogated by the uPA inhibitor,
anti-MMPs or anti-ανβ6, suggesting that the inhibitory effects of
downregulated ανβ6 expression was via the plasminogen activation
cascade. Therefore, our results also demonstrated that inhibition
of ανβ6 expression in breast cancer MCF-7 cells suppresses the
plasminogen-dependent degradation of the extracellular matrix.
The MAPK signaling pathway, a family of
protein-serine/threonine kinases, plays a fundamental role in
regulating cellular proliferation, differentiation, migration and
apoptosis (34). MAPKs are involved
in the regulation of MMP expression associated with invasion of
malignant tumor cells (35). It is
now becoming clear that activation of epidermal growth factor
receptor (EGFR) and its subsequent regulation of extracellular
signal-regulated kinases (ERKs) for cell survival is dependent on
integrin-mediated, ligand-independent signal transduction involving
MAPK cascades (36). Moreover, it
is important that integrin ligation induces activation of ERKs when
the concentration of growth factors available to the cell is
limited. Either deletion of the ERK2 binding site on the β6
cytoplasmic domain or downregulation of β6 expression inhibits
tumor growth and may be due to an association between ERK and the
β6 subunit (37). This is also
supported by the finding that integrin-mediated MMP-9 secretion is
dependent upon direct binding between the β6 integrin subunit and
ERK2 (31). The data in this study
show that downregulation of integrin ανβ6 expression by
pSUPER-β6shRNAs significantly reduced the levels of phosphorylated
and unphosphorylated ERK1/2. Furthermore, plasminogen-dependent ECM
degradation of untreated and pSUPER-control treated cells was
almost completely abolished by the specific MEK1/2 inhibitor U0126,
indicating that suppression of ECM degradation was induced by
inhibition of integrin ανβ6 expression dependent on MAPK
activity.
In summary, our findings thus demonstrated that RNA
technology-based approach for suppression of the ανβ6 gene in
vitro can efficiently downregulate ανβ6 expression in breast
cancer, and in the future may offer a useful therapeutic strategy
to block invasion and migration for the treatment of breast cancer.
In addition, our study revealed that inhibition of the ανβ6 gene
markedly decreased the activity of MMP-9, MMP-3, uPA and the
ERK1/2-dependent degradation of ECM. These data therefore provide
new insight into the regulatory mechanism of integrin ανβ6 in ECM
degradation, which might be involved in inactivation of the MAP
kinase pathway in the progression of human breast cancer.
Acknowledgements
This study was supported in part by the Youth
Science Foundation of Beijing Tiantan Hospital, Capital Medical
University, P.R. China (no. ky2008-08) and the National Natural
Science Foundation of China (NSFC, no. 30300124).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Liotta LA and Stetler-Stevenson WG: Tumor
invasion and metastasis: an imbalance of positive and negative
regulation. Cancer Res. 51:S5054–S5059. 1991.PubMed/NCBI
|
|
3
|
Yoon WH, Jung YJ, Kim TD, Li G, Park BJ,
Kim JY, Lee YC, Kim JM, Park JI, Park HD, No ZS, Lim K, Hwang BD
and Kim YS: Gabexate mesilate inhibits colon cancer growth,
invasion, and metastasis by reducing matrix metalloproteinases and
angiogenesis. Clin Cancer Res. 10:4517–4526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McGowan PM and Duffy MJ: Matrix
metalloproteinase expression and outcome in patients with breast
cancer: analysis of a published database. Ann Oncol. 19:1566–1572.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blasi F and Carmeliet P: uPAR: a versatile
signalling orchestrator. Nat Rev Mol Cell Biol. 3:932–943. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bates RC, Bellovin DI, Brown C, Maynard E,
Wu B, Kawakatsu H, Sheppard D, Oettgen P and Mercurio AM:
Transcriptional activation of integrin beta6 during the
epithelial-mesenchymal transition defines a novel prognostic
indicator of aggressive colon carcinoma. J Clin Invest.
