Introduction
MicroRNAs (miRNAs) are a class of small cellular
RNAs of 18–25 nucleotides in length that can play important
regulatory roles in many cell events, including tumorigenesis and
organ development (1–3). The biogenesis pathway of miRNAs
contain more than one process, and there are six steps from miRNA
genes to mature miRNA in miRISC, including transcription, cropping,
export, dicing, strand selection and miRISC assembly. A variety of
enzyme proteins are involved in these biological processes, such as
RNA polymerase II (Pol II) and Drosha-DGCR8. Exp5-RanGTP exhibited
a biological reaction within the nucleus, whereas Dicer-TRBP and
Dicer-TRBP-Ago achieved their function in the cytoplasm (4). miRNAs can direct RISC to downregulate
gene mRNA expression by mRNA cleavage or translational repression
(1). Various calculation methods
have been used to predict the targeted genes and target sites of
miRNA, and it is estimated that over one third of human genes
appear to be conserved miRNA targets (5).
The role of miRNA has been increasingly emphasized
in the development and progression of liver disease. Results of
previous studies have shown that miRNAs were associated with
numerous types of liver disease, including non-alcoholic fatty
liver disease, viral hepatitis and hepatocellular carcinoma (HCC)
(6). HCC is the most common primary
malignancy of the liver (7). The
abnormal expression of miRNA in HCC has been previously identified
and verified, with miR-21, miR-135a, miR-146a, miR-151, miR-221,
miR-222, miR-192, miR-801, miR-194 and miR-618 being upregulated,
and let-7g, miR-22, miR-26a, miR-27a, miR-29c, miR-122, miR-320,
miR-491, miR-650 and miR-145 being downregulated in HCC.
Additionally, the disorder of miRNA expression occurs in HCC
associated with HBV or HCV (6,8). A
lower expression of miR-145 is also associated with many other
tumors, including breast, colon, prostate, lung, bladder,
pituitary, B-cell malignancies and ovarian. In addition, miR-145 is
known to be important in the development of tumor, including cell
growth, invasion and metastasis (9). However, the underlying mechanism
involved remains to be determined.
A previous study demonstrated that some miRNAs may
function as oncogenes or tumor suppressors (10). miR-145 is usually considered to be a
tumor-suppressor gene. It can inhibit the occurrence and
development of cancer by regulating oncogenes and/or genes that
control cell growth, differentiation or apoptosis (11–13). A
handful of target genes of miR-145 have been identified and
validated, including YES and STAT1 (14), mucin 1 (13), HDAC2 (15), EGFR and NUDT1 (16), and CTGF (17). It has been found that miR-145 can
repress the migration and invasion of tumor cells by targeting A
disintegrin and metalloproteinase 17 (ADAM17) in several malignant
tumors (18–20). ADAM17, also known as
TNF-α-converting enzyme (TACE), plays a vital role during the
ectodomain shedding of TNF-α (21,22),
and contributes to the progression of breast cancer and glioma
(23,24). TNF-α induces the NF-κB signaling
pathway to mediate the matrix metalloproteinase-9 (MMP-9)
expression (25,26). Findings of a recent study showed
that ADAM17 can mediate MMP-9 expression in lung epithelial cells
(27). Therefore, we hypothesized
that miR-145 is able to inhibit the HCC cell growth by moderating
the ADAM17/MMP-9 pathway. In the present study, we aimed to
identify the association between this pathway and
hepatocarcinogenesis and determine whether miR-145 is a promising
biomarker in the diagnosis and treatment of HCC.
Materials and methods
Cell culture
SMMC-7721, BEL-7402, Huh-7 and HEK-293T cells were
purchased from the Cell Bank of the Chinese Academy of Sciences.
SMMC-7721 and BEL-7402 cells were cultured in RPMI-1640 medium,
supplemented with 10% (v/v) fetal bovine serum (FBS), while Huh-7
and HEK-293T cells were maintained in DMEM high-glucose medium
supplemented with 10% (v/v) FBS (all from Gibco, Invitrogen Life
Technologies, Grand Island, NY, USA). The media contained 100 U/ml
penicillin and 100 μg/ml streptomycin. The cells were incubated at
37°C and supplemented with 5% CO2 in the humidified
chamber.
