Introduction
Schwannomas are benign tumors that arise from
Schwann cells. They typically appear in the vestibulocochlear nerve
and are considered to be grade I tumors; approximately 95% are
unilateral and present sporadically, whereas 5% are associated with
neurofibromatosis type 2 syndrome (NF2). Patients with NF2 present
with bilateral schwannomas and other tumors, frequently
meningiomas, which originate from arachnoid cells, and account for
20% of all primary intracranial tumors. The current classification
of meningiomas by the World Health Organization (WHO) includes
three grades: 90% are classified as grade I tumors; ~8–9% are
atypical grade II tumors; and 1–2% are anaplastic/malignant grade
III tumors (1). Meningiomas have a
recurrence rate of 18, 40 and 80% for grade I, II and III,
respectively.
Preliminary cytogenetic studies have demonstrated
the absence of one chromosome 22 in both neoplasms (2,3), thus
suggesting a common genetic origin for at least some subgroups of
these neurogenic tumors. Subsequently, NF2 gene (located at
22q12.2) inactivation was found to be due to several mechanisms,
such as mutations or allelic loss due to monosomy or deletion of
chromosome 22, accounting for up to 66% in schwannomas (4) and 18–50% in sporadic meningiomas,
depending on the histopathological subtypes (5). In addition to the characteristic
chromosome 22 loss, secondary alterations such as 1p deletions have
been described in both tumor types, and these alterations appear to
be related to tumor progression in meningiomas (6–8).
Although DNA methylation studies on these neurogenic tumors have
revealed the non-random involvement of this mechanism in the
inactivation of some tumor-related genes (9–11),
controversial data are available concerning the epigenetic (through
CpG island aberrant methylation) NF2 inactivation in both
neoplasms (12–16). Indeed, recent studies on genome-wide
methylation suggest that this mechanism is associated with
malignant transformation in meningiomas, and allows for the
epigenetic subclassification of this tumor (17,18).
Global exome sequencing in meningiomas showed that,
in grade I tumors, NF2 gene alteration (by mutation and/or
loss of chromosome 22) is mutually exclusive with other gene
mutations such as AKT1, TRAF7, KLF4 and SMO (19), but not with others such as
NF1 and NEGR1 (20).
In schwannomas, no alternative mutation has been found for those
samples lacking hits over NF2 and, however, Merlin (the NF2
protein) does not seem to be present in the cases analyzed to date
(21).
The expression analysis of tumor-related genes in
meningiomas and schwannomas suggests a possible molecular subgroup
classification in both tumors (22)
with the involvement of differential regulatory pathways (23,24)
related to the allelic losses at 1p and 14q in meningiomas
(25). Whole genome expression
analysis has been performed on schwannomas (26–28)
and meningiomas (29–32). Whereas meningiomas have shown
differential expression patterns based on progression and
recurrence, but not strictly supported by grade (31), in schwannomas no distinctive pattern
has been found using clinical correlations (28). However, a critical deregulation of
microRNAs, including the upregulation of those located at the 14q32
chromosomal region, was a characteristic feature of vestibular
tumors (33).
Intracranial non-recurrent WHO grade I meningiomas
and schwannomas represent similar problems for patients, depending
on the brain structures affected by their non-invasive growth.
Currently, treatment options for patients with grade I meningiomas
or schwannomas include surgery resection, radio-surgery and a ‘wait
and see’ strategy. Thus, there is no available chemotherapeutic
treatment for these tumors besides surgery, a situation especially
traumatic for patients suffering bilateral vestibular schwannomas
and several meningiomas such as those affected by NF2. Due to the
common genetic origin of these tumors (NF2 inactivation),
previous studies have attempted to identify targets with which to
inhibit both schwannoma and meningioma progression. AR42, a histone
deacetylase inhibitor, was found to suppress the proliferation of
meningioma and schwannoma cell lines in vitro (34), and the same effect was shown by
cucurbitacin D and goyazensolide in primary cultures (35).
In the present study, we used microarray technology
to compare gene-expression patterns and identify genes and pathways
of potential interest as key targets for the combined treatment of
vestibular schwannomas and grade I meningiomas.
Materials and methods
Statementofethicsandsamples
The local Ethics Review Board of La Paz University
Hospital approved the study protocol according to the principles of
the Declaration of Helsinki. All patients received detailed
information concerning the study and provided their written
informed consent prior to their inclusion. In this study, we used
RNA from 22 meningiomas, 31 schwannomas and, as non-tumoral
controls, 3 healthy meningeal tissues, 8 non-tumoral nerves and 1
primary Schwann cell culture. The three control non-tumoral
meningeal RNAs derived from two healthy males and one female and
were purchased from BioChain® (cat. no. R1234043-10-D03;
lot nos. B108134, A602330 and B501146).
RNA extraction and microarray
experiments
The RNA was extracted with the RNeasy®
Mini kit (Qiagen, Valencia, CA, USA) as indicated previously
(28). For global gene expression,
the Affymetrix Human Gene 1.0 ST was used. The expression profile
of the meningiomas and the meninges samples can be accessed at the
gene expression Omnibus (GEO) database GSE54934. The arrays of
schwannomas and control nerves were previously published (28) and are available at the GEO database
GSE39645. The arrays were processed at the Institute for Research
in Biomedicine (IRB), Barcelona, Spain.
Statistical analysis
The normalization and summarization were performed
using the robust multichip average (RMA). In order to reduce the
batch effect among tumors (schwannomas and meningiomas) and
controls (healthy nerves and meninges), a critical aspect for our
analysis, we used ComBat (36). For
data analysis, the genes were considered deregulated between groups
when at least a 2-fold change in expression and a P<0.05 cut-off
(ANOVA) was identified, as previously recommended by the MAQC
consortium (37). For the
comparison between schwannomas and meningiomas in order to obtain a
list of genes with no changes among both tumor types, we used a
more restrictive fold (<1.5) exclusively for this purpose, since
the ComBat effect could have lowered these values and
false-positives could appear. For comparative purposes, a list of
differentially expressed genes and fold-change was obtained with
the Geo2R web tool (http://www.ncbi.nlm.nih.gov/geo/geo2r/) in the series
GSE43290, which includes 4 meninges as controls and 47 tumors
(29). As our meningioma series
mainly included grade I tumors, only the 33 WHO grade I meningiomas
and the 4 controls included in this report were used for
comparison.