115:339–347. 2005. View Article : Google Scholar
|
|
7
|
Breuss JM, Gallo J, De Lisser HM,
Klimanskaya IV, Folkesson HG, Pittet JF, Nishimura SL, Aldape K,
Landers DV and Carpenter W: Expression of the beta 6 integrin in
development, neoplasia and tissue repair suggests a role in
epithelial remodelling. J Cell Sci. 108:2241–2251. 1995.PubMed/NCBI
|
|
8
|
Niu J, Gu X, Ahmed N, Andrews S, Turton J,
Bates R and Agrez M: The alphaVbeta6 integrin regulates its own
expression with cell crowding: Implications for tumour progression.
Int J Cancer. 1:40–48. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arihiro K, Kaneko M, Fujii S, Inai K and
Yokosaki Y: Significance of alpha 9 beta 1 and alpha v beta 6
integrin expression in breast carcinoma. Breast Cancer. 7:19–26.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Azare J, Leslie K, Al-Ahmadie H, Gerald W,
Weinreb PH, Violette SM and Bromberg J: Constitutively activated
Stat3 induces tumorigenesis and enhances cell motility of prostate
epithelial cells through integrin beta 6. Mol Cell Biol.
27:4444–4453. 2007. View Article : Google Scholar
|
|
11
|
Hausner SH, Abbey CK, Bold RJ, Gagnon MK,
Marik J, Marshall JF, Stanecki CE and Sutcliffe JL: Targeted in
vivo imaging of integrin alphavbeta6 with an improved radiotracer
and its relevance in a pancreatic tumor model. Cancer Res.
69:5843–5850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hazelbag S, Kenter GG, Gorter A, Dreef EJ,
Koopman LA, Violette SM, Weinreb PH and Fleuren GJ: Overexpression
of the alpha v beta 6 integrin in cervical squamous cell carcinoma
is a prognostic factor for decreased survival. J Pathol.
212:316–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eberlein C, Kendrew J, McDaid K, Alfred A,
Kang JS, Jacobs VN, Ross SJ, Rooney C, Smith NR, Rinkenberger J,
Cao A, Churchman A, Marshall JF, Weir HM, Bedian V, Blakey DC,
Foltz IN and Barry ST: A human monoclonal antibody 264RAD targeting
αvβ6 integrin reduces tumour growth and metastasis, and modulates
key biomarkers in vivo. Oncogene. 32:4406–4416. 2013.
|
|
14
|
Allen MD, Thomas GJ, Clark SE, Dawoud MM,
Vallath S, Payne SJ, Gomm JJ, Dreger SA, Dickinson S, Edwards DR,
Pennington CJ, Sestak I, Cuzick J, Marshall JF, Hart IR and Jones
JL: Altered microenvironment promotes progression of pre-invasive
breast cancer: myoepithelial expression of αvβ6 integrin in DCIS
identifies high-risk patients and predicts recurrence. Clin Cancer
Res. 20:344–357. 2014.PubMed/NCBI
|
|
15
|
Kundu P, Mukhopadhyay AK, Patra R,
Banerjee A, Berg DE and Swarnakar S: Cag pathogenicity
island-independent up-regulation of matrix metalloproteinases-9 and
-2 secretion and expression in mice by Helicobacter pylori
infection. J Biol Chem. 281:34651–34662. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Niu J, Dorahy DJ, Gu X, Scott RJ, Draganic
B, Ahmed N and Agrez MV: Integrin expression in colon cancer cells
is regulated by the cytoplasmic domain of the beta6 integrin
subunit. Int J Cancer. 99:529–537. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agrez M, Gu X, Turton J, Meldrum C, Niu J,
Antalis T and Howard EW: The alpha v beta 6 integrin induces
gelatinase B secretion in colon cancer cells. Int J Cancer.
81:90–97. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kähäri VM and Saarialho-Kere U: Matrix
metalloproteinases and their inhibitors in tumour growth and
invasion. Ann Med. 31:34–45. 1999.PubMed/NCBI
|
|
19
|
Burkhardt BR, Lyle R, Qian K, Arnold AS,
Cheng H, Atkinson MA and Zhang YC: Efficient delivery of siRNA into
cytokine-stimulated insulinoma cells silences Fas expression and
inhibits Fas-mediated apoptosis. FEBS Lett. 580:553–560. 2006.