RNA oligonucleotides and
transfection
The pre-miR™ miR-145 precursors (miR-145) and
pre-miR™ miRNA precursor negative control (NC) were obtained from
Ambion, Inc. (Austin, TX, USA). Cell transfection was performed
using siPORT™ NeoFX™ Transfection Agent (Ambion) according to the
manufacturer’s instructions.
Plasmid construction and
transfection
The human ADAM17 mRNA sequence (NM_003183.4) was
used to design siRNA target sequences according to the design
principle of shRNA (28). Four
siRNA target sequences (971–991, 1658–1678, 1864–1884 and
2540–2560) were selected. A negative control (shNC) siRNA sequence
was used as a control for ADAM17 siRNA (Table I). CACC was added at the end of the
sense strand template, which is complementary with the formed
sticky end after BbsI enzyme digestion. GATC was added at
the end of the antisense strand template, which is complementary
with the formed sticky end after BamHI enzyme digestion. A
stem-loop-stem conformation was selected to avoid the formation of
termination signal. The structure of T6 was selected as the shRNA
transcription termination sequence. DNA oligonucleotides chemically
were purchased by the Shanghai GenePharma Co., Ltd. (Shanghai,
China). The shRNA-annealed oligonucleotides were ligated into
pGPU6/GFP/Neo siRNA expression vectors (GenePharma) between the
BbsI and BamHI sites by T4 DNA ligase (Takara Bio
Inc., Shiga, Japan). The orientation of the inserted shRNA
cassettes was verified by restriction enzyme analysis and DNA
sequencing. Cell transfection was performed using Lipofectamine
2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according
to the manufacturer’s instructions.
 | Table ITarget sequences for RNA interference
in ADAM17 gene. |
Table I
Target sequences for RNA interference
in ADAM17 gene.
| shRNA vector | Target
sequence |
|---|
| 971-ADAM17 |
GGAACACTTCATGGGATAATG |
| 1658-ADAM17 |
GCATCATGTATCTGAACAACG |
| 1864-ADAM17 |
GCTGAAGATGACACTGTTTGC |
| 2540-ADAM17 |
GCACAGCTGCCAAGTCATTTG |
| Negative
control |
GTTCTCCGAACGTGTCACGT |
Quantitative PCR and semi-quantitative
RT-PCR
Total RNA was extracted from cultured cells using
TRIzol (Invitrogen). Quantitative PCR (qPCR) was carried out to
quantify the expression of miR-145. The TaqMan MicroRNA
Transcription kit, TaqMan Universal Master Mix II and TaqMan assays
were used according to the manufacturer’s instructions (Invitrogen
Life Technologies). U6 snRNA was used as the normalization control.
Each sample was tested in triplicate and the relative gene
expression was determined. The data were calculated using the
2−ΔΔCt method.
Semi-quantitative RT-PCR was performed to detect the
expression of ADAM17. Total RNA (2 μg) was used for synthesis of
Oligo(dT)-primed single-stranded cDNA using a First Strand cDNA
Synthesis kit (Fermentas, Burlington, Canada). cDNA products were
then amplified using Platinum PCR SuperMix, with GAPDH serving as
an internal control for the total cDNA content. The oligonucleotide
sequences used as PCR primers were: ADAM17 (forward), 5′-ATGACA
TCTATCGGAACAC-3′, and (reverse), 5′-GCTATAATAAGC CTTTGGAC-3′; GAPDH
(forward), 5′-AATCCCATCACC ATCTTCC-3′ and (reverse),
5′-CATCACGCCACAGTTTCC-3′. PCR products were visualized by
electrophoresis on ethidium bromide-stained 1.5% agarose gels, and
the intensities of the bands were measured with the aid of Bio-Rad
Image Analysis software. ADAM17 expression was evaluated by
determining the ADAM17 mRNA/GAPDH mRNA ratio. Experiments were
repeated at least three times.
