Introduction
The S100 proteins comprise a family of more than 25
different members (1). Found
exclusively in vertebrates, these proteins are involved in
activating specific biochemical pathways to regulate various
cellular functions, including proliferation, survival,
differentiation and motility (2).
The S100 protein family has attracted increasing attention in the
field of cancer research; in particular, S100A4 reportedly
contributes to various aspects of tumor progression, including cell
motility, metastasis and angiogenesis (3–5).
S100A4 was first isolated as the product of a gene that is
differentially expressed in highly metastatic mouse mammary
adenocarcinoma cells (6). It has
since been shown to be specifically upregulated in aggressive and
advanced metastatic tumors relative to non-invasive, non-metastatic
tumors (7,8). Elevated expression of S100A4 has been
found in numerous cancer types; its expression in non-metastatic
cell lines was shown to trigger a more metastatic phenotype
(9,7), whereas decreased S100A4 expression was
associated with a lower metastatic capacity (10,11).
Furthermore, transgenic animal studies have established positive
associations between S100A4 and both metastasis and tumor
development (12–15). Clinical studies have convincingly
demonstrated that significant expression of S100A4 in primary
tumors is indicative of poor prognosis (16–18),
and that S100A4 may be a useful marker for predicting the
development, progression and metastasis of human gastric cancer
(19).
Despite these findings, however, we do not yet fully
understand the exact mechanisms through which S100A4 executes its
pro-metastatic functions. Intracellular S100A4 has been shown to
interact with non-muscle myosin IIA at the leading edge of
migrating cells, which may promote cell migration (20). In addition, S100A4 may modulate the
expression levels of MMP9 and MMP13, thereby regulating invasion
and metastasis in human prostate and breast cancer cells (21,22),
respectively. In esophageal squamous cell carcinoma, the ability of
S100A4 to promote tumor invasion and metastasis is associated with
the upregulation of MMP2 and the down-regulation of E-cadherin
(23). Moreover, Lo et al
showed that S100A4 induced epithelial-mesenchymal transition (EMT)
to maintain the stemness of cancer cells and the tumorigenic
properties of head and neck cancers (24).
In addition to acting intracellularly, some of the
S100 proteins demonstrate extracellular activity by acting as
chemo-attractants. S100A4 can be secreted, and several lines of
evidence suggest that it can induce cytokine networks, such as
those mediated by the inflammatory cytokines IL8, CCL2 and SAA,
thereby enabling tumor cells to engage with angiogenic and
inflammatory stromal cells (25,26).
In this regard, S100A4 is believed to have potential as a highly
prognostic molecular biomarker for metastatic potential, as already
shown for breast, colorectal, gallbladder, pancreatic as well as
other types of cancer (3,18).
However, although data indicate that high-level
expression of S100A4 is associated with increased metastatic
capacity, we are only just beginning to unravel the potential roles
of this protein in chemoresistance. Moderate S100A4 overexpression
was found in a doxorubicin-resistant colon cancer cell line
compared to doxorubicin-sensitive cells (27), whereas S100A4 knockdown was
associated with upregulation of BNIP3, increased sensitivity of
pancreatic ductal adenocarcinoma cell lines to gemcitabine
treatment, and enhanced apoptosis (28). Furthermore, S100A4 mRNA and protein
levels were found to be upregulated in methotrexate (MTX)-resistant
cancer cells and to contribute to MTX resistance (29). Other S100 family proteins have also
been demonstrated to contribute to chemoresistance (30,27).
Despite these previous findings, however, the involvement of S100A4
in the drug responsiveness of gastric cancer remains less well
understood.
Considering the upregulation of S100A4 in metastatic
tumors and the literature correlating its expression with poor
prognosis, we investigated whether S100A4 may mediate
chemotherapeutic resistance in gastric cancer. Here, we reported
that ectopic expression of S100A4 did not promote anticancer drug
resistance in gastric cancer cells, and S100A4 knockdown had little
effect on the survival of drug-treated cells. These data strongly
suggest that, depending on the cell context, the
metastasis-promoting effect of S100A4 may not be positively
correlated with anticancer drug resistance in the clinic.
Materials and methods
Cell culture
The human gastric carcinoma cell lines, AGS, TMC-1,
SNU-1, TMK-1, SCM-1, MKN-45, and KATO III, were cultured in
RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10%
fetal bovine serum (FBS), sodium bicarbonate (2%, w/v), L-glutamine
(0.29 mg/ml), penicillin (100 U/ml), and streptomycin (100 μg/ml)
(Invitrogen) at 37°C in a humidified 5% CO2
incubator.
Antibodies and chemicals
Specific antibodies against S100A4, ribophorin II
(RPN2) and β-actin were obtained from Santa Cruz Biotechnology
(Santa Cruz, CA, USA). Anti-PARP and anti-caspase-3 were obtained
from Cell Signaling Technology (Beverly, MA, USA). Anti-Myc was
purchased from Millipore (Millipore Corporation, Bedford, MA, USA).
Cisplatin was purchased from Sigma (St. Louis, MO, USA).
MTS assays
Cells (5×103) were seeded in 96-well
culture plates, incubated overnight at 37°C in medium containing
10% FBS, and then treated with the indicated concentrations of
anticancer drugs for 48 h. Cell viability was determined using an
MTS colorimetric assay (CellTiter 96® cell proliferation
assay kit; Promega, Madison, WI, USA) as described by the
manufacturer. All experiments were performed at least in
triplicate, on three separate occasions. A dose-response curve was
plotted, and the drug concentration that decreased color
development by 50% (i.e., the IC50 value) was calculated
for each drug. The data are presented as means ± SDs.
RNA interference
For small-interfering RNA (siRNA) knockdown of
S100A4, ON-TARGET plus SMART pool siRNAs against S100A4 were
purchased from Dharmacon Research (Lafayette, CO, USA).
Non-targeting siRNA duplexes were used as negative controls
(Dharmacon Research). Cells were transfected with siRNA using
Lipofectamine RNAiMAX (Invitrogen) and incubated in glucose-free
Opti-MEM (Invitrogen) according to the manufacturer’s
recommendations.
Western blot analysis
Cell extracts were prepared in lysis buffer (50 mM
HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF,
1 mM Na3VO4, 10% glycerol, and a protease
inhibitor cocktail). Equal amounts of proteins were separated by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF)
membranes (Millipore, Billerica, MA, USA). The membranes were
blocked, washed, probed with the indicated primary antibodies,
washed again, and incubated with horseradish peroxidase-conjugated
secondary antibodies for 1 h. Finally, the blots were washed, and
then developed using enhanced chemiluminescence (ECL) reagents
(Millipore) according to the manufacturer’s protocol.
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
RNA was isolated from cultured cells using the
TRIzol reagent (Invitrogen) according to the manufacturer’s
instructions, and cDNA was synthesized from 2 μg of total RNA by
reverse transcription using the ImProm-II Reverse Transcriptase kit
(Promega) with oligo(dT)12–18 primers. The resulting
cDNA was used for subsequent PCR using specific PCR primers for
S100A4 (forward, 5′-ATGGCGTGCCCTCTGGAG-3′ and reverse,
5′-TTTCTTCCTGGGCTGC-3′).
Site-directed mutagenesis
The full-length human S100A4 cDNA was inserted into
pcDNA3.1 (Invitrogen) to express the S100A4-Myc recombinant
protein. Asp-10 was substituted with a valine residue using a
QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla,
CA, USA) using specific DNA oligonucleotides (D10V forward,
5′-GAGAAGGCCCTGGTTGTGATGGTGTCC-3′ and D10V reverse,
5′-GGACACCATCACAACCAGGGCCTTCTC-3′).
Real-time cell analysis (RTCA)
system
For continuous monitoring of cell migration, cells
(1×104 cells/well) suspended in serum-free medium were
seeded into the upper compartment of CIM-plates 16 (Roche,
Mannheim, Germany). The lower compartment was then filled with
medium containing 10% FBS and incubated in the RTCA station
(xCELLigence System, Roche). Cell migration was monitored for 24 h
with impedance measured every 15 min. Cell impedance was
represented as cell index (CI) = (Zi-Z0)
[Ohm]/15[Ohm], where Z0 was the background resistance
and Zi was the resistance at a given time-point. A
normalized cell index was determined as the cell index at a given
time-point (CIti) divided by the cell index at the
normalization time-point (CInml_time).
Cell migration assay
The in vitro cell migration assay was
performed in Transwell chambers (Millipore), using 8.0-μm pore-size
filters according to the manufacturer’s recommendations. Briefly,
1×105 cells were suspended in serum-free DMEM and seeded
to the upper compartment of a Transwell insert, while the lower
compartment was loaded with a 24-well dish containing medium
supplemented with 10% FBS. After incubation at 37°C in 5%
CO2 for 18 h, a cotton swab was used to remove the
non-migrated cells from the upper surface of the membrane. The
cells that had migrated through the membrane and adhered to the
lower surface were fixed with methanol and then stained with
crystal violet (1% crystal violet in 75% ethanol). The cells were
examined under a microscope and counted. Experiments were performed
in triplicate, and the results were calculated by averaging the
total number of cells from three membranes.
Statistical analyses
All experiments were performed in triplicate. The
significances of between-group differences were determined using
the Student’s t-test. A P-value <0.05 was considered to indicate
a statistically significant difference.
Results
S100A4 expression and cell migration
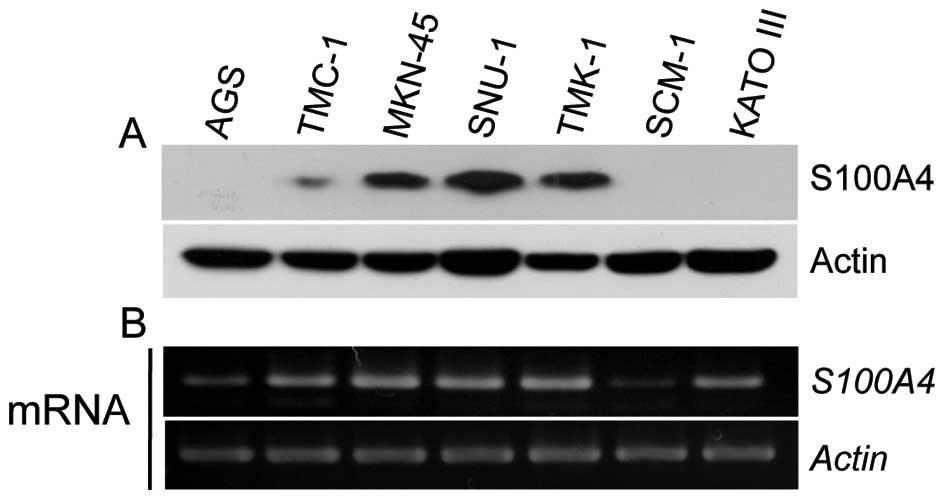
First, we examined the expression levels of S100A4
in seven gastric cancer cell lines. S100A4 was found to be highly
expressed in MKN-45, SNU-1 and TMK-1 cells at both the protein and
mRNA levels, whereas AGS and SCM-1 cells exhibited much lower
expression of S100A4 at the protein and mRNA levels (Fig. 1). We therefore used AGS, MKN-45,
TMK-1, and SCM-1 cells in our subsequent studies. To examine the
functional significance of S100A4 in the cell migration of these
gastric cancer cell lines, we used the RTCA system, which is a
label-free, real-time automated continuous-monitoring platform that
assesses cell migration by measuring changes in the electrical
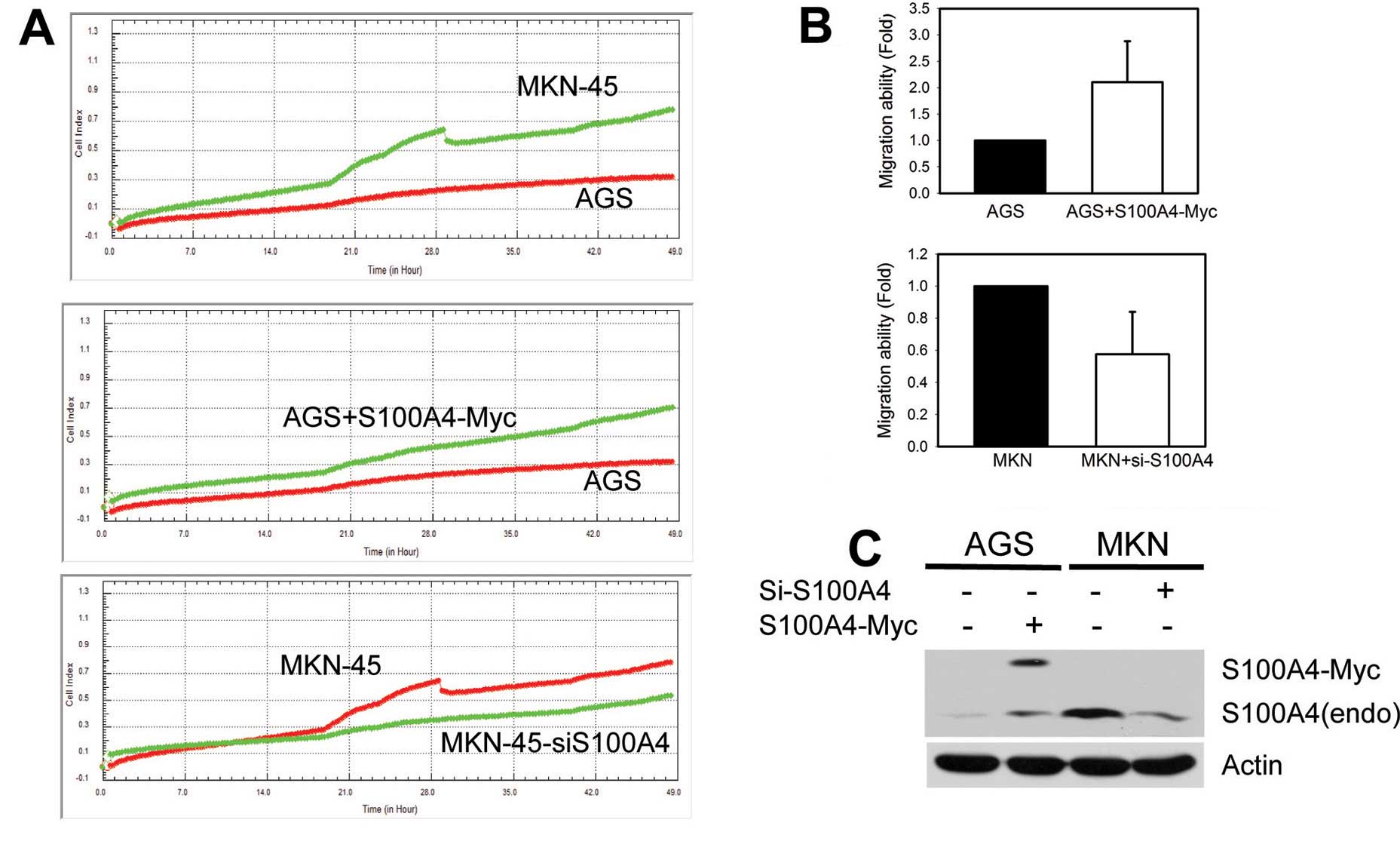
impedance at the electrode/cell interface. As shown in Fig. 2, higher expression levels of
endogenous S100A4 were significantly associated with the higher
migration ability of MKN-45 cells compared to AGS cells. Moreover,
the migration ability of AGS cells was markedly enhanced by ectopic
expression of S100A-Myc, whereas siRNA-mediated knockdown of
endogenous S100A4 expression attenuated the cell migration of
MKN-45 cells (Fig. 2A–C).
Consistent with the results from previous reports, our data
confirmed that S100A4 plays a critical role in the cell migration
of gastric cancer cell lines.
S100A4 expression and drug
responsiveness
To investigate the role of S100A4 in anticancer drug
resistance, AGS cells expressing exogenous S100A4-Myc were exposed
to the half-maximal inhibitory concentrations (IC50) of
six conventional anticancer drugs for 48 h. Immunoblot analysis of
apoptosis-related protein markers revealed that overexpression of
S100A4 slightly decreased the cisplatin-induced cleavage of
caspase-3 and poly(ADP-ribose) polymerase (PARP) in AGS cells
(Fig. 3A). MTS-based cell viability
analyses showed that overexpression of S100A4 slightly (but not
significantly) increased cell viability relative to empty-vector
controls in AGS cells treated with any of the tested drugs
(Fig. 3B). Next, MKN-45 cells were
subjected to siRNA-mediated knockdown of S100A4 (siS100A4) for 24 h
and then challenged with anticancer drugs for 48 h. S100A4
knockdown slightly increased the cisplatin-induced cleavage of
caspase-3 and PARP, and decreased BCL2 protein levels (Fig. 4A). However, our MTS-based analysis
of cell viability indicated that knockdown of S100A4 did not have
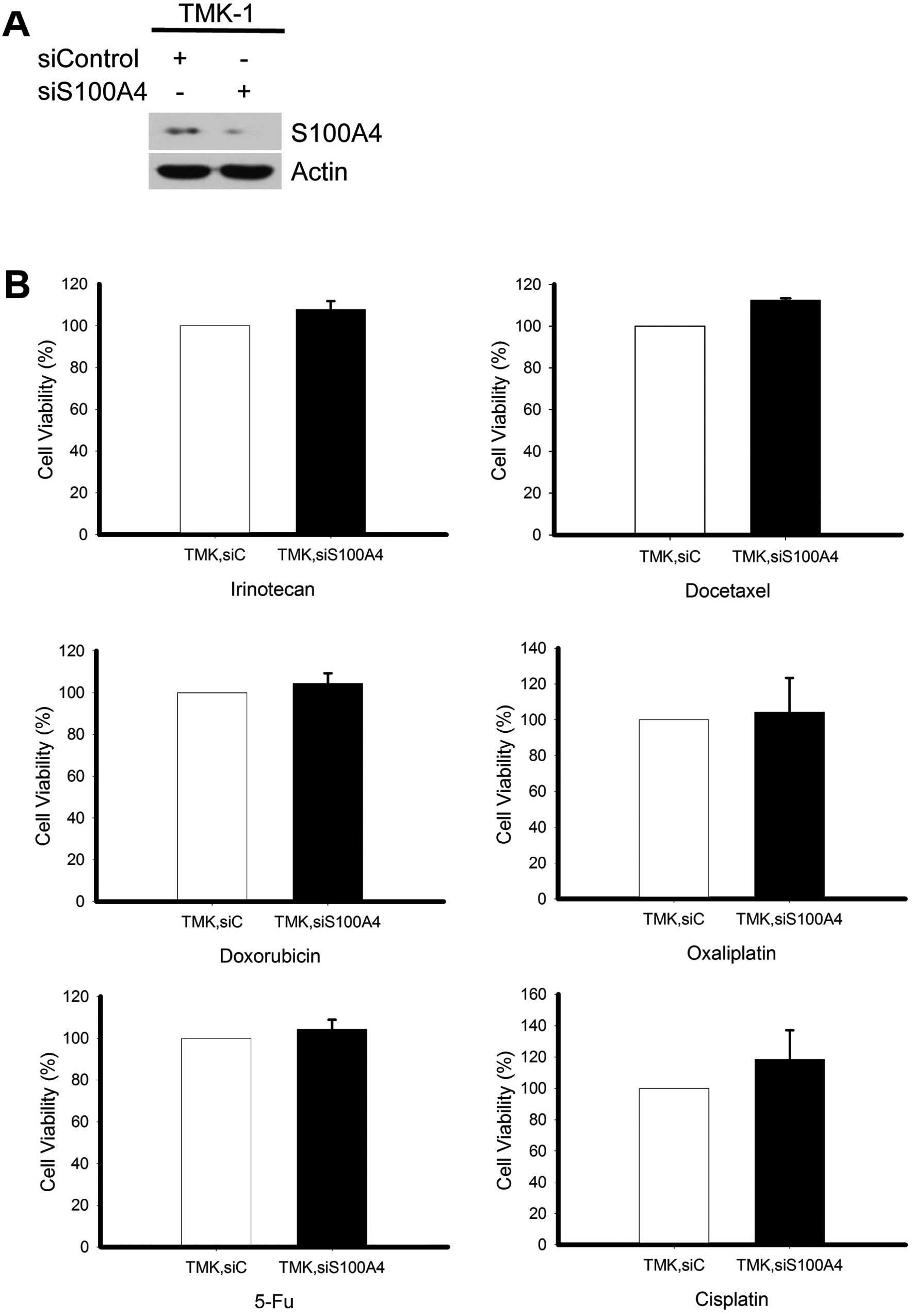
any discernible effect on cell survival (Fig. 4B). A similar pattern of drug
responsiveness was obtained in TMK-1 cells, wherein significant
knockdown of S100A4 (Fig. 5A) did
not alter the resistance to various anticancer drugs (Fig. 5B).
The S100A4 D10V polymorphism affects the
cell migration ability of gastric cancer cells
Numerous proteins have been identified as
potentially interacting with S100 proteins, including Annexin II.
(18,31) We selected nonsynonymous
single-nucleotide polymorphisms (SNPs) in S100A4 from dbSNP, and
examined whether they alter the biological function of the protein.
We identified an SNP of S100A4 (NM_002961.2: c.29A>T, rs1803245)
that resulted in the substitution of an Asp residue with a Val
residue (NP002952.1:p.Asp10Val), and further found that it
localizes within a sequence that is conserved among the members of
the S100A protein family (Fig. 6A).
Computer modeling of the S100A4 protein structure showed that the
D10 residue is localized within the binding surface for Annexin II
(Fig. 6B). To examine the potential
significance of this SNP in cancer cell migration and anticancer
drug resistance, we next examined whether cancer cells expressing
this S100A4 protein variant showed changes in their
metastasis-related properties. AGS cells were transfected with
wild-type S100A4D10 (A allele) or S100A4D10V
(T allele), and cell motility was assessed by an in vitro
cell migration assay. Compared to cells expressing
S100A4D10, those expressing S100A4D10V showed
significantly fewer instances of traversing the membrane
(P<0.01) (Fig. 6C and D). This
suggests that S100A4D10V is negatively correlated with
cancer cell motility compared to the wild-type.
The S100A4 D10V polymorphism does not
affect anticancer drug responsiveness in gastric cancer cells
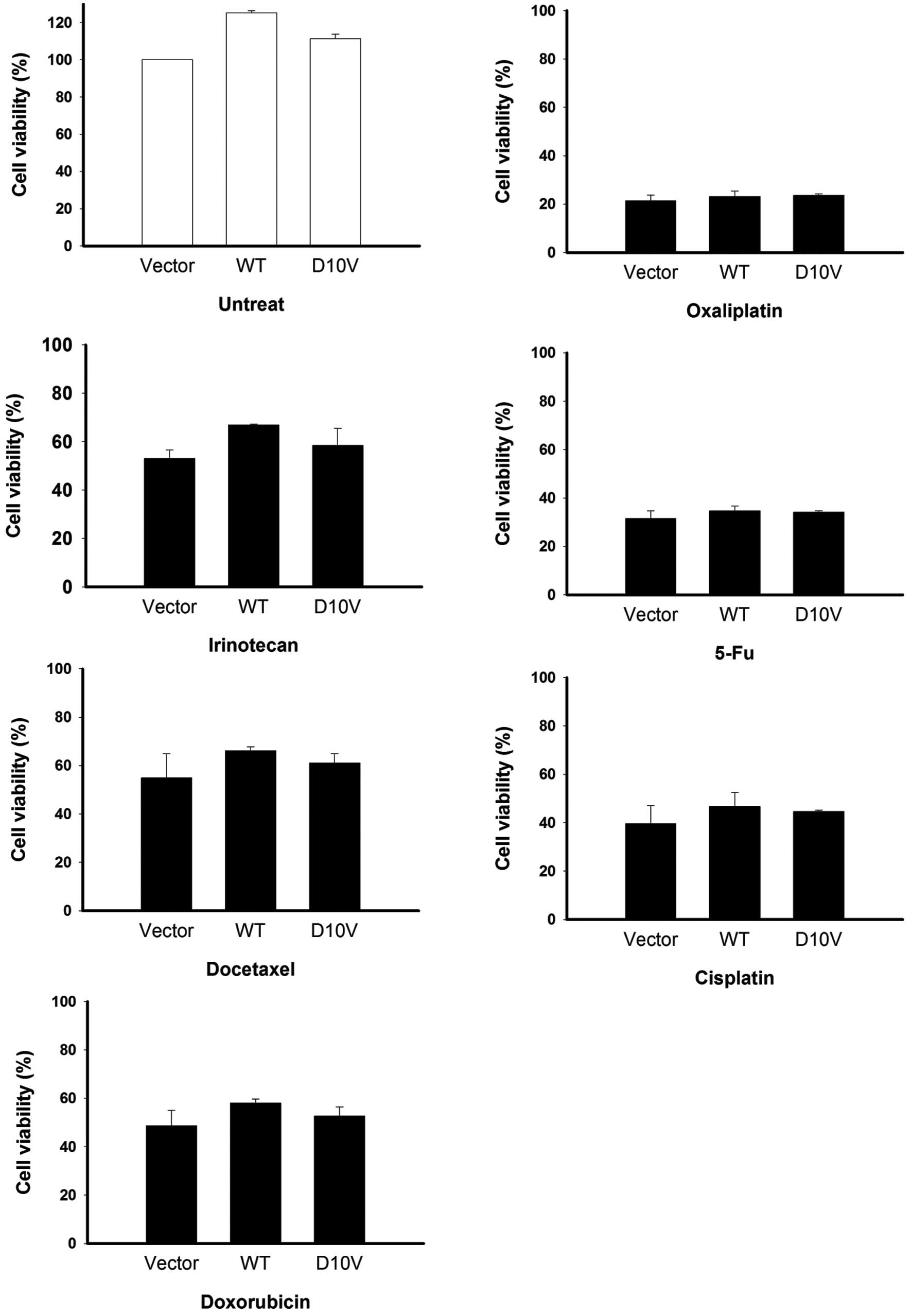
The drug responsiveness of cells expressing
S100A4D10V was further investigated. AGS cells
overexpressing S100A4D10V were individually treated with
the six tested anticancer drugs for 48 h, and cell viability was
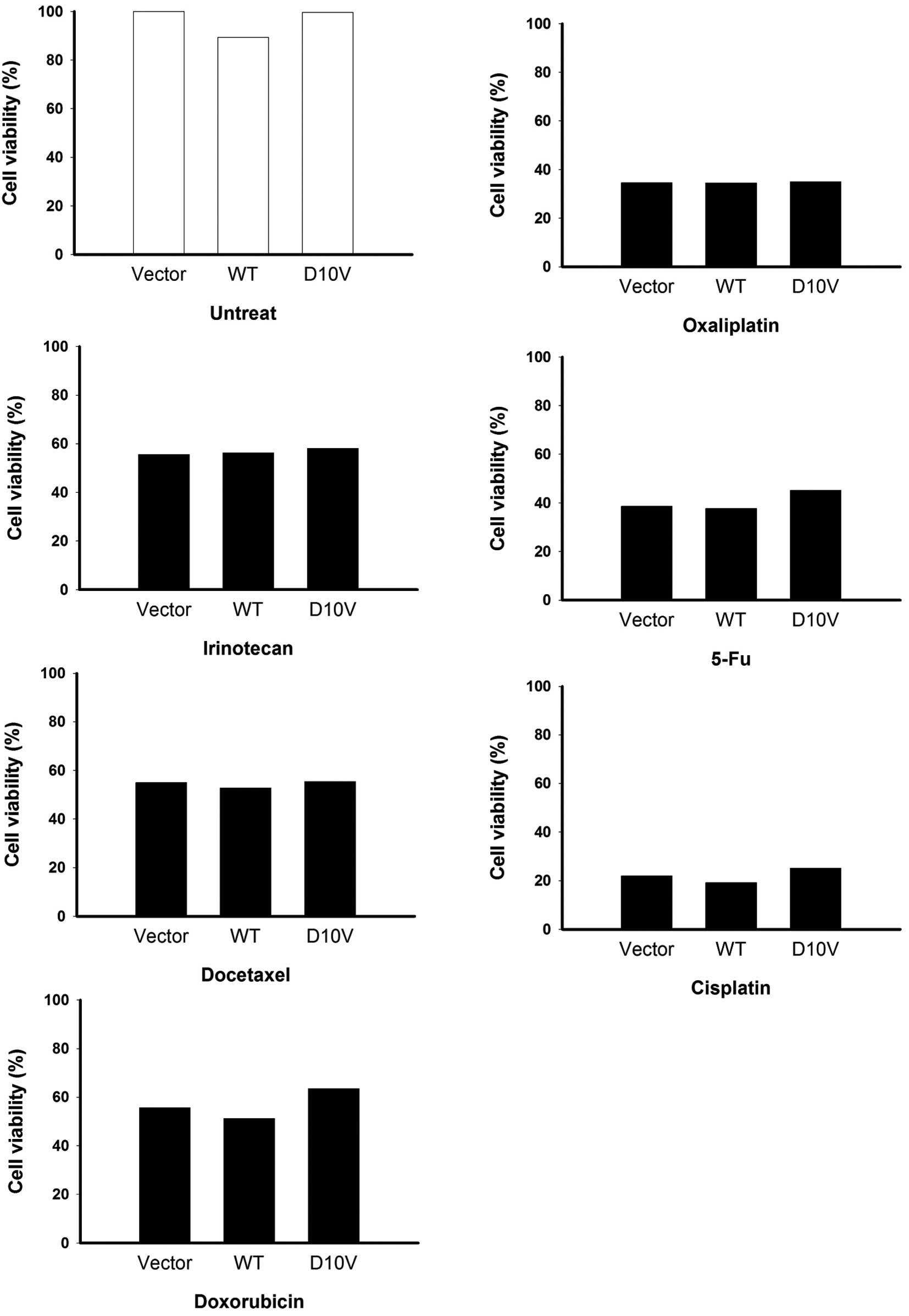
determined. As shown in Fig. 7, the
D10V substitution was not found to correlate with drug
responsiveness. Similar results were obtained in SCM-1 cells
expressing S100A4D10V (Fig.
8). Overexpression of both S100A4 versions did not have any
discernible effect on cell growth in the absence of drugs (Figs. 7 and 8), with the exception of a slight
enhancement of viability among AGS cells (Fig. 7). Taken together, our results
suggest that S100A4 contributes to cancer cell migration but not
the resistance to anticancer drugs.
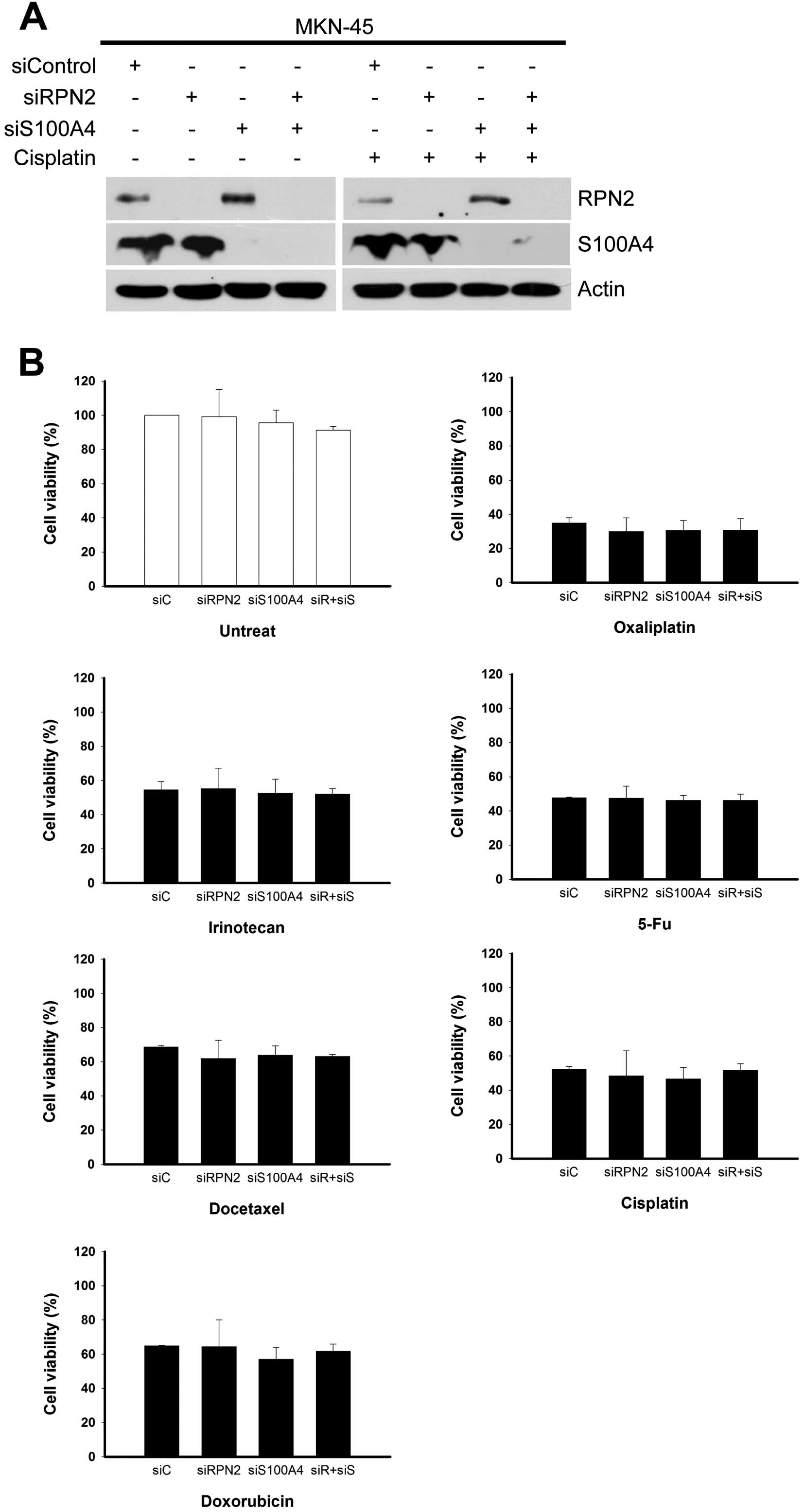
Dual knockdown of S100A4 and RPN2 does
not affect drug responsiveness
Ribophorin II (RPN2) is a prognostic marker and has
been shown to contribute to resistance against chemotherapeutic
agents in human breast tumors and animal models of breast cancer
(32–36). Therefore, we tested whether RPN2
depletion could have a synergistic effect in S100A4-knockdown
gastric cancer cells. MKN-45 cells were subjected to simultaneous
siRNA-mediated depletion of RPN2 and S100A4 for 24 h, and
individually challenged with the six tested anticancer drugs for
another 48 h. MTS-based assays of cell survival revealed that there
was no change in cell viability among cultures subjected to RPN2
knockdown, S100A4 knockdown, or dual knockdown of RPN2 plus S100A4,
regardless of drug treatment (Fig.
9).
Discussion
Researchers have investigated S100A4 for its
involvement in multiple aspects of tumor progression, such as
proliferation, apoptosis, cell motility, extracellular matrix
remodeling and angiogenesis. Recent studies have shown that
elevated S100A4 protein levels are positively correlated with
various human tumors and are associated with poor prognosis in
human gastric, colorectal, pancreatic, thyroid, breast, lung,
prostate and renal cell cancer (18). However, the potential involvement of
S100A4 in chemoresistance is not yet fully understood. To further
explore the potential role of S100A4 in the drug resistance of
gastric cancers, we herein investigated the correlation of S100A4
expression levels with the survival of gastric cancer cell lines
exposed to anticancer drug treatment. We found that siRNA-mediated
knockdown of S100A4 in gastric cancer cell lines significantly
reduced cell migration but did not alter cell viability following
administration of the tested drugs. Moreover, migration assays
showed that cells overexpressing the S100A4D10V variant
had significantly fewer instances of traversing the membrane,
suggesting that the D10V (rs1803245) polymorphism may interfere
with the interaction between S100A4 and Annexin II. Taken together,
our data indicate that there is no significant correlation between
S100A4 status and chemoresistance in the tested gastric cancer cell
lines.
Several recent studies have evaluated S100A4 as a
therapeutic target in preclinical models (37), and some have found that S100A4 is
negatively correlated with cell growth in tumor cells. For example,
the metastatic phenotype of human osteosarcoma cells was
significantly inhibited by a ribozyme directed against the S100A4
gene transcript, but no effect was observed on cell proliferation
or tumorigenicity in vitro and in vivo (14). A whole-human-genome microarray
analysis identified S100A4 as being differentially expressed
between MTX-sensitive and -resistant cells (29), showing overexpression in five of the
seven tested MTX-resistant cell lines. Knockdown of S100A4
increased the sensitivity of HT29 cells toward MTX, but knockdown
of S100A4 had no effect on cell viability in HT29 MTX-resistant
cells, suggesting that other proteins may coordinately fulfill the
drug resistance (29). Conversely,
S100A4 depletion was found to suppress cell proliferation and
invasiveness in pancreatic cancer cell lines characterized by
high-level expression of endogenous S100A4 (38). Moreover, forced expression of S100A4
markedly accelerated the cell migration of pancreatic cancer cell
lines with relatively low-level endogenous expression of S100A4,
but did not affect their cell growth or invasion ability (5).
Based on the above somewhat contradictory findings,
we herein analyzed the potential involvement of S100A4 in the cell
motility and proliferation of gastric cancer cell lines. We failed
to obtain convincing evidence supporting the involvement of S100A4
in the survival of the tested cell lines. The complex phenomenon of
tumor chemoresistance likely requires both gains and losses of
various functions, enabling escape from the cytotoxicity induced by
chemotherapeutic agents in tumor cells. Our present results suggest
that in spite of S100A4, the acquisition of cell survival and
growth ability in response to chemotherapy may result from the
integration of other factors, at least among the tested gastric
cancer cell lines.
Although the clinical correlation between S100A4
expression and tumor metastasis is more conclusive, future studies
are warranted to define the relevant interactions between S100A4
and its binding partners. Several downstream targets and
interaction partners of S100A4 are involved in S100A4-mediated
tumor migration and invasion (18),
and S100A4 reportedly interacts with multiple molecular proteins
involved in the metastatic processes of cytoskeletal rearrangement
and cell motility, including F-actin, tropomyosin and the heavy
chain of nonmuscle myosin II (18).
By surveying an SNP database and performing computer modeling, for
the first time, we identified an SNP in S100A4 (NM_002961.2:
c.29A>T, rs1803245); encoding a valine substitution at D10, it
is localized within the potential binding surface to Annexin II.
Cell migration assays showed that the D10V substitution reduced the
migration ability of the tested cell lines, as compared to cells
overexpressing wild-type S100A4. Future studies are required to
examine whether the interaction between S100A4 and Annexin II has
functional effects, perhaps contributing to the metastatic
phenotype.
In conclusion, it is likely that multiple
dysregulated pathways synergistically contribute to chemoresistance
in cancer cells. Here, we report that S100A4 appears to have little
effect on drug responsiveness in various gastric cancer cell lines,
suggesting that we should identify other relevant factors in order
to improve our therapeutic strategies against gastric cancer.
Acknowledgements
This study was supported by the Ministry of Health
and Welfare, Feng Yuan Hospital Research Project 101-002,
Taiwan.
References
|
1
|
Santamaria-Kisiel L, Rintala-Dempsey AC
and Shaw GS: Calcium-dependent and -independent interactions of the
S100 protein family. Biochem J. 396:201–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fritz G, Botelho HM, Morozova-Roche LA and
Gomes CM: Natural and amyloid self-assembly of S100 proteins:
structural basis of functional diversity. FEBS J. 277:4578–4590.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Helfman DM, Kim EJ, Lukanidin E and
Grigorian M: The metastasis associated protein S100A4: role in
tumour progression and metastasis. Br J Cancer. 92:1955–1958. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boye K and Maelandsmo GM: S100A4 and
metastasis: a small actor playing many roles. Am J Pathol.
176:528–535. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sekine H, Chen N, Sato K, Saiki Y, Yoshino
Y, Umetsu Y, Jin G, Nagase H, Gu Z, Fukushige S, Sunamura M and
Horii A: S100A4, frequently overexpressed in various human cancers,
accelerates cell motility in pancreatic cancer cells. Biochem
Biophys Res Commun. 429:214–219. 2012. View Article : Google Scholar
|
|
6
|
Ebralidze A, Tulchinsky E, Grigorian M,
Afanasyeva A, Senin V, Revazova E and Lukanidin E: Isolation and
characterization of a gene specifically expressed in different
metastatic cells and whose deduced gene product has a high degree
of homology to a Ca2+-binding protein family. Genes Dev.
3:1086–1093. 1989. View Article : Google Scholar
|
|
7
|
Grigorian M, Ambartsumian N, Lykkesfeldt
AE, Bastholm L, Elling F, Georgiev G and Lukanidin E: Effect of
mts1 (S100A4) expression on the progression of human breast cancer
cells. Int J Cancer. 67:831–841. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yonemura Y, Endou Y, Kimura K, Fushida S,
Bandou E, Taniguchi K, Kinoshita K, Ninomiya I, Sugiyama K,
Heizmann CW, Schafer BW and Sasaki T: Inverse expression of S100A4
and E-cadherin is associated with metastatic potential in gastric
cancer. Clin Cancer Res. 6:4234–4242. 2000.PubMed/NCBI
|
|
9
|
Davies BR, Davies MP, Gibbs FE,
Barraclough R and Rudland PS: Induction of the metastatic phenotype
by transfection of a benign rat mammary epithelial cell line with
the gene for p9Ka, a rat calcium-binding protein, but not with the
oncogene EJ-ras-1. Oncogene. 8:999–1008. 1993.PubMed/NCBI
|
|
10
|
Takenaga K, Nakamura Y and Sakiyama S:
Expression of antisense RNA to S100A4 gene encoding an S100-related
calcium-binding protein suppresses metastatic potential of
high-metastatic Lewis lung carcinoma cells. Oncogene. 14:331–337.
1997. View Article : Google Scholar
|
|
11
|
Takenaga K, Nakanishi H, Wada K, Suzuki M,
Matsuzaki O, Matsuura A and Endo H: Increased expression of S100A4,
a metastasis-associated gene, in human colorectal adenocarcinomas.
Clin Cancer Res. 3:2309–2316. 1997.PubMed/NCBI
|
|
12
|
Ambartsumian NS, Grigorian MS, Larsen IF,
Karlstrom O, Sidenius N, Rygaard J, Georgiev G and Lukanidin E:
Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic
for the mts1 gene. Oncogene. 13:1621–1630. 1996.PubMed/NCBI
|
|
13
|
Davies MP, Rudland PS, Robertson L, Parry
EW, Jolicoeur P and Barraclough R: Expression of the
calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice
induces metastasis of mammary tumours. Oncogene. 13:1631–1637.
1996.PubMed/NCBI
|
|
14
|
Maelandsmo GM, Hovig E, Skrede M,
Engebraaten O, Florenes VA, Myklebost O, Grigorian M, Lukanidin E,
Scanlon KJ and Fodstad O: Reversal of the in vivo metastatic
phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme.
Cancer Res. 56:5490–5498. 1996.PubMed/NCBI
|
|
15
|
Saleem M, Adhami VM, Ahmad N, Gupta S and
Mukhtar H: Prognostic significance of metastasis-associated protein
S100A4 (Mts1) in prostate cancer progression and chemoprevention
regimens in an autochthonous mouse model. Clin Cancer Res.
11:147–153. 2005.
|
|
16
|
Lee WY, Su WC, Lin PW, Guo HR, Chang TW
and Chen HH: Expression of S100A4 and Met: potential predictors for
metastasis and survival in early-stage breast cancer. Oncology.
66:429–438. 2004. View Article : Google Scholar
|
|
17
|
Boye K, Nesland JM, Sandstad B, Maelandsmo
GM and Flatmark K: Nuclear S100A4 is a novel prognostic marker in
colorectal cancer. Eur J Cancer. 46:2919–2925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mishra SK, Siddique HR and Saleem M:
S100A4 calcium-binding protein is key player in tumor progression
and metastasis: preclinical and clinical evidence. Cancer
Metastasis Rev. 31:163–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang YY, Ye ZY, Zhao ZS, Tao HQ and Chu
YQ: High-level expression of S100A4 correlates with lymph node
metastasis and poor prognosis in patients with gastric cancer. Ann
Surg Oncol. 17:89–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim EJ and Helfman DM: Characterization of
the metastasis-associated protein, S100A4. Roles of calcium binding
and dimerization in cellular localization and interaction with
myosin. J Biol Chem. 278:30063–30073. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saleem M, Kweon MH, Johnson JJ, Adhami VM,
Elcheva I, Khan N, Bin Hafeez B, Bhat KM, Sarfaraz S, Reagan-Shaw
S, Spiegelman VS, Setaluri V and Mukhtar H: S100A4 accelerates
tumorigenesis and invasion of human prostate cancer through the
transcriptional regulation of matrix metalloproteinase 9. Proc Natl
Acad Sci USA. 103:14825–14830. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Wang X, Liang Y, Diao X and Chen
Q: S100A4 promotes invasion and angiogenesis in breast cancer
MDA-MB-231 cells by upregulating matrix metalloproteinase-13. Acta
Biochim Pol. 59:593–598. 2012.PubMed/NCBI
|
|
23
|
Zhang HY, Zheng XZ, Wang XH, Xuan XY, Wang
F and Li SS: S100A4 mediated cell invasion and metastasis of
esophageal squamous cell carcinoma via the regulation of MMP-2 and
E-cadherin activity. Mol Biol Rep. 39:199–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lo JF, Yu CC, Chiou SH, Huang CY, Jan CI,
Lin SC, Liu CJ, Hu WY and Yu YH: The epithelial-mesenchymal
transition mediator S100A4 maintains cancer-initiating cells in
head and neck cancers. Cancer Res. 71:1912–1923. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bettum IJ, Vasiliauskaite K, Nygaard V,
Clancy T, Pettersen SJ, Tenstad E, Maelandsmo GM and Prasmickaite
L: Metastasis-associated protein S100A4 induces a network of
inflammatory cytokines that activate stromal cells to acquire
pro-tumorigenic properties. Cancer Lett. 344:28–39. 2014.
View Article : Google Scholar
|
|
26
|
Hansen MT, Forst B, Cremers N, Quagliata
L, Ambartsumian N, Grum-Schwensen B, Klingelhofer J, Abdul-Al A,
Herrmann P, Osterland M, Stein U, Nielsen GH, Scherer PE, Lukanidin
E, Sleeman JP and Grigorian M: A link between inflammation and
metastasis: serum amyloid A1 and A3 induce metastasis, and are
targets of metastasis-inducing S100A4. Oncogene. Jan 27–2014.(Epub
ahead of print). View Article : Google Scholar
|
|
27
|
Bertram J, Palfner K, Hiddemann W and
Kneba M: Elevated expression of S100P, CAPL and MAGE 3 in
doxorubicin-resistant cell lines: comparison of mRNA differential
display reverse transcription-polymerase chain reaction and
subtractive suppressive hybridization for the analysis of
differential gene expression. Anticancer Drugs. 9:311–317.
1998.
|
|
28
|
Mahon PC, Baril P, Bhakta V, Chelala C,
Caulee K, Harada T and Lemoine NR: S100A4 contributes to the
suppression of BNIP3 expression, chemoresistance, and inhibition of
apoptosis in pancreatic cancer. Cancer Res. 67:6786–6795. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mencia N, Selga E, Rico I, de Almagro MC,
Villalobos X, Ramirez S, Adan J, Hernandez JL, Noe V and Ciudad CJ:
Overexpression of S100A4 in human cancer cell lines resistant to
methotrexate. BMC Cancer. 10:2502010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, Norton L, Brogi E and Massague J: A CXCL1 paracrine
network links cancer chemoresistance and metastasis. Cell.
150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Semov A, Moreno MJ, Onichtchenko A,
Abulrob A, Ball M, Ekiel I, Pietrzynski G, Stanimirovic D and
Alakhov V: Metastasis-associated protein S100A4 induces
angiogenesis through interaction with Annexin II and accelerated
plasmin formation. J Biol Chem. 280:20833–20841. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kaushal M, Mishra AK, Sharma J, Zomawia E,
Kataki A, Kapur S and Saxena S: Genomic alterations in breast
cancer patients in betel quid and non betel quid chewers. PLoS One.
7:e437892012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu J, He J, Liu Y, Simeone DM and Lubman
DM: Identification of glycoprotein markers for pancreatic cancer
CD24+CD44+ stem-like cells using
nano-LC-MS/MS and tissue microarray. J Proteome Res. 11:2272–2281.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Honma K, Iwao-Koizumi K, Takeshita F,
Yamamoto Y, Yoshida T, Nishio K, Nagahara S, Kato K and Ochiya T:
RPN2 gene confers docetaxel resistance in breast cancer. Nat Med.
14:939–948. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
De Souza R, Zahedi P, Badame RM, Allen C
and Piquette-Miller M: Chemotherapy dosing schedule influences drug
resistance development in ovarian cancer. Mol Cancer Ther.
10:1289–1299. 2011.PubMed/NCBI
|
|
36
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Nagai Y, Ishimoto T, Baba Y, Mimori K and
Baba H: RPN2 expression predicts response to docetaxel in
oesophageal squamous cell carcinoma. Br J Cancer. 107:1233–1238.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hernandez JL, Padilla L, Dakhel S, Coll T,
Hervas R, Adan J, Masa M, Mitjans F, Martinez JM, Coma S, Rodriguez
L, Noe V, Ciudad CJ, Blasco F and Messeguer R: Therapeutic
targeting of tumor growth and angiogenesis with a novel anti-S100A4
monoclonal antibody. PLoS One. 8:e724802013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tabata T, Tsukamoto N, Fooladi AA,
Yamanaka S, Furukawa T, Ishida M, Sato D, Gu Z, Nagase H, Egawa S,
Sunamura M and Horii A: RNA interference targeting against S100A4
suppresses cell growth and motility and induces apoptosis in human
pancreatic cancer cells. Biochem Biophys Res Commun. 390:475–480.
2009. View Article : Google Scholar : PubMed/NCBI
|