View Article : Google Scholar
|
|
20
|
Chu CY and Rana TM: Potent RNAi by short
RNA triggers. RNA. 14:1714–1719. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mungall BA, Schopman NC, Lambeth LS and
Doran TJ: Inhibition of Henipavirus infection by RNA interference.
Antiviral Res. 80:324–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Devi GR: siRNA-based approaches in cancer
therapy. Cancer Gene Ther. 13:819–829. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Delgado R and Regueiro BJ: The future of
HIV infection: gene therapy and RNA interference. Enferm Infecc
Microbiol Clin. 23:76–83. 2005. View Article : Google Scholar
|
|
24
|
Chen Y, Chen H, Hoffmann A, Cool DR, Diz
DI, Chappell MC, Chen AF and Morris M: Adenovirus-mediated
small-interference RNA for in vivo silencing of angiotensin AT1a
receptors in mouse brain. Hypertension. 47:145–146. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matters GL, Harms JF, McGovern CO,
Jayakumar C, Crepin K, Smith ZP, Nelson MC, Stock H, Fenn CW,
Kaiser J, Kester M and Smith JP: Growth of human pancreatic cancer
is inhibited by down-regulation of gastrin gene expression.
Pancreas. 38:151–161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stein U, Walther W, Stege A, Kaszubiak A,
Fichtner I and Lage H: Complete in vivo reversal of the multidrug
resistance phenotype by jet-injection of anti-MDR1 short hairpin
RNA-encoding plasmid DNA. Mol Ther. 16:178–186. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agrez M, Chen A, Cone RI, Pytela R and
Sheppard D: The alpha v beta 6 integrin promotes proliferation of
colon carcinoma cells through a unique region of the beta 6
cytoplasmic domain. J Cell Biol. 127:547–556. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cassoni P, Marrocco T, Bussolati B, Allia
E, Munaron L, Sapino A and Bussolati G: Oxytocin induces
proliferation and migration in immortalized human dermal
microvascular endothelial cells and human breast tumor-derived
endothelial cells. Mol Cancer Res. 4:351–359. 2006. View Article : Google Scholar
|
|
30
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu X, Niu J, Dorahy DJ, Scott R and Agrez
MV: Integrin alpha(v) beta6-associated ERK2 mediates MMP-9
secretion in colon cancer cells. Br J Cancer. 87:348–351. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Godier A and Hunt BJ: Plasminogen
receptors and their role in the pathogenesis of inflammatory,
autoimmune and malignant disease. J Thromb Haemost. 11:26–34. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ahmed N, Oliva K, Wang Y, Quinn M and Rice
G: Downregulation of urokinase plasminogen activator receptor
expression inhibits Erk signalling with concomitant suppression of
invasiveness due to loss of uPAR-beta1 integrin complex in colon
cancer cells. Br J Cancer. 89:374–384. 2003. View Article : Google Scholar
|
|
34
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koul HK, Pal1 M and Koul S: Role of p38
MAP Kinase signal transduction in solid tumors. Genes Cancer.
4:11–12. 2013.PubMed/NCBI
|
|
36
|
Edick MJ, Tesfay L, Lamb LE, Knudsen BS
and Miranti CK: Inhibition of integrin-mediated crosstalk with
epidermal growth factor receptor/Erk or Src signaling pathways in
autophagic prostate epithelial cells induces caspase-independent
death. Mol Biol Cell. 18:2481–2490. 2007. View Article : Google Scholar
|
|
37
|
Ahmed N, Niu J, Dorahy DJ, Gu XH, Andrews
S, Meldrum CJ, Scott RJ, Baker MS, Macreadie IG and Agrez MV:
Direct integrin alphavbeta6-ERK binding: implications for tumour
growth. Oncogene. 21:1370–1380. 2002. View Article : Google Scholar : PubMed/NCBI
|