Introduction
Hepatocellular carcinoma (HCC) is the third most
common and aggressive malignancy in the world, estimated to cause
more than half a million deaths annually (1). Although surgery is the best available
treatment option for patients with resectable HCCs, high
postoperative recurrence remains a major obstacle that influences
long-term survival, while most recurrences are due to
invasion-related spreading (2). In
addition, chemotherapy and radiotherapy offer unsatisfactory
response rates. Therefore, it is necessary to explore new
strategies to treat HCC. Gene therapy represents an attractive
alternative compared with conventional therapies.
A number of studies have identified a disintegrin
and metalloproteinase 10 (ADAM10) as a rational target for
anticancer therapy (3,4). ADAM10, a member of the ADAM family,
has been found to have upregulated expression in a range of tumors
including melanoma, pancreatic carcinoma, gastric cancer and oral
squamous cell carcinoma (5–8). Consistent with these findings, we
recently showed that ADAM10 is overexpressed in HCC tissues and
there were significant associations between protein levels of
ADAM10 and tumor grade, amount of tumor differentiation, tumor size
and the presence of metastasis (9).
ADAM10 has been reported to play important roles in cell migration,
tumor development and metastasis by proteolytic shedding of cell
surface proteins (10,11). Recent reports showed that ADAM10
RNAi-based gene therapy was also a suitable strategy to treat
ADAM10-overexpressed cancers, including liver cancer (12).
The gene associated with retinoid-interferon-induced
mortality (GRIM) apoptosis-related gene family potentially
represents a novel type of tumor suppressors that could act as
candidates for use as biological markers and new targets for drug
development (13). GRIM-19, a new
member of the GRIM family located on human chromosome 19p13.1,
encodes for a 16-kDa protein that is present in both nuclear and
cytoplasmic compartments (14). It
was originally isolated and identified as a growth suppressive gene
product involved in the interferon-β (IFNβ)-/all-trans retinoic
acid (RA)-induced cell death pathway using a genetic screen
(13). GRIM-19 expression has been
found to be lost in renal cell cancers, certain prostate and
colonic tumors, prostate carcinoma and HCC (15–17).
It has also been shown that GRIM-19 suppresses signal transducer
and activator of transcription 3 (Stat3) transcriptional activation
by inhibiting its functional interaction (18,19).
Numerous reports showed that overexpression of GRIM-19 could
inhibit cell proliferation, migration and invasion, as well as
induce apoptosis in human prostate, breast and gastric cancer, and
renal carcinoma cells (15,20,21).
Notably, a recent report showed that downregulation of GRIM-19 in
the hepatic and HCC cell lines enhanced adhesive and invasive
potential of these cells by modulating epithelial mesenchymal
transition (EMT) and cell contact inhibition (22). These studies suggested that GRIM-19
may act as a new potential target for treatment of HCCs.
The onset and progression of tumors is a highly
complex process and it is very difficult to cure the tumor by using
a single therapeutic gene. It has been shown that both ADAM10 and
GRIM-19 are ideal targets for cancer gene therapy. However, to
date, there is no study adopting a therapeutic strategy targeting
the two targets simultaneously to treat tumor. Therefore, the aim
of the present study was to test the hypothesis that combinational
gene therapy with ADAM10-specific siRNA and GRIM19 gene could
result in a better therapeutic outcome. Thus, in the present study,
the plasmid carrying GRIM-19 and ADAM10-specific short hairpin RNA
(p-Si-ADAM10-GRIM-19) was constructed, identified and transfected
into HepG2 cells to examine the effect of co-expression GRIM-19 and
ADAM10-specific short hairpin RNA on cell proliferation, cycle,
apoptosis, migration and invasion in an HCC cell line (HepG2 cells)
in vitro, as well as on tumor growth in xenograft mouse
models.