DNA extraction
The DNA was extracted by standard methods, as
previously described (22). The
data regarding the NF2 status, including loss of
heterozygosity of 22q (LOH), Multiplex ligation-dependent probe
amplification (MLPA) of NF2 (SALSA P044) and sequence
analysis by dHPLC were reported previously in detail (22,28),
and were performed as described (22). Clinical and NF2 status data
from the meningiomas correspond to cases M02, M04, M05, M07, M09,
M10, M12, M14, M24, M25, M28, M29, M30, M31, M32, M33, M34, M38,
M39, M40, M41 and M42, as previously reported (22). The complete case series of
schwannomas from our previous report (28) was included.
Results and Discussion
Comparison with respect to previous
analyses of meningiomas and the summary of results in
schwannomas
Meningioma profiling was analyzed extensively in
previous studies, and up to five expression subgroups were
characterized (31), although this
classification did not represent the actual WHO classification.
Recurrence and progression appear to play a relevant role in the
expression pattern of these tumors (32), and two meningioma groups were
identified showing different clinical and pathological behaviors,
more related to clinical outcome than to WHO grade per se.
Furthermore, depending on the cytogenetic aberrations, differential
expression patterns have been described (25,29).
Tumors that presented monosomy of chromosome 22 and cases with
multiple karyotype alterations had a differential expression
pattern, whereas those cases with deletion of chromosome 1 alone
showed random behavior (29). In
summary, previous analyses of gene-expression patterns in
meningiomas do not seem to accurately represent the current WHO
classification, although recurrence and progression status might be
reflected in these studies. We used 20 grade I meningiomas, 2 grade
II meningiomas and 3 healthy meninges. Practically the same values
were obtained when meningiomas grade II were removed from the study
(data not shown). When we compared our results in meningiomas with
those obtained from the dataset GSE43290 (29), we found a high consistency in our
results, such as the downregulation of diverse genes such as
SNAP25, MBP, TTR and VSNL1, and the upregulation of
FBN2, FGF9 and SULF1 (full data available upon
request). As in previous studies, our 2 cases of meningioma grade
II did not show a different trend. The schwannoma expression
profile was previously explained (28). In brief, the upregulation of
SPP1, MET and associated genes or LATS2 was reported,
whereas the downregulation of CAV1, AR and PAWR was
found. In general, myelinization genes were overexpressed,
suggesting that schwannoma cells could resemble a previous state of
mature Schwann cells.
Gene co-overexpression in meningiomas and
schwannomas
Using the ANOVA test at P<0.05 significance
across the four groups (all meningiomas, schwannomas, control
healthy meninges and control nerves), we obtained a list of 12,395
genes with differential expression among these four groups. Of
those, 346 (data not shown; available upon request) did not meet
the criteria established for accepting deregulation differences
between both tumor groups, which was ≤1.5-fold of the differential
expression between the schwannomas and the meningiomas, a limit
value selected as deregulated between these two groups due to the
correction effect of ComBat. Among those 346 genes with similar
expression in tumors, 47 showed co-overexpression in schwannomas
and meningiomas when compared with their respective controls at
2-fold (as ComBat correction would only be based on batch effect)
(Table I). These genes included
E-cadherin (CDH1), which is usually silenced by several
mechanisms, such as the Wnt signaling pathway in human cancer,
including meningiomas (38), and
platelet-derived growth factor D (PDGFD), an activator for
PDGFR-β (39). This pathway has
been reported as overexpressed in multiple cancer types such as
pancreatic cancer and brain tumors, including schwannomas (39). Another gene reported as expressed
(and protein present) in meningiomas and schwannomas is tyrosine
kinase receptor MET (40),
which is responsible for cell migration, anchorage-independent
growth and many other functions. High levels of this receptor have
been found in a wide variety of tumors, such as breast cancer,
renal cell carcinoma and head and neck tumors (41). Mechanisms such as point mutations,
alternative splicing, genomic amplification and transcript
amplification appear to participate in overexpression of
c-MET (reviewed in ref. 42). Accordingly, we found MET
upregulation in both neoplasms compared with their respective
control tissues, and again, a similar level of expression between
both tumor types was detected. SLIT2 is a member of the Slit
family that modulates cell migration by binding with the Robo
family. This gene has been found expressed in the development of
several malignancies such as colorectal epithelial cell
carcinogenesis (43). The findings
in this report (43), suggest that
Slit2-Robo1 causes E-cadherin degradation, and although our results
show an upregulation of the E-cadherin gene, the former mechanism
should not be ruled out in the tumors we studied. In other
neoplasms, although expressed, SLIT2 does not seem to play
any role (44). In agreement with
the data from the study selected for validation (29), several genes, including CDH1,
PDGFD, CX3CR1, CCND1 and SLIT2, were also upregulated,
as shown in data obtained from meningioma dataset GSE43290
(29); in contrast, MET
showed a trend of upregulation but did not reach 2-fold. Functional
annotation using DAVID showed enrichment in inflammatory response,
cell migration and defense response (data not shown; available upon
request).
 | Table IGenes overexpressed in meningioma and
schwannoma when compared with their respective control tissue. |
Table I
Genes overexpressed in meningioma and
schwannoma when compared with their respective control tissue.
| Gene | Database | Chromosome | C-M | N-S | M-S | P-value |
|---|
| CDH1 | NM_004360 | 16q22.1 | 5.4 | 5.8 | −1.0 | 8.35E-09 |
| PDGFD | NM_025208 | 11q22.3 | 4.4 | 6.2 | −1.1 | 7.48E-13 |
| SLIT2 | NM_004787 | 4p15.2 | 3.6 | 5.8 | −1.1 | 2.02E-13 |
|
HLA-DPA1 | NM_033554 | 6p21.3 | 2.9 | 3.9 | −1.1 | 5.24E-07 |
| PAPPA | NM_002581 | 9q33.2 | 2.8 | 3.8 | −1.1 | 3.68E-06 |
| TREM2 | NM_018965 | 6p21.1 | 2.7 | 5.5 | −1.3 | 2.8E-12 |
|
HLA-DPA1 | NM_033554 | 6p21.3 | 2.6 | 4.1 | −1.2 | 5.24E-07 |
| HPGDS | NM_014485 | 4q22.3 | 2.6 | 2.4 | −1.1 | 3.83E-07 |
| GPR34 | NM_001097579 | Xp11.4 | 2.6 | 12.0 | −1.5 | 9.9E-11 |
| CX3CR1 | NM_001337 | 3p21|3p21.3 | 2.5 | 5.7 | −1.2 | 2.15E-07 |
| ANKRD22 | NM_144590 | 10q23.31 | 2.5 | 7.3 | −1.4 | 1.04E-06 |
| C3 | NM_000064 | 19p13.3-p13.2 | 2.4 | 2.6 | −1.2 | 5.77E-05 |
| CYBB | NM_000397 | Xp21.1 | 2.4 | 4.1 | −1.2 | 1.11E-07 |
|
LGALS3BP | NM_005567 | 17q25 | 2.4 | 2.9 | −1.1 | 4.67E-13 |
| WIPI1 | NM_017983 | 17q24.2 | 2.4 | 2.0 | −1.0 | 2.55E-12 |
|
APOBEC3C | NM_014508 | 22q13.1 | 2.4 | 2.5 | −1.1 | 3.35E-08 |
| C3AR1 | NM_004054 | 12p13.31 | 2.4 | 4.8 | −1.2 | 4.43E-09 |
| FCGBP | NM_003890 | 19q13.1 | 2.3 | 9.7 | −1.3 | 1.03E-11 |
| FRAS1 | NM_025074 | 4q21.21 | 2.3 | 3.2 | −1.2 | 7.42E-08 |
| FLRT3 | NM_198391 | 20p11 | 2.3 | 3.8 | −1.2 | 1.76E-05 |
| FCGR1A | NM_000566 | 1q21.2–q21.3 | 2.3 | 4.3 | −1.3 | 1.66E-08 |
| MET | NM_001127500 | 7q31 | 2.3 | 4.9 | −1.3 | 1.22E-08 |
| ITPR3 | NM_002224 | 6p21 | 2.3 | 3.7 | −1.1 | 3.69E-12 |
| FCGR1B | NM_001017986 | 1p11.2 | 2.3 | 2.9 | −1.2 | 4.14E-08 |
| ALCAM | NM_001627 | 3q13.1 | 2.2 | 2.5 | −1.0 | 4.26E-09 |
|
HLA-DPB1 | NM_002121 | 6p21.3 | 2.2 | 4.6 | −1.3 | 1.75E-07 |
| LAMB1 | NM_002291 | 7q22 | 2.2 | 2.0 | −1.2 | 4.57E-07 |
| C8orf84 | NM_153225 | 8q21.11 | 2.2 | 2.6 | −1.2 | 8.11E-06 |
| SLFN12 | NM_018042 | 17q12 | 2.2 | 2.3 | −1.2 | 4.14E-10 |
| FCGR1A | NM_000566 | 1q21.2–q21.3 | 2.2 | 3.1 | −1.2 | 1.66E-08 |
| LHFPL2 | NM_005779 | 5q14.1 | 2.1 | 2.3 | −1.1 | 8.13E-09 |
| MS4A6A | NM_152852 | 11q12.1 | 2.1 | 4.5 | −1.3 | 4.8E-08 |
| CD84 | NM_001184879 | 1q24 | 2.1 | 2.7 | −1.2 | 3.38E-09 |
| TRIM22 | NM_006074 | 11p15 | 2.1 | 2.2 | −1.1 | 2.09E-09 |
| CD4 | NM_000616 | 12pter-p12 | 2.1 | 2.5 | −1.1 | 1.69E-07 |
| CSF1R | NM_005211 | 5q32 | 2.1 | 3.8 | −1.2 | 5.21E-08 |
| GFRA1 | NM_005264 | 10q26.11 | 2.1 | 4.9 | −1.4 | 1.34E-07 |
|
HLA-DPB1 | NM_002121 | 6p21.3 | 2.1 | 4.5 | −1.3 | 1.75E-07 |
| CD86 | NM_175862 | 3q21 | 2.1 | 2.9 | −1.3 | 1.03E-06 |
| C1QA | NM_015991 | 1p36.12 | 2.1 | 4.3 | −1.2 | 1.24E-07 |
| TLR7 | NM_016562 | Xp22.3 | 2.0 | 3.6 | −1.3 | 7E-08 |
| CCND1 | NM_053056 | 11q13 | 2.0 | 2.7 | −1.1 | 1.48E-11 |
|
HLA-DQA1 | NM_002122 | 6p21.3 | 2.0 | 2.6 | −1.2 | 4.19E-05 |
| FAM105A | NM_019018 | 5p15.2 | 2.0 | 2.6 | −1.1 | 9.2E-08 |
|
C6orf138 | NM_001013732 | 6p12.3 | 2.0 | 3.4 | −1.2 | 1.99E-10 |
| P2RY13 | NM_176894 | 3q24 | 2.0 | 2.4 | −1.2 | 3.08E-06 |
| PROS1 | NM_000313 | 3q11.2 | 2.0 | 5.0 | −1.4 | 9.1E-14 |
Gene co-infraexpression in meningiomas
and schwannomas
A total of 35 genes (Table II) with no difference in expression
between schwannomas and meningiomas were underexpressed when
compared with their respective controls in both neoplasms. Among
them are selectin E (SELE) and Rho family GTPase 1
(RND1), which is linked to semaphorins (45,46)
and cytoskeleton organization in axons. The chemokine (C-X-C motif)
ligand 2 (CXCL2) was significantly downregulated in
schwannomas and meningiomas, whereas the opposite trend has been
shown in malignant neoplasms such as ovarian and endometrial cancer
and oral squamous cell carcinoma (47). As schwannomas and meningiomas are
usually non-invasive, this fact could explain the different trend
in deregulation of CXCL2. Stathmin-like 2 (STMN2)
showed the same pattern: upregulation in hepatoma cells but
downregulation in schwannomas and meningiomas. Notably,
STMN2 interacts with Rho family GTPase 1 (RND1) in
axon extension (48), another gene
that was downregulated in both tumors. Other downregulated genes in
both tumors were E-selectin (SELE) and vascular adhesion
protein 1 (AOC3), related to the tethering and rolling of
leukocytes (49); thus, the
non-invasive nature of grade I meningiomas and schwannomas could
explain the downregulation of these genes. Validation with the
dataset GSE43290 was performed, and included, among others,
downregulation of AOC3, STMN2, SELE, RGS4, THBS4 and
RND1. Functional analysis with DAVID included leukocyte and
cell migration, heparin binding or membrane fraction (data
available upon request).
 | Table IIGenes infraexpressed in meningioma
and schwannoma when compared with their respective control
tissue. |
Table II
Genes infraexpressed in meningioma
and schwannoma when compared with their respective control
tissue.
| Gene | Database | Chromosome | C-M | N-S | M-S | P-value |
|---|
| SAA1 | NM_000331 | 11p15.1 | −2.0 | −2.7 | 1.1 | 0.000414 |
| INHBA | NM_002192 | 7p15-p13 | −2.0 | −4.1 | 1.2 | 2.42E-09 |
| PCDH18 | NM_019035 | 4q31 | −2.0 | −2.6 | 1.2 | 0.000183 |
| PTGIS | NM_000961 | 20q13.13 | −2.1 | −3.4 | 1.3 | 1.72E-06 |
| HHIP | NM_022475 | 4q28–q32 | −2.1 | −2.9 | 1.1 | 1.44E-06 |
| AQP9 | NM_020980 | 15q | −2.1 | −4.5 | 1.0 | 2.78E-08 |
| TCEAL2 | NM_080390 | Xq22.1–q22.3 | −2.1 | −2.4 | 1.4 | 0.000135 |
| S100A12 | NM_005621 | 1q21 | −2.2 | −5.6 | 1.1 | 9.51E-07 |
| PDE3A | NM_000921 | 12p12 | −2.2 | −2.0 | 1.2 | 2.03E-08 |
| S100A9 | NM_002965 | 1q21 | −2.2 | −4.6 | −1.0 | 2.63E-05 |
| PAK3 | NM_002578 | Xq23 | −2.3 | −4.1 | 1.3 | 5.36E-11 |
| SLC16A7 | NM_004731 | 12q13 | −2.3 | −2.2 | 1.1 | 4.89E-12 |
| PI16 | NM_153370 | 6p21.2 | −2.3 | −5.1 | 1.2 | 1.76E-09 |
| MGST1 | NM_145792 | 12p12.3-p12.1 | −2.3 | −3.6 | 1.1 | 0.001079 |
| FGFR2 | NM_000141 | 10q26 | −2.3 | −2.6 | 1.3 | 1.69E-09 |
| TRPM3 | NM_206946 | 9q21.12 | −2.4 | −2.0 | 1.1 | 3.65E-11 |
| PDZRN4 | NM_013377 | 12q12 | −2.5 | −3.0 | 1.1 | 1.14E-08 |
| THBS4 | NM_003248 | 5q13 | −2.5 | −3.5 | 1.0 | 1.71E-09 |
| STEAP4 | NM_024636 | 7q21.12 | −2.7 | −4.2 | 1.1 | 1.38E-07 |
| DCLK1 | NM_004734 | 13q13 | −2.8 | −2.4 | 1.1 | 2.22E-07 |
| ZNF385D | NM_024697 | 3p24.3 | −3.0 | −2.0 | 1.1 | 1.34E-05 |
| CXCL2 | NM_002089 | 4q21 | −3.1 | −3.3 | −1.0 | 7.52E-09 |
| FABP4 | NM_001442 | 8q21 | −3.1 | −13.5 | 1.2 | 1.29E-12 |
| IL6 | NM_000600 | 7p21 | −3.1 | −4.6 | −1.0 | 0.000397 |
| SELE | NM_000450 | 1q22–q25 | −3.3 | −6.6 | 1.0 | 2.29E-09 |
| SLC14A1 | NM_001128588 | 18q11–q12 | −4.0 | −3.3 | 1.0 | 2.7E-09 |
| APLNR | NM_005161 | 11q12 | −4.8 | −3.1 | −1.0 | 8.05E-11 |
| ADH1B | NM_000668 | 4q23 | −4.9 | −3.6 | −1.1 | 6.95E-07 |
|
ADCYAP1R1 | NM_001118 | 7p14 | −5.1 | −3.3 | 1.0 | 6.98E-11 |
| RND1 | NM_014470 | 12q12 | −5.4 | −3.1 | −1.0 | 3.5E-09 |
| ADAMTS1 | NM_006988 | 21q21.2 | −6.0 | −2.4 | −1.1 | 4.64E-11 |
| HSPB8 | NM_014365 | 12q24.23 | −6.0 | −3.5 | 1.0 | 5.3E-07 |
| AOC3 | NM_003734 | 17q21 | −6.3 | −2.5 | −1.1 | 3.64E-11 |
| RGS4 | NM_001102445 | 1q23.3 | −6.7 | −2.1 | −1.1 | 2.55E-10 |
| STMN2 | NM_007029 | 8q21.13 | −9.2 | −2.9 | −1.1 | 4.92E-11 |
Gene expression differences between
meningiomas and schwannomas
The main goal of our study was to test gene
expression profiles common to schwannomas and meningiomas in regard
to their respective controls, and having taken into account their
relative expression. However, we also studied the gene expression
differences between both neurogenic neoplasms. As samples were
processed in various batches, we used a Bayesian method to reduce
the batch effect. Because of this effect, the differential
expression of certain genes in schwannomas and meningiomas could
have decreased. This issue, although it limits our information, is
vital to our study since the batch effect was very marked; 192
genes were upregulated at 1.5-fold differences and P<0.05 (data
available upon request) in schwannomas as compared to meningioma
expression. Most of these genes are related to neuron migration and
the myelin sheath, such as the following: peripheral myelin protein
2 (PMP2), expressed in the cytoplasmic side of myelin in the
peripheral nervous system (50);
myelin protein zero (MPZ), representing 50% of the total
myelin protein in the peripheral nervous system (51); neurexin 1 (NRXN1), which
mediates formation and maintenance of synaptic junctions (52); and neural cell adhesion molecule 2
(NCAM2), which is involved in axonal projection (53).