Western blot analysis
The cells were treated in lysis buffer (50 mM
Tris-HCl, 0.5% sodium deoxycholate, 25 mM EDTA) containing protease
inhibitor cocktail (Roche Diagnostics GmbH Mannheim, Germany) for
30 min on ice. Equal amounts of protein were separated by 10%
SDS-PAGE gels, and transferred onto polyvinylidene difluoride
membranes. After blocking with 5% skim milk solution for 2 h, the
membrane was incubated with antibodies against ADAM17 (Abcam,
Cambridge, UK) or MMP-9 (Cell Signaling, Danvers, MA, USA) or
β-actin (Proteintech, Chicago, IL, USA) at 4°C overnight. The
appropriate horseradish-conjugated secondary antibody was incubated
for 1 h at room temperature, the protein signal was then identified
using the chemiluminescence method (ECL; Auragene Bioscience
Corporation, Hunan, China). Band intensities were quantified using
Bio-Rad image analysis software. Experiments were repeated at least
three times.
Luciferase activity assay
A 500-bp sequence containing the predicted miR-145
binding site at the 3′UTRs of ADAM17 or a 500-bp sequence
containing a scrambled sequence was cloned into the
XhoI/NotI site of the psiCHECK-2 vector (Promega,
Madison, WI, USA) to generate PsiCHECK-2-ADAM17-WT and
PsiCHECK-2-ADAM17-mut vectors, respectively. SMMC-7721 and HEK-293T
cells were cultured in 24-well plates, and co-transfected with
PsiCHECK-2-ADAM17-WT and PsiCHECK-2-ADAM17-mut, and miR-145
precursor or the negative control using Lipofectamine 2000
(Invitrogen). Luciferase assays were performed using a luciferase
assay kit (Promega) according to the manufacturer’s instructions.
Firefly luciferase was used for normalization. Experiments were
repeated at least three times.
Cell proliferation assay
SMMC-7721 cells (4×103 cells/well) were
plated in 96-well plates and incubated for 24 h. An MTT assay was
performed to measure cell viability at 24, 48 and 72 h after
transfection. The absorbance at 490 nm was measured using a
microplate reader (Bio-Tek Instruments, Winooski, VT, USA). Each
experiment was performed in six replicate wells and independently
repeated three times.
Colony formation assay
SMMC7721 cells were collected at 48 h after
transfection, counted and seeded in 6-well plates at
4×102 cells/well. During colony growth, fresh culture
medium was replaced every 2–3 days and then cells were cultured for
2 weeks. The cells were washed twice in phosphate-buffered saline
(PBS) and fixed by methanol for 15 min. The fixed cells were
stained with appropriate Giemsa solution for 30 min. The cells were
subsequently washed slowly in running water and air-dried. Colonies
were counted only when they contained >50 cells. The ability of
colony formation was assessed by the colony formation rate,
calculated as the number of colonies/number of seeded cells × 100%.
Experiments were repeated at least three times.
Cell cycle analysis
SMMC7721 cells were trypsinized at 48 h after
transfection, and washed in PBS, fixed with 500 μl of 70% ethanol
at −20°C overnight. Subsequently, the cells were stained with
propidium iodide (PI) solution at 37°C for 30 min. Analysis was
performed on a FACS flow cytometer (BD Biosciences, San Jose, CA,
USA). Experiments were repeated at least three times.
Cell invasion and migration assay
A transwell chamber (8-μM pore diameter; Corning
Costar) was used to perform the assay. The filter was coated with
Matrigel (BD Biosciences) for examination of cell invasion, while
the filter without coated Matrigel was used for the migration
assay. The transfected cells were resuspended in serum-free medium
and 200 μl cell resuspension solution (cell invasion,
1×105 cells/well; cell migration, 1×106
cells/well) was added to the hydrated Transwell chambers.
Simultaneously, 500 μl complete medium with 10% FBS was added to
the lower chambers. The cells were cultured in the incubator for 12
h (cell migration) or 24 h (cell invasion). Subsequently, the cells
on the upper surface of the membrane were removed with cotton swab.