Materials and methods
Plasmid construction
The ADAM10 siRNA sequence (CAGTGTGCATTCAAGTCAA) and
the sequence of scrambled control (AATTCTCCGAACGTGTCACGT) were
designed and synthesized (Shanghai GeneChem Co., Ltd., China) and
cloned into the pSuper siRNA expression vector according to the
manufacturer’s instructions. Then, a serial of the eukaryotic
expression plasmid was used to construct by pcDNA3.1 (Invitrogen,
Carlsbad, CA, USA) as follows: the pcDNA3.1-siRNA-ADAM10 (plasmid
pSi-ADAM10) encoding siRNA specific to ADAM10; the pSi-Scramble
vector (containing scrambled siRNA sequence) which served as a
negative control; pcDNA3.1-GRIM19 containing GRIM19 coding region
(plasmid pGRIM-19); and the co-expression plasmid
pcDNA3.1-SiRNA-ADAM10-GRIM-19 (plasmid pSi-ADAM10-GRIM19)
expressing both siRNA-ADAM10 and GRIM-19 tumor suppressor
protein.
Cell culture and transfection
The human HCC cell line HepG2 was purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA). HepG2
cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)
(Sigma-Aldrich, St. Louis, MO, USA) supplemented with 1%
penicillin/streptomycin (Gibco-BRL, Grand Island, NY, USA) and 10%
heat-inactivated fetal calf serum (FCS; Invitrogen).
HepG2 cells were transfected with various plasmids
using the Lipofectamine 2000 reagent (Invitrogen), according to the
manufacturer’s instructions.
Semi-quantitative reverse
transcription-PCR (RT-PCR)
The mRNA expression levels of ADAM10 and GRIM-19
were examined using semi-quantitative RT-PCR. Cells were collected
48 h after transfection with pSi-ADAM10, pGRIM-19 or
pSi-ADAM10-GRIM-19 plasmids. Total RNA was extracted using the
TRIzol reagent (Invitrogen). Then, RNA was reverse-transcribed into
cDNA using a PrimeScript™ RT reagent kit according to the
manufacturer’s instructions (Takara, Dalian, China). According to
the cDNA sequences of ADAM10 and GRIM-19 gene in GenBank database,
corresponding primers were designed and synthesized by Shanghai
Biological Engineering Company (Shanghai, China). For ADAM10, the
forward primer was 5′-TCTGAGAAATGTCGGGATGAT-3′ and the reverse
primer was 5′-TCTGTAAAGTTGGGCT TGGG-3′. The mixture for PCR
included cDNA (4 μl), 25 mM dNTP (1 μl), 10X PCR buffer (5 μl),
forward and reverse primer (50 pmol) (2 μl), Taq enzyme (1
U) (2.0 μl) and ddH2O (34 μl). The conditions for PCR
were: 94°C for 3 min; 32 cycles of 94°C for 20 sec, 55°C for 20
sec, 72°C for 30 sec and a final 72°C for 5 min. GRIM-19 primers
were: forward, 5′-GAGTCACGCACTGTCTGGG-3′ and reverse,
5′-CGGTCGGTTTCTGCCTGTA-3′. The components of mixture for PCR were
the same as those above. The conditions for PCR were: 32 cycles of
94°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min and a final
72°C for 5 min. The PCR products were subjected to 1% agarose gel
electrophoresis. GAPDH served as an internal reference to normalize
the expression of target gene. The experiments were repeated thrice
and t-test was employed for analysis.
Western blotting
For western blot analyses, after 48 h transfection,
cells were harvested and rinsed once with ice-cold
phosphate-buffered saline (PBS) and lysed in ice-cold cell lysis
buffer (Walterson, London, UK) containing complete protease
inhibitor cocktail (Sigma-Aldrich, Germany). The protein
concentration was determined using the BCA protein assay kit
(KeyGen Biotech) using a c-globulin standard curve.