Upregulation in meningiomas compared with
schwannomas gave us 88 genes (data available upon request) and
included cellular retinoic acid binding protein 2 (CRABP2),
a chaperon downregulated in high-grade gliomas (54), and secreted frizzled-related protein
2 gene (SFRP2), a gene identified as a tumor suppressor in a
renal cell carcinoma cell line (55).
Another comparison concerned those genes that were
upregulated in schwannomas with respect to nerves, and
downregulated in meningiomas with respect to healthy meninges.
These findings included genes such as hepatocyte cell adhesion
molecule (HEPACAM), neuritin 1 (NRN1) and kinesin
family member 1A (KIF1A).
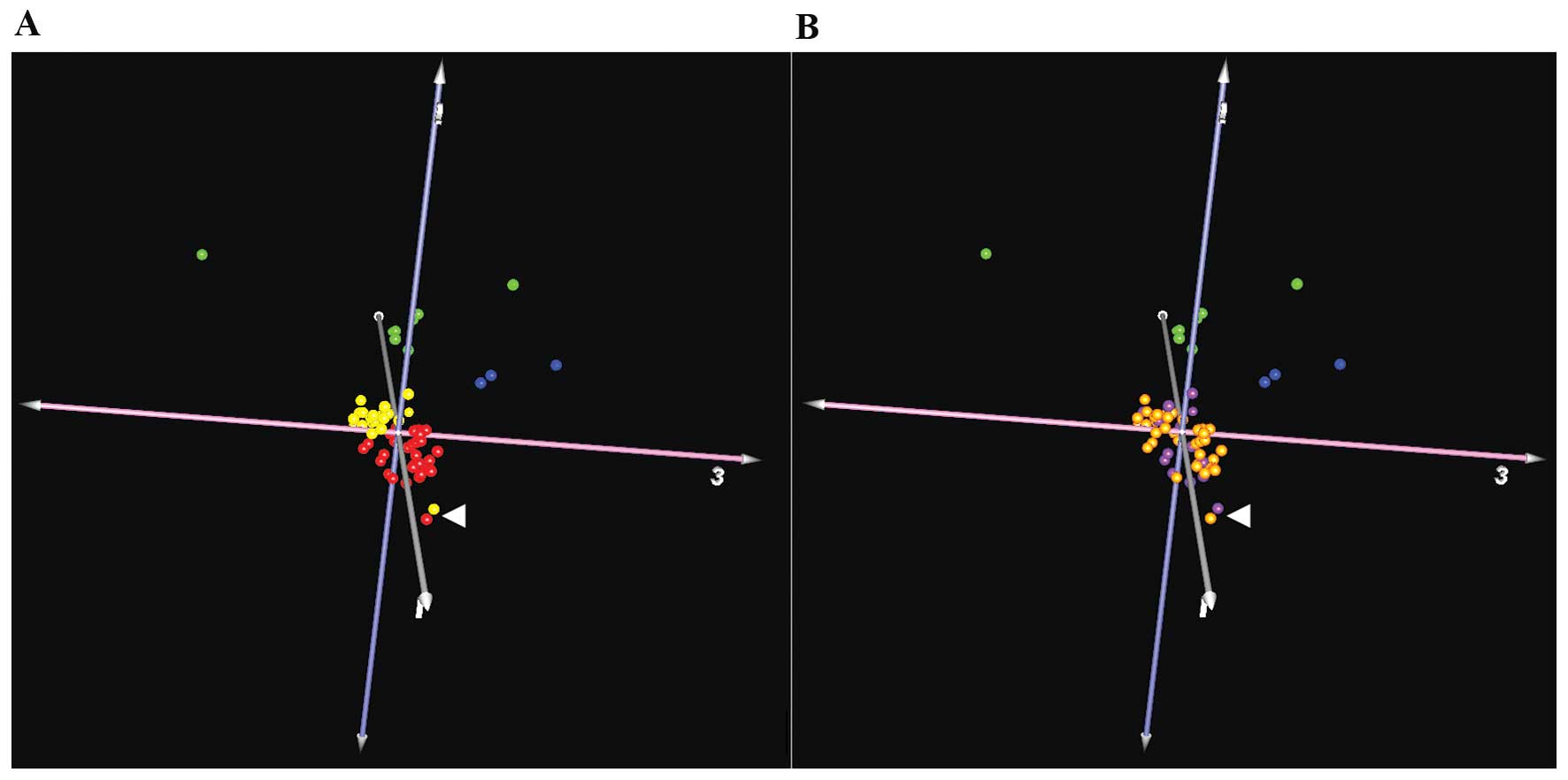
The NF2 mutation rate (determined by
sequencing, MLPA and chromosome 22q LOH analyses) in this series
was 74% for schwannomas and 68% for meningiomas. We compared the
expression patterns in samples from both tumor types, and with
respect to the presence or lack of any alteration in the NF2
gene (38 samples with alteration and 15 without any). Using these
groups, we identified 2 genes with differential expression levels.
The natriuretic peptide receptor C/guanylate cyclase C
(atrionatriuretic peptide receptor C) (NPR3) was
downregulated in those samples without NF2 alterations. This
gene codes for a receptor coupled to various signaling transduction
cascades in several tissues such as cardiac myocytes and
fibroblasts (56). On the other
hand, the G antigen 12J (GAGE12J) gene, transcribed in human
fetal and tumoral tissues (57),
was also downregulated, but on this occasion in tumors with
NF2 alteration. As only 2 genes were detected, based on our
microarray results in both neoplasms, it would seem that there is
no differentiated subset of expression profiles of genes between
samples with or without alteration over NF2 in grade I
meningiomas and schwannomas (Fig.
1). Nevertheless, single genes could be altered in tumors with
or without NF2 alteration, although such a reduced number
could be due to outlier values.
At present, there is no chemotherapeutic treatment
available for either meningiomas or schwannomas, thus research for
a combined solution could be of great value to those patients
affected with both tumor types, primarily patients with
neurofibromatosis type 2. In this study, we found a set of genes
with aberrant expression in both entities compared with their
respective control tissue, but with similar expression levels
between these tumors, including PDGF, c-Met or Slit2 pathways.
Thus, these and the other genes identified in this study, and their
regulatory pathways, might be of interest for further experiments
in the search for common solutions for patients affected by
schwannomas and meningiomas.
Acknowledgements
The authors would like to thank Carolina
Peña-Granero for her excellent technical assistance. This study was
supported by grants PI10/1972 and PI13/00055 from Fondo de
Investigaciones Sanitarias, Ministerio de Ciencia e Innovación,
Spain; and PI13/00800, from the Fundación Sociosanitaria de
Castilla-La Mancha, Spain.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD and
Cavenee WK: WHO Classification of Tumors of the Central Nervous
System. IARC Press; Lyon: 2007
|
|
2
|
Zankl H and Zang KD: Cytological and
cytogenetical studies on brain tumors. 4. Identification of the
missing G chromosome in human meningiomas as no. 22 by fluorescence
technique. Humangenetik. 14:167–169. 1972.PubMed/NCBI
|
|
3
|
Rey JA, Bello MJ, De Campos JM, Kusak ME
and Moreno S: Cytogenetic analysis in human neurinomas. Cancer
Genet Cytogenet. 28:187–188. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hadfield KD, Smith MJ, Urquhart JE,
Wallace AJ, Bowers NL, King AT, Rutherford SA, Trump D, Newman WG
and Evans DG: Rates of loss of heterozygosity and mitotic
recombination in NF2 schwannomas, sporadic vestibular schwannomas
and schwanno-matosis schwannomas. Oncogene. 29:6216–6221. 2010.
View Article : Google Scholar
|
|
5
|
Hansson CM, Buckley PG, Grigelioniene G,
Piotrowski A, Hellström AR, Mantripragada K, Jarbo C, Mathiesen T
and Dumanski JP: Comprehensive genetic and epigenetic analysis of
sporadic meningioma for macro-mutations on 22q and micro-mutations
within the NF2 locus. BMC Genomics. 8:162007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leone PE, Bello MJ, de Campos JM, Vaquero
J, Sarasa JL, Pestaña A and Rey JA: NF2 gene mutations and
allelic status of 1p, 14q and 22q in sporadic meningiomas.
Oncogene. 18:2231–2239. 1999. View Article : Google Scholar
|
|
7
|
Leone PE, Bello MJ, Mendiola M, Kusak ME,
De Campos JM, Vaquero J, Sarasa JL, Pestana A and Rey JA: Allelic
status of 1p, 14q and 22q and NF2 gene mutations in sporadic
schwannomas. Int J Mol Med. 1:889–892. 1998.PubMed/NCBI
|
|
8
|
Bello MJ, de Campos JM, Kusak ME, Vaquero
J, Sarasa JL, Pestaña A and Rey JA: Allelic loss at 1p is
associated with tumor progression of meningiomas. Genes Chromosomes
Cancer. 9:296–298. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bello MJ, Martinez-Glez V,
Franco-Hernandez C, Pefla-Granero C, de Campos JM, Isla A,
Lassaletta L, Vaquero J and Rey JA: DNA methylation pattern in 16
tumor-related genes in schwannomas. Cancer Genet Cytogenet.
172:84–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bello MJ, Amiñoso C, Lopez-Marin I, Arjona
D, Gonzalez-Gomez P, Alonso ME, Lomas J, de Campos JM, Kusak ME,
Vaquero J, Isla A, Gutierrez M, Sarasa JL and Rey JA: DNA
methylation of multiple promoter-associated CpG islands in
meningiomas: relationship with the allelic status at 1p and 22q.
Acta Neuropathol. 108:413–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Pang JC, Dong S, Mao B, Poon WS and
Ng HK: Aberrant CpG island hypermethylation profile is associated
with atypical and anaplastic meningiomas. Hum Pathol. 36:416–425.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kino T, Takeshima H, Nakao M, Nishi T,
Yamamoto K, Kimura T, Saito Y, Kochi M, Kuratsu J, Saya H and Ushio
Y: Identification of the cis-acting region in the NF2 gene
promoter as a potential target for mutation and
methylation-dependent silencing in schwannoma. Genes Cell.
6:441–454. 2001.
|
|
13
|
Gonzalez-Gomez P, Bello MJ, Alonso ME,
Lomas J, Arjona D, de Campos JM, Vaquero J, Isla A, Lassaletta L,
Gutierrez M, Sarasa JL and Rey JA: CpG island methylation in
sporadic and neurofibromatis type 2-associated schwannomas. Clin
Cancer Res. 9:5601–5606. 2003.PubMed/NCBI
|
|
14
|
Lomas J, Bello MJ, Arjona D, Alonso ME,
Martinez-Glez V, Lopez-Marin I, Amiñoso C, de Campos JM, Isla A,
Vaquero J and Rey JA: Genetic and epigenetic alteration of the NF2
gene in sporadic meningiomas. Genes Chromosomes Cancer. 42:314–319.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kullar PJ, Pearson DM, Malley DS, Collins
VP and Ichimura K: CpG island hypermethylation of the
neurofibromatosis type 2 (NF2) gene is rare in sporadic vestibular
schwannomas. Neuropathol Appl Neurobiol. 36:505–514. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Koutsimpelas D, Ruerup G, Mann WJ and
Brieger J: Lack of neurofibromatosis type 2 gene promoter
methylation in sporadic vestibular schwannomas. ORL J
Otorhinolaryngol Relat Spec. 74:33–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kishida Y, Natsume A, Kondo Y, Takeuchi I,
An B, Okamoto Y, Shinjo K, Saito K, Ando H, Ohka F, Sekido Y and
Wakabayashi T: Epigenetic subclassification of meningiomas based on
genome-wide DNA methylation analyses. Carcinogenesis. 33:436–441.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao F, Shi L, Russin J, Zeng L, Chang X,
He S, Chen TC, Giannotta SL, Weisenberger DJ, Zada G, Mack WJ and
Wang K: DNA methylation in the malignant transformation of
meningiomas. PloS One. 8:e541142013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clark VE, Erson-Omay EZ, Serin A, Yin J,
Cotney J, Ozduman K, Avşar T, Li J, Murray PB, Henegariu O, Yilmaz
S, Günel JM, Carrión-Grant G, Yilmaz B, Grady C, Tanrikulu B,
Bakircioğlu M, Kaymakçalan H, Caglayan AO, Sencar L, Ceyhun E, Atik
AF, Bayri Y, Bai H, Kolb LE, Hebert RM, Omay SB, Mishra-Gorur K,
Choi M, Overton JD, et al: Genomic analysis of non-NF2 meningiomas
reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science.
339:1077–1080. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brastianos PK, Horowitz PM, Santagata S,
Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen
P, Ducar MD, Raza A, Sunkavalli A, Macconaill LE,
Stemmer-Rachamimov AO, Louis DN, Hahn WC, Dunn IF and Beroukhim R:
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1
mutations. Nat Genet. 45:285–289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stemmer-Rachamimov AO, Xu L,
Gonzalez-Agosti C, Burwick JA, Pinney D, Beauchamp R, Jacoby LB,
Gusella JF, Ramesh V and Louis DN: Universal absence of merlin, but
not other ERM family members, in schwannomas. Am J Pathol.
151:1649–1654. 1997.PubMed/NCBI
|
|
22
|
Martinez-Glez V, Franco-Hernandez C,
Alvarez L, De Campos JM, Isla A, Vaquero J, Lassaletta L,
Casartelli C and Rey JA: Meningiomas and schwannomas: molecular
subgroup classification found by expression arrays. Int J Oncol.
34:493–504. 2009.PubMed/NCBI
|
|
23
|
Torres-Martín M, Martinez-Glez V,
Peña-Granero C, Isla A, Lassaletta L, De Campos JM, Pinto GR,
Burbano RR, Meléndez B, Castresana JS and Rey JA: Gene expression
analysis of aberrant signaling pathways in meningiomas. Oncol Lett.
6:275–279. 2013.PubMed/NCBI
|
|
24
|
Torres-Martín M, Martinez-Glez V,
Peña-Granero C, Lassaletta L, Isla A, de Campos JM, Pinto GR,
Burbano RR, Meléndez B, Castresana JS and Rey JA: Expression
analysis of tumor-related genes involved in critical regulatory
pathways in schwannomas. Clin Transl Oncol. 15:409–411.
2013.PubMed/NCBI
|
|
25
|
Martínez-Glez V, Alvarez L,
Franco-Hernández C, Torres-Martin M, de Campos JM, Isla A, Vaquero
J, Lassaletta L, Castresana JS, Casartelli C and Rey JA: Genomic
deletions at 1p and 14q are associated with an abnormal cDNA
microarray gene expression pattern in meningiomas but not in
schwannomas. Cancer Genet Cytogenet. 196:1–6. 2010.PubMed/NCBI
|
|
26
|
Aarhus M, Bruland O, Sætran HA, Mork SJ,
Lund-Johansen M and Knappskog PM: Global gene expression profiling
and tissue microarray reveal novel candidate genes and
downregulation of the tumor suppressor gene CAV1 in sporadic
vestibular schwannomas. Neurosurgery. 67:998–1019. 2010. View Article : Google Scholar
|
|
27
|
Cayé-Thomasen P, Borup R, Stangerup S-E,
Thomsen J and Nielsen FC: Deregulated genes in sporadic vestibular
schwannomas. Otol Neurotol. 31:256–266. 2010.
|
|
28
|
Torres-Martin M, Lassaletta L,
San-Roman-Montero J, De Campos JM, Isla A, Gavilan J, Melendez B,
Pinto GR, Burbano RR, Castresana JS and Rey JA: Microarray analysis
of gene expression in vestibular schwannomas reveals SPP1/MET
signaling pathway and androgen receptor deregulation. Int J Oncol.
42:848–862. 2013.
|
|
29
|
Tabernero MD, Maillo A, Gil-Bellosta CJ,
Castrillo A, Sousa P, Merino M and Orfao A: Gene expression
profiles of meningiomas are associated with tumor cytogenetics and
patient outcome. Brain Pathol. 19:409–420. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Keller A, Ludwig N, Backes C, Romeike BFM,
Comtesse N, Henn W, Steudel W-I, Mawrin C, Lenhof H-P and Meese E:
Genome wide expression profiling identifies specific deregulated
pathways in meningioma. Int J Cancer. 124:346–351. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee Y, Liu J, Patel S, Cloughesy T, Lai A,
Farooqi H, Seligson D, Dong J, Liau L, Becker D, Mischel P, Shams S
and Nelson S: Genomic landscape of meningiomas. Brain Pathol.
20:751–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pérez-Magán E, Campos-Martín Y, Mur P,
Fiaño C, Ribalta T, García JF, Rey JA, Rodríguez de Lope A, Mollejo
M and Meléndez B: Genetic alterations associated with progression
and recurrence in meningiomas. J Neuropathol Exp Neurol.
71:882–893. 2012.PubMed/NCBI
|
|
33
|
Torres-Martin M, Lassaletta L, de Campos
JM, Isla A, Gavilan J, Pinto GR, Burbano RR, Latif F, Melendez B,
Castresana JS and Rey JA: Global profiling in vestibular
schwannomas shows critical deregulation of microRNAs and
upregulation in those included in chromosomal region 14q32. PloS
One. 8:e658682013. View Article : Google Scholar
|
|
34
|
Bush ML, Oblinger J, Brendel V, Santarelli
G, Huang J, Akhmametyeva EM, Burns SS, Wheeler J, Davis J, Yates
CW, Chaudhury AR, Kulp S, Chen CS, Chang LS, Welling DB and Jacob
A: AR42, a novel histone deacetylase inhibitor, as a potential
therapy for vestibular schwannomas and meningiomas. Neuro Oncol.
13:983–999. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Spear SA, Burns SS, Oblinger JL, Ren Y,
Pan L, Kinghorn AD, Welling DB and Chang LS: Natural compounds as
potential treatments of NF2-deficient schwannoma and meningioma:
cucurbitacin D and goyazensolide. Otol Neurotol. 34:1519–1527.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Johnson WE, Li C and Rabinovic A:
Adjusting batch effects in microarray expression data using
empirical Bayes methods. Biostatistics. 8:118–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
MAQC Consortium. Shi L, Reid LH, Jones WD,
Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F,
Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA,
Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM,
Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U,
Thierry-Mieg J, Wang C, Wilson M, Wolber PK, et al: The MicroArray
Quality Control (MAQC) project shows inter- and intraplatform
reproducibility of gene expression measurements. Nat Biotechnol.
24:1151–1161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou K, Wang G, Wang Y, Jin H, Yang S and
Liu C: The potential involvement of E-cadherin and beta-catenins in
meningioma. PloS One. 5:e112312010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Kong D, Li Y and Sarkar FH: PDGF-D
signaling: a novel target in cancer therapy. Curr Drug Targets.
10:38–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moriyama T, Kataoka H, Kawano H, Yokogami
K, Nakano S, Goya T, Uchino H, Koono M and Wakisaka S: Comparative
analysis of expression of hepatocyte growth factor and its
receptor, c-met, in gliomas, meningiomas and schwannomas in humans.
Cancer Lett. 124:149–155. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cipriani NA, Abidoye OO, Vokes E and
Salgia R: MET as a target for treatment of chest tumors. Lung
Cancer. 63:169–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lai AZ, Abella JV and Park M: Crosstalk in
Met receptor oncogenesis. Trends Cell Biol. 19:542–551. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou WJ, Geng ZH, Chi S, Zhang W, Niu XF,
Lan SJ, Ma L, Yang X, Wang LJ, Ding YQ and Geng JG: Slit-Robo
signaling induces malignant transformation through Hakai-mediated
E-cadherin degradation during colorectal epithelial cell
carcinogenesis. Cell Res. 21:609–626. 2011. View Article : Google Scholar
|
|
44
|
Dai CF, Jiang YZ, Li Y, Wang K, Liu PS,
Patankar MS and Zheng J: Expression and roles of Slit/Robo in human
ovarian cancer. Histochem Cell Biol. 135:475–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zanata SM, Hovatta I, Rohm B and Püschel
AW: Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor
activity in Semaphorin 3A-induced cytoskeletal collapse. J
Neurosci. 22:471–477. 2002.PubMed/NCBI
|
|
46
|
Hota PK and Buck M: Thermodynamic
characterization of two homologous protein complexes: associations
of the semaphorin receptor plexin-B1 RhoGTPase binding domain with
Rnd1 and active Rac1. Protein Sci. 18:1060–1071. 2009. View Article : Google Scholar
|
|
47
|
Oue E, Lee JW, Sakamoto K, Iimura T, Aoki
K, Kayamori K, Michi Y, Yamashiro M, Harada K, Amagasa T and
Yamaguchi A: CXCL2 synthesized by oral squamous cell carcinoma is
involved in cancer-associated bone destruction. Biochem Biophys Res
Commun. 424:456–461. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li YH, Ghavampur S, Bondallaz P, Will L,
Grenningloh G and Püschel AW: Rnd1 regulates axon extension by
enhancing the microtubule destabilizing activity of SCG10. J Biol
Chem. 284:363–371. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jalkanen S, Karikoski M, Mercier N,
Koskinen K, Henttinen T, Elima K, Salmivirta K and Salmi M: The
oxidase activity of vascular adhesion protein-1 (VAP-1) induces
endothelial E- and P-selectins and leukocyte binding. Blood.
110:1864–1870. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Eylar EH, Szymanska I, Ishaque A, Ramwani
J and Dubiski S: Localization of the P2 protein in peripheral nerve
myelin. J Immunol. 124:1086–1092. 1980.PubMed/NCBI
|
|
51
|
Everly JL, Brady RO and Quarles RH:
Evidence that the major protein in rat sciatic nerve myelin is a
glycoprotein. J Neurochem. 21:329–334. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bottos A, Rissone A, Bussolino F and Arese
M: Neurexins and neuroligins: synapses look out of the nervous
system. Cell Mol Life Sci. 68:2655–2666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alenius M and Bohm S: Identification of a
novel neural cell adhesion molecule-related gene with a potential
role in selective axonal projection. J Biol Chem. 272:26083–26086.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Campos B, Warta R, Chaisaingmongkol J,
Geiselhart L, Popanda O, Hartmann C, von Deimling A, Unterberg A,
Plass C, Schmezer P and Herold-Mende C: Epigenetically mediated
downregulation of the differentiation-promoting chaperon protein
CRABP2 in astrocytic gliomas. Int J Cancer. 131:1963–1968. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Konac E, Varol N, Yilmaz A, Menevse S and
Sozen S: DNA methyltransferase inhibitor-mediated apoptosis in the
Wnt/β-catenin signal pathway in a renal cell carcinoma cell line.
Exp Biol Med. 238:1009–1016. 2013.PubMed/NCBI
|
|
56
|
Rose RA and Giles WR: Natriuretic peptide
C receptor signalling in the heart and vasculature. J Physiol.
586:353–366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gjerstorff MF and Ditzel HJ: An overview
of the GAGE cancer/ testis antigen family with the inclusion of
newly identified members. Tissue Antigens. 71:187–192. 2008.
View Article : Google Scholar : PubMed/NCBI
|