Filters were fixed in 4% formaldehyde for 30 min and stained with
0.1% crystal violet for 20 min. The cells were counted under a
microscope. Experiments were repeated at least three times.
Statistical analysis
Data are presented as the mean and standard error
(means ± SEM). The two-tailed Student’s t-test, one-way ANOVA,
Mann-Whitney U or Kruskal-Wallis tests were used for data analysis.
The statistical analyses were carried out using SPSS 19.0 for
windows. P<0.05 was considered to indicate a statistically
significant result.
Results
ADAM17 was directly regulated by miR-145
in HCC
The identification of an appropriate target gene for
miRNA using miRNA-target gene prediction software is important in
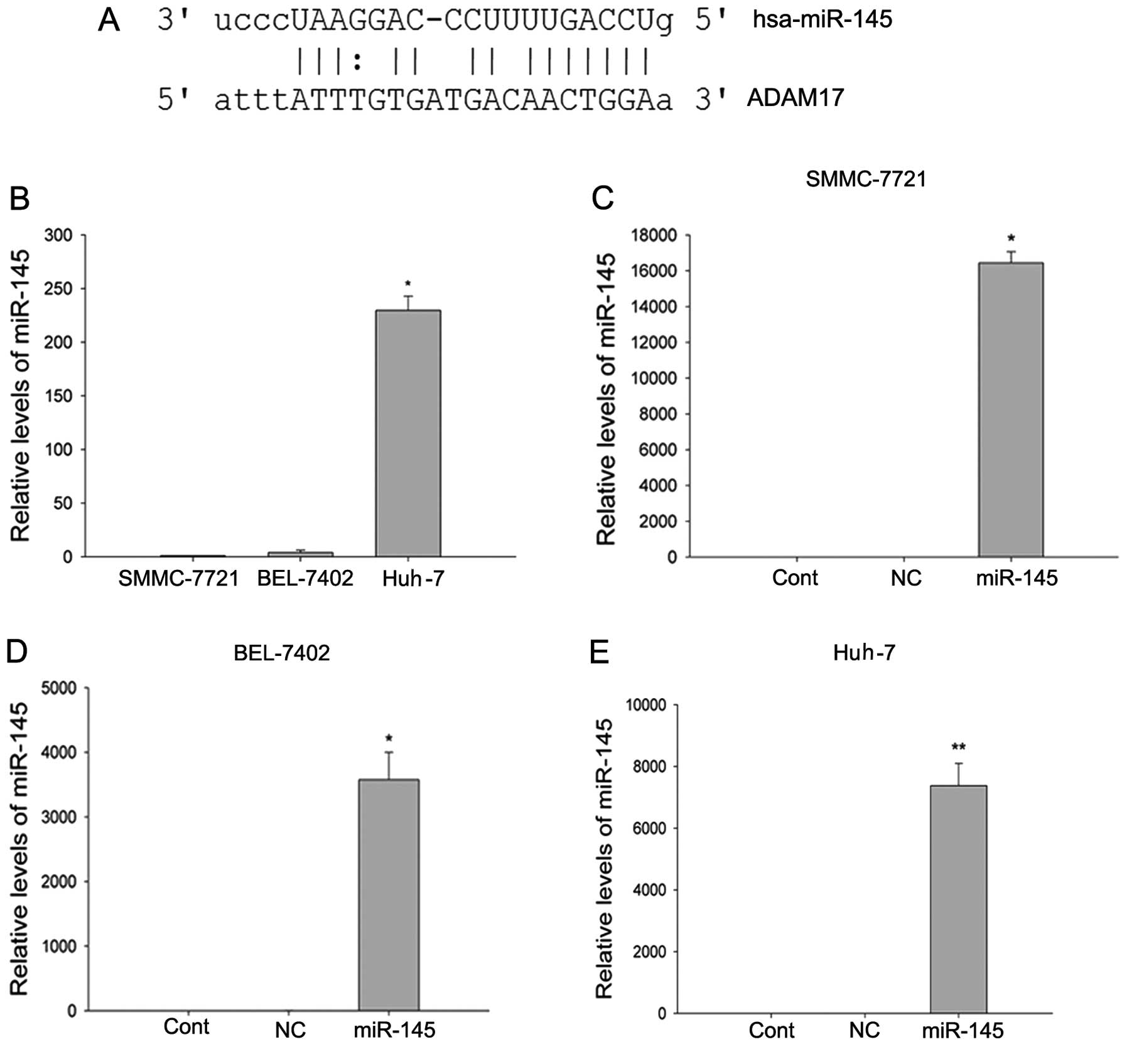
understanding its function. ADAM17 was predicted by TargetScan
(5), PicTar (29) and miRanda (30) (Fig.
1A), and assayed as a target of miR-145. Expression levels of
miR-145 were determined by qPCR in the SMMC-7721, BEL-7402 and
Huh-7 cell lines (Fig. 1B), and
found to be significantly lower in the SMMC-7721 and BEL-7402 cell
lines as compared to the Huh-7 cell lines (P<0.05,
Kruskal-Wallis test). miR-145 precursor and a negative control RNA
were used to transfect the HCC cell line with SMMC7721, BEL-7402
and Huh-7 (Fig. 1C–E). Following
transfection, miR-145 was significantly overexpressed in the three
cell lines (P<0.01, the two-tailed Student’s t-test).
The expression of ADAM17 was detected by RT-PCR and
western blotting, respectively. ADAM17 mRNA expression was
decreased significantly in BEL-7402 and Huh-7 cells, however, it
did not exhibit any change in SMMC-7721 cells (Fig. 2A). ADAM17 protein expression was
markedly decreased in SMMC-7721 and Huh-7 cells, however, no change
was observed in the BEL-7402 cells (Fig. 2B). To determine whether miR-145
directly regulates ADAM17 expression, a luciferase activity assay
was performed. miR-145 overexpression reduced the activity of the
luciferase reporter gene fused to the wild-type ADAM17 3′UTR in
SMMC-7721 (P<0.01, Kruskal-Wallis test) and HEK-293T cells
(P<0.05, one-way ANOVA) (Fig.
2C). These resutlts showed that miR-145 targets ADAM17
directly.
Effect of miR-145 on the cell
proliferation, colony formation and cell cycle in HCC cells
To investigate whether the ectopic expression of
miR-145 inhibits the viability and growth of cells in liver cancer
tumorigenesis, miR-145 precursor or the negative control was
transfected into SMMC-7721 cells. The MTT assay showed that the
overexpression of miR-145 resulted in a significantly decreased
cell proliferation at 24 h (P<0.01, the two-tailed Student’s
t-test), 48 h (P<0.01, the two-tailed Student’s t-test) and 72 h
(P<0.01, Mann-Whitney U test) (Fig.
3A). The colony-forming efficiency of SMMC-7721 cells
transfected with miR-145 precursor was markedly lower than that of
the control group (P<0.05, Mann-Whitney U test) (Fig. 3B). These results demonstrated that
miR-145 overexpression is able to suppress cell growth in HCC.
Distribution of the cell cycle was determined by flow cytometry.
The results showed that miR-145 overexpression resulted in a
slightly increase in the number of cells in the G2 phase of the
cell cycle, although this result was not statistically significant
(P=0.693, the two-tailed Student’s t-test). Similarly, there was no
significant difference for the amount of G1-phase (P=0.798, the
two-tailed Student’s t-test) and S-phase (P=0.877, the two-tailed
Student’s t-test) (Fig. 3C).
miR-145 does not affect cell migration
and invasion ability
Migration and invasion ability of SMMC-7721 cells
transfected with miR-145 precursor or negative control were
determined by Matrigel chamber assays. The results demonstrated
that the overexpression of miR-145 did not exert any significant
effect on the migration (P=0.886, the two-tailed Student’s t-test)
(Fig. 4A) and invasion ability
(P=0.404, the two-tailed Student’s t-test) (Fig. 4B) of SMMC-7721 cell lines.
Silencing of ADAM17 inhibited the growth
of cells and reduced the expression of MMP-9 in SMMC-7721
cells
To clarify the mechanisms by which ADAM17 decreases
cell proliferation, we suppressed ADAM17 expression using siRNA.
RT-PCR and western blot analysis showed that the expression levels
of ADAM17 mRNA and protein were significantly lower (Fig. 5A and B). The MTT assay showed that
this suppression of ADAM17 markedly reduced the proliferation
ability of SMMC-7721 cells relative to the control cells at 72 h
(P<0.01, Mann-Whitney U test), however it did not exhibit any
change at 24 h (P=0.762, the two-tailed Student’s t-test) and 48 h
(P=0.083, the two-tailed Student’s t-test) (Fig. 5C). The colony-forming test assay
demonstrated that the overexpression of miR-145 markedly inhibited
cell proliferation in SMMC-7721 cells (P<0.05, Mann-Whitney U
test) (Fig. 5D). At the same time,
ADAM17 siRNA caused th edownregulation of MMP-9 (Fig. 5E). It has been reported that MMP-9
proteolytically activates TGF-β and promotes tumor growth and
invasion (31). Therefore, these
results suggested that suppression of cell growth may be connected
to ADAM17 siRNA reducing the expression of MMP-9.
Discussion
Accumulating evidence suggests a role for microRNAs
(miRNAs) whereby they regulate cancer development and progression
(32–34). It is of vital importance to
understand the functions and mechanisms of cancer-specific miRNAs
for cancer therapy and prevention. In the present study, we found
that ADAM17 is a target gene of miR-145. The experimental results
showed that the overexpression of miR-145 led to a marked reduction
of ADAM17 expression in different HCC cells. Moreover, the ability
of miR-145 to downregulate ADAM17 expression was performed through
a drect combination with the complimentary sequence in the 3′UTR of
ADAM17 mRNA to the miR-145. Notably, miR-145 significantly
inhibited the growth activity of SMMC-7721 cells. These findings
suggest that miR-145 plays a tumor-suppressor role in
hepatocarcinogenesis by targeting ADAM17.
ADAM17 is one of the members of the family of ADAMs
(a disintegrin and metalloproteinase). Previous studies have
confirmed that ADAM17 is important in cancer formation and
progression (35–37). Notably, apart from miR-145, various
miRNA molecules can regulate the expression of ADAM17. For example,
miR-26 mediated the observed effects on white and brite
adipogenesis by targeting ADAM17 (38). Chen et al reported that
hydroquinone-induced miR-122 downregulation resulted in the
upregulation of ADAM17 expression in human leukemic cells (39). miR-122 affects intrahepatic
metastasis in HCC by regulating the expression of ADAM17 (40). Thus, the downregulation of ADAM17 is
important in miR-145-mediated suppression of cell growth and
metastasis. In the present study, the ADAM17 siRNA can reduce the
protein expression of ADAM17 in SMMC-7721 cells and suppress the
growth and proliferation activity of SMMC-7721 cells, which serves
as evidence for our previous hypothesis.
The molecular mechanism of ADAM17 has been shown to
be responsible for the growth inhibition of cancer cells. Das et
al reported that ADAM17 silencing decreased the expression of
vascular endothelial growth factor-A (VEGF-A) and MMP-9 in murine
colon carcinoma cells (MC38CEA) and exhibited its antitumor immune
response (41). Xiao et al
found that the knockdown of ADAM17 can deactivate the EGFR-MEK-ERK
signaling pathway, resulting in the downregulation of MMP-2 and
MMP-9, therby reducing the invasive ability of prostate cancer
cells (42). ADAM17 siRNA may
suppress cancer growth by decreasing the expression of MMP-9. MMP-9
is expressed as a 92-kDa proenzyme and is converted to the active
83-kDa mature enzyme (43). MMP-9
plays a significant role in different stages of tumor progression
and metastasis, and its inhibitory role in cell signaling pathways
can block MMP-9-mediated cell migration and metastasis (44–46).
Results of the present study have shown that the silencing of
ADAM17 reduces the expression of MMP-9 in SMMC-7721 cells.
Therefore, we suggest that there is likely a link between the two
proteins in tumorigenesis. In addition, it is possible that the
miR-145-mediated suppression of cell proliferation was performed
through the ADAM17/MMP-9 pathway.
As a promising miRNA, the therapeutic potential of
miR-145 is important in examining the relationship between miR-145
and other mRNAs in hepatocarcinogenesis. The present study has
demonstrated that miR-145 suppressed the proliferation of HCC cells
by targeting ADAM17, suggesting miR-145 has the potential to be
applied in tumor treatment. This result led to investigatation of
the anti-neoplastic effect of miR-145. In general, the results
suggest that miR-145 may be crucial in miRNA-base anticancer
therapies in the future.
Acknowledgements
We would like to thank Dr Zhaojun Duan for her
excellent technical assistance. This study was supported by the
National Natural Science Foundation of China (no. 81270501), and
the Exploring Program of Central South University
(2012QNZT080).
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar
|
|
6
|
Kerr TA, Korenblat KM and Davidson NO:
MicroRNAs and liver disease. Transl Res. 157:241–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
8
|
Takahashi K, Yan I, Wen HJ and Patel T:
microRNAs in liver disease: from diagnostics to therapeutics. Clin
Biochem. 46:946–952. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
10
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao C, Xu Y, Zhang Y, et al:
Downregulation of miR-145 contributes to lung adenocarcinoma cell
growth to form brain metastases. Oncol Rep. 30:2027–2034.
2013.PubMed/NCBI
|
|
12
|
Havelange V, Stauffer N, Heaphy CC, et al:
Functional implications of microRNAs in acute myeloid leukemia by
integrating microRNA and messenger RNA expression profiling.
Cancer. 117:4696–4706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gregersen LH, Jacobsen AB, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-145 targets YES and
STAT1 in colon cancer cells. PLoS One. 5:e88362010.
View Article : Google Scholar
|
|
15
|
Noh JH, Chang YG, Kim MG, et al: MiR-145
functions as a tumor suppressor by directly targeting histone
deacetylase 2 in liver cancer. Cancer Lett. 335:455–462. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho WC, Chow AS and Au JS: MiR-145
inhibits cell proliferation of human lung adenocarcinoma by
targeting EGFR and NUDT1. RNA Biol. 8:125–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee HK, Bier A, Cazacu S, et al:
MicroRNA-145 is downregulated in glial tumors and regulates glioma
cell migration by targeting connective tissue growth factor. PLoS
One. 8:e546522013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doberstein K, Steinmeyer N, Hartmetz AK,
et al: MicroRNA-145 targets the metalloprotease ADAM17 and is
suppressed in renal cell carcinoma patients. Neoplasia. 15:218–230.
2013.PubMed/NCBI
|
|
19
|
Lu Y, Chopp M, Zheng X, Katakowski M,
Buller B and Jiang F: MiR-145 reduces ADAM17 expression and
inhibits in vitro migration and invasion of glioma cells.
Oncol Rep. 29:67–72. 2013.PubMed/NCBI
|
|
20
|
Yang XW, Zhang LJ, Huang XH, et al:
miR-145 suppresses cell invasion in hepatocellular carcinoma cells:
miR-145 targets ADAM17. Hepatol Res. 44:551–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Black RA, Rauch CT, Kozlosky CJ, et al: A
metalloproteinase disintegrin that releases tumour-necrosis
factor-α from cells. Nature. 385:729–733. 1997.
|
|
22
|
Mohan MJ, Seaton T, Mitchell J, et al: The
tumor necrosis factor-α converting enzyme (TACE): a unique
metalloproteinase with highly defined substrate selectivity.
Biochemistry. 41:9462–9469. 2002.
|
|
23
|
Zheng X, Jiang F, Katakowski M, Zhang ZG,
Lu QE and Chopp M: ADAM17 promotes breast cancer cell malignant
phenotype through EGFR-PI3K-AKT activation. Cancer Biol Ther.
8:1045–1054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng X, Jiang F, Katakowski M, Lu Y and
Chopp M: ADAM17 promotes glioma cell malignant phenotype. Mol
Carcinog. 51:150–164. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee SO, Jeong YJ, Yu MH, et al: Wogonin
suppresses TNF-α-induced MMP-9 expression by blocking the NF-κB
activation via MAPK signaling pathways in human aortic smooth
muscle cells. Biochem Biophys Res Commun. 351:118–125. 2006.
|
|
26
|
Balasubramanian S, Fan M, Messmer-Blust
AF, et al: The interferon-γ-induced GTPase, mGBP-2, inhibits tumor
necrosis factor α (TNF-α) induction of matrix metalloproteinase-9
(MMP-9) by inhibiting NF-κB and Rac protein. J Biol Chem.
286:20054–20064. 2011.
|
|
27
|
Li YQ, Yan JP, Xu WL, et al: ADAM17
mediates MMP9 expression in lung epithelial cells. PLoS One.
8:e517012013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Elbashir SM, Harborth J, Weber K and
Tuschl T: Analysis of gene function in somatic mammalian cells
using small interfering RNAs. Methods. 26:199–213. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krek A, Grün D, Poy MN, et al:
Combinatorial microRNA target predictions. Nat Genet. 37:495–500.
2005. View
Article : Google Scholar
|
|
30
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar
|
|
31
|
Yu Q and Stamenkovic I: Cell
surface-localized matrix metalloproteinase-9 proteolytically
activates TGF-β and promotes tumor invasion and angiogenesis. Genes
Dev. 14:163–176. 2000.PubMed/NCBI
|
|
32
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kusenda B, Mraz M, Mayer J and Pospisilova
S: MicroRNA biogenesis, functionality and cancer relevance. Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub. 150:205–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McGowan PM, Ryan BM, Hill AD, McDermott E,
O’Higgins N and Duffy MJ: ADAM-17 expression in breast cancer
correlates with variables of tumor progression. Clin Cancer Res.
13:2335–2343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kenny PA and Bissell MJ: Targeting
TACE-dependent EGFR ligand shedding in breast cancer. J Clin
Invest. 117:337–345. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Borrell-Pagès M, Rojo F, Albanell J,
Baselga J and Arribas J: TACE is required for the activation of the
EGFR by TGF-α in tumors. EMBO J. 22:1114–1124. 2003.
|
|
38
|
Karbiener M, Pisani DF, Frontini A, et al:
MicroRNA-26 family is required for human adipogenesis and drives
characteristics of brown adipocytes. Stem Cells. 32:1578–1590.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen YJ and Chang LS: Hydroquinone-induced
miR-122 down-regulation elicits ADAM17 up-regulation, leading to
increased soluble TNF-α production in human leukemia cells with
expressed Bcr/Abl. Biochem Pharmacol. 86:620–631. 2013.PubMed/NCBI
|
|
40
|
Tsai WC, Hsu PW, Lai TC, et al:
MicroRNA-122, a tumor suppressor microRNA that regulates
intrahepatic metastasis of hepatocellular carcinoma. Hepatology.
49:1571–1582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Das S, Czarnek M, Bzowska M, et al: ADAM17
silencing in mouse colon carcinoma cells: the effect on tumoricidal
cytokines and angiogenesis. PLoS One. 7:e507912012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao LJ, Lin P, Lin F, et al: ADAM17
targets MMP-2 and MMP-9 via EGFR-MEK-ERK pathway activation to
promote prostate cancer cell invasion. Int J Oncol. 40:1714–1724.
2012.PubMed/NCBI
|
|
43
|
Mattu TS, Royle L, Langridge J, et al:
O-glycan analysis of natural human neutrophil gelatinase B using a
combination of normal phase-HPLC and online tandem mass
spectrometry: implications for the domain organization of the
enzyme. Biochemistry. 39:15695–15704. 2000. View Article : Google Scholar
|
|
44
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.
|
|
46
|
Klein T and Bischoff R: Physiology and
pathophysiology of matrix metalloproteases. Amino Acids.
41:271–290. 2011. View Article : Google Scholar : PubMed/NCBI
|