Equal amounts of protein (15 μg/lane) from the cell
lysates were separated on an 8–12% SDS-polyacrylamide gel
(SDS-PAGE) and transferred onto nitrocellulose membranes (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The membrane was
blocked with 5% skim milk in TBS-T (10 mM Tris-HCl, 150 mM NaCl,
0.25% Tween-20, pH 7.5) at room temperature for 2 h followed by
appropriate primary antibody in TBS containing 5% skim milk
overnight at 4°C. After washing with TBS-T, the membrane was
incubated with a secondary antibody in TBS-T buffer for 3 h at room
temperature, followed by three washes with TBS-T. The
antibody-bound bands were visualized using ECL reagents (ECL,
Amersham, GE Healthcare, Velizy-Villacoublay, France), and
quantified using the VisionWorksLS software (UVP, LLC Upland, CA,
USA). All primary antibodies were used at 1:1,000 and secondary
antibodies at 1:5,000. The primary and secondary antibodies used in
the western blot analyses were: antibodies against ADAM10, GRIM-19,
survivin and Bcl-2 (Cell Signaling Technology, Beverley, MA, USA),
p21, cyclin D1, matrix metalloproteinase-2 (MMP-2), MMP-9 and
β-actin (Santa Cruz Biotechnology, Inc.). Anti-mouse secondary
horseradish peroxidase-conjugated antibody (Amersham Biosciences,
Uppsala, Sweden).
Cell proliferation
To measure the effect of plasmid pSi-ADAM10,
pGRIM-19 or co-expression plasmids pSi-ADAM10-GRIM-19 on cell
proliferation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) assay was performed. In brief, HepG2 cells
transfected with the indicated plasmid, together with untreated
cells, were seeded in 96-well plates at a density of
5×103 cells/well. At indicated time points, 20 μl
methylthiazoletetrazolium (MTT) solution (5 mg/ml) was added into
each well. After 4 h of incubation at 37°C, 150 μl dimethyl
sulfoxide (DMSO) was added to dissolve the crystals. After 10 min
at room temperature, the absorbance was recorded at 570 nm with a
microplate reader (Molecular Devices Corp., Sunnyvale, CA, USA).
Each cell viability assay was performed in quadruplicate and
repeated three times.
The proliferation rate of cells was determined by
measuring the incorporation of bromodeoxyuridine (BrdU) into the
genomic DNA. In brief, HepG2 cells transfected with the indicated
plasmid, along with untreated cells, were seeded in 96-well plates
at a density of 2×103 cells/well. A 5-BrdU incorporation
assay was performed using the BrdU cell proliferation assay kit
(Chemicon, Temecula, CA, USA) according to the manufacturer’s
instructions. The growth rate of cells was calculated as previously
described (23).
Cell cycle analysis
HepG2 cells were seeded at a density of
1×106 on 60 mm dishes in DMEM. Cells were transfected
with plasmid pSi-ADAM10, pGRIM-19, pSi-Scramble and co-expression
plasmids pSi-ADAM10-GRIM-19 for 8 h, respectively, and then the old
medium was removed and fresh medium was added to cells. After 72 h,
the cells were harvested, fixed in 70% ethanol, and stained with 40
μg/ml propidium iodide (PI; Sigma, USA) staining for cell cycle
progression analysis
At least 10,000 cells were analyzed per sample using
a FACSCalibur machine (BD, San Jose, CA, USA). In addition, we also
detected cyclin D1 and P21 protein expression by western blotting
as an additional indicator of cell cycle.
Apoptosis analysis
To determine the number of apoptotic cells, TUNEL
assay was performed. In brief, cellular DNA fragmentation was
measured with the ApoTag Red In Situ Apoptosis Detection kit
(Chemicon), according to the manufacturer’s instructions, after
HepG2 cells were transfected with the indicated plasmid for 48 h.
To quantify the apoptotic cells, the TUNEL-positive cells were
counted using confocal microscopy (Olympus, Tokyo, Japan). In
addition, in the present study, we also detected caspase-3, -8 and
-9 activity by ELISA as an additional indicator of apoptosis.
Caspase activity
The activity of caspase-3, -8 and -9 was determined
by caspases colorimetric protease assay kits (Millipore
Corporation, Billerica, MA, USA) according to the manufacturer’s
instructions. In brief, HepG2 cells were transfected with the
indicated plasmid for 24 h. After treatment, cells were washed
twice with ice-cold PBS and corrected by centrifugation. The cell
pellets were then lysed in 150 μl buffer provided in the kit.
Protein concentrations of lysates were measured by the Lowry
method. An aliquot of lysates (80 μl) was incubated with 10 μl
substrate of each caspase at 37°C for 2 h. Samples were analyzed at
405 nm in a microplate reader (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The relative caspase activity of the control
group was referred as 100.
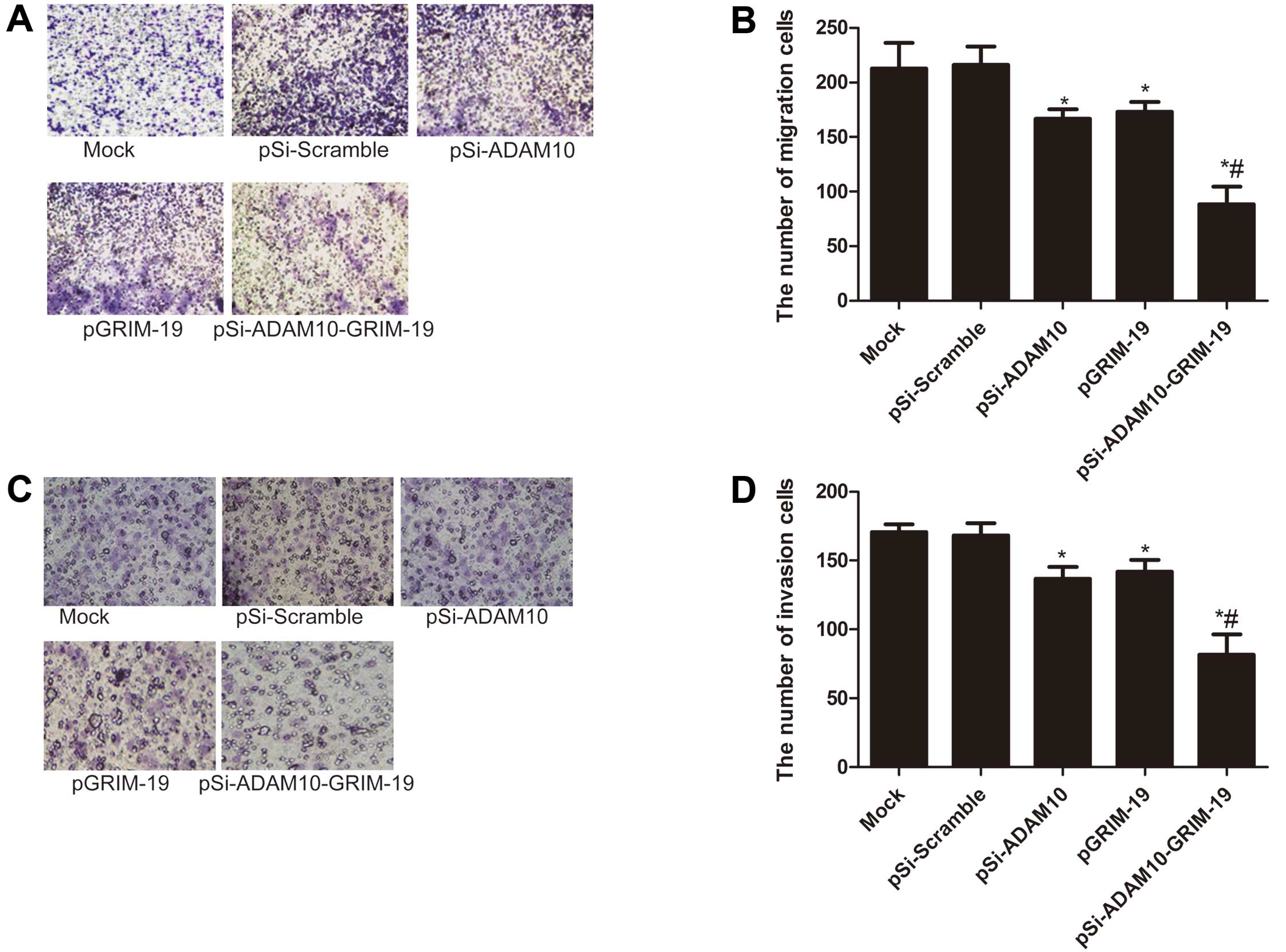
Cell migration and invasion assay
At 48 h after transfection with the indicated
plasmid, HepG2 cells were cultured in serum-free DMEM medium for 24
h. The migration and invasion of cells were assessed using a
Transwell chamber with 8-mm pore filters (Millipore). In brief,
HepG2 cells were detached with trypsin and resuspended in
serum-free medium. Then, 200 μl of cell suspensions
(3.0×105 cells/ml) was added to the upper chamber with
non-coated membrane (24-well insert; 8-mm pore size; Millipore) or
coated with Matrigel (BD Biosciences, Bedford, MA, USA) for the
Transwell migration or invasion assays, respectively. Culture
medium containing 10% FBS was added to the bottom wells of the
chambers. The cells were incubated for 24 h (migration assay) or 48
h (invasion assay). After 24 or 48 h, the cells that had remained
in the upper surface of the filters were removed with cotton swabs,
and the cells that had migrated to the lower surface of the filters
were fixed with 100% methanol and stained with 0.2% crystal violet
and counted. The mean of triplicate assays for each experimental
condition was used.
Tumor growth in vivo
All animal experiments were performed following the
standards of animal care as outlined in the Guide for the Care and
Use of Experimental Animals of Jilin University, following a
protocol approved by the Ethics Committees of the Disease Model
Research Center, The First Hospital of Jilin University.
Approximately 5–6-week-old female BALB/c nude mice (Jilin Institute
of Experimental Animals) were maintained under specific
pathogen-free (SPF) conditions and provided with food and water
ad libitum.
In vitro cultured HepG2 cells were harvested
and a tumorigenic dose of 2×106 cells was injected
intraperitoneally into the right flank of BALB/c mice. Tumor size
was measured every 2–3 days, and tumor volume was calculated as:
0.5236 × width2 × length. When tumors grew to an average
volume of 100 mm3, 50 mice were randomly divided into
five groups: i) control, ii) pSi-Scramble, iii) pSi-ADAM10, iv)
pGRIM-19 and v) pSi-ADAM10-GRIM-19 group. Mice in the control group
were inoculated with 50 μl injection of PBS and mice in the other
groups were inoculated with 20 μg/50 μl per mouse via i.t.
injection of plasmids pSi-Scramble, pSi-ADAM10, pGRIM-19 and
pSi-ADAM10-GRIM-19, respectively. Injection was performed once
every 3 days. Immediately after injection, tumors were pulsed with
an electroporation generator (ECM 830, BTX). Pulses were delivered
at a frequency of 1/s, 150 V/cm, with a length of 50 ms. Mice were
sacrificed on day 35; tumors were resected and weighed, and part of
the tumors was fixed in 10% PBS to TUNEL. We also measured the
ADAM10 and GRIM-19 protein expression in tumor tissue by western
blotting. Then, TUNEL staining was performed on 5 μm sections of
the excised tumors using ApoTag Red In Situ Apoptosis
Detection kit according to the manufacturer’s protocol. The number
of apoptotic cells in five random high power fields was counted and
expressed as a percentage of total cells (apoptotic fraction).
Statistical analysis
Data from at least three independent experiments are
expressed as the means ± SD. The significant differences were
tested either by one-way analysis of variance (ANOVA) or by a
two-sided Student’s t-test. All data were analyzed using the
SPSS® statistical package, version 19.0 (SPSS Inc.,
Chicago, IL, USA) and the GraphPad Prism version 5.01 (GraphPad
Software, San Diego, CA, USA) for Windows. A P-value <0.05 was
considered to indicate a statistically significant difference.
Results
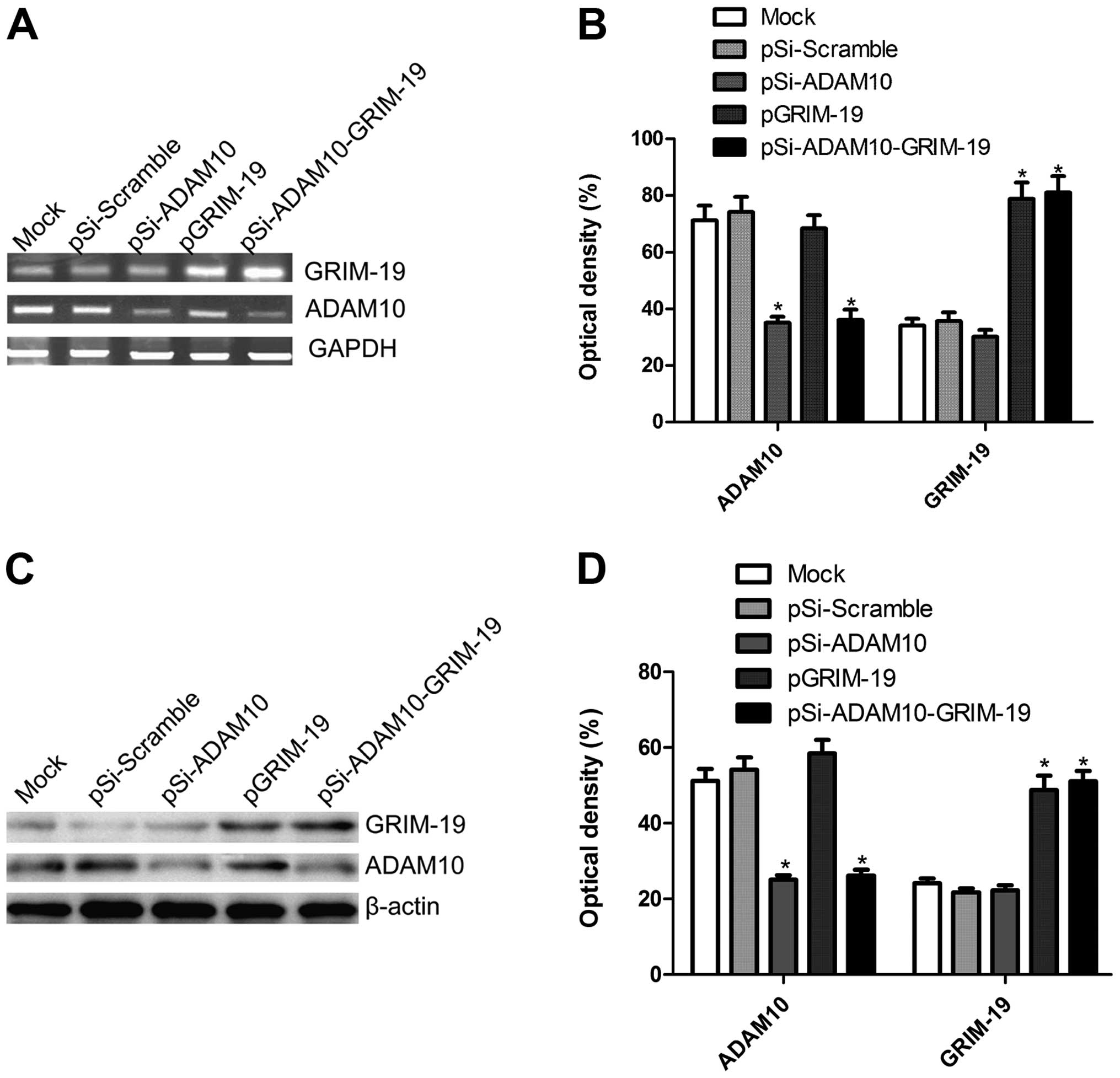
Effect of co-expression plasmid
pSi-ADAM10-GRIM-19 on mRNA and protein expression of ADAM10 and
GRIM-19 in HepG2 cells
We constructed three plasmids (pSi-ADAM10, pGRIM-19
and pSi-ADAM10-GRIM-19) that were capable of expressing a shRNA
that targets the ADAM10, tumor suppressor GRIM-19 either alone or
in combination. These plasmids were transfected into HepG2 cells,
and then their expression on mRNA and protein level were determined
by RT-PCR and western blotting, respectively. Our results showed
that ADAM10 expression on mRNA level and protein level
significantly decreased after transfection of expression plasmid
pSi-ADAM10 and pSi-ADAM10-GRIM-19, and that GRIM-19 expression at
the mRNA and protein level presented significant upregulation after
transfection with plasmids pGRIM-19, pSi-ADAM10-GRIM-19 compared to
those that received mock or pSi-Scramble via transfection (RT-PCR
results are shown in Fig. 1A and B
and western blotting data are presented in Fig. 1C and D).
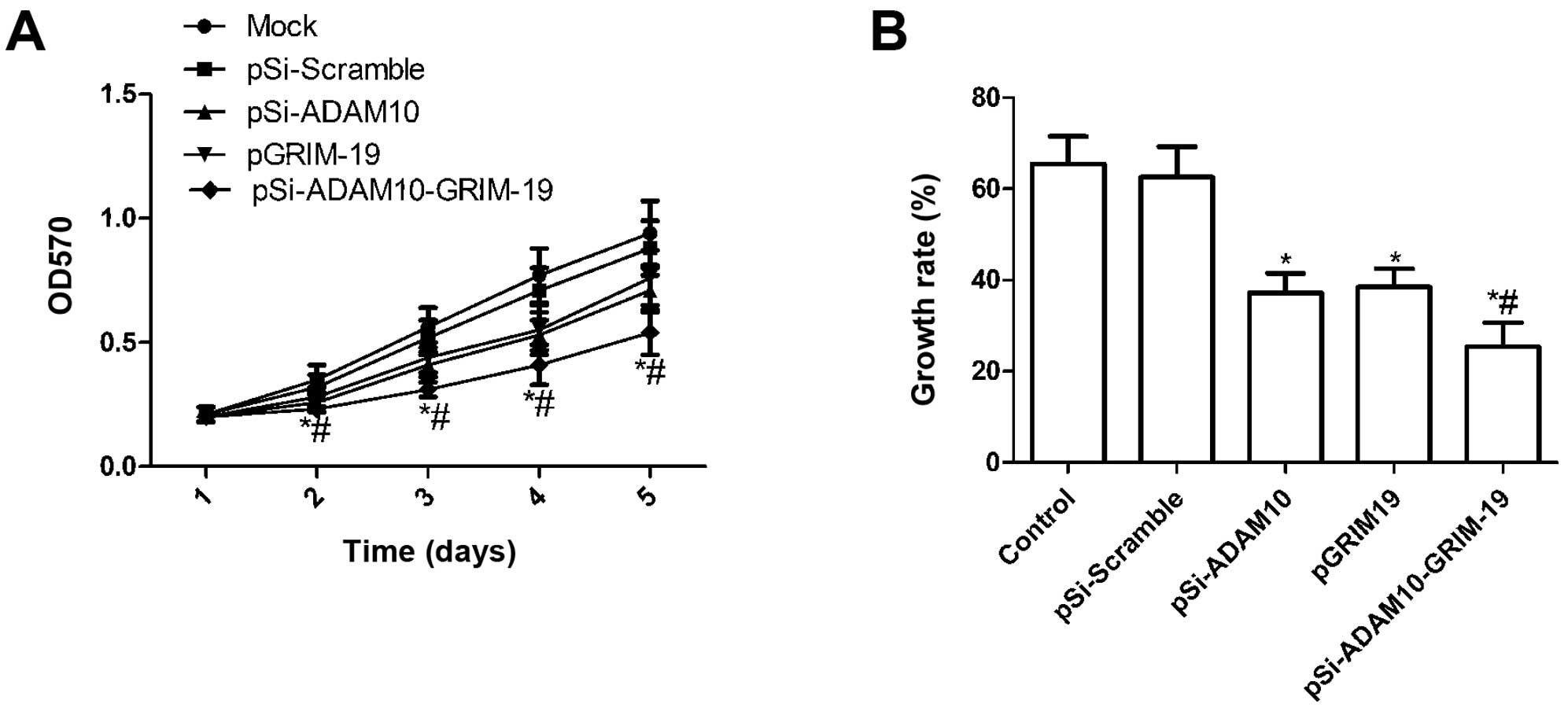
Effect of co-expression plasmid
pSi-ADAM10-GRIM-19 on cell proliferation in HepG2 cells
To investigate if plasmid pSi-ADAM10, pGRIM-19 and
pSi-ADAM10-GRIM-19 exert significantly different effects on cell
proliferation, MTT assay was performed when HepG2 were transfected
with individual expression plasmid. Fig. 2A shows that there were no
statistically significant differences in viability between mock
cells and cells transfected with pSi-Scramble, while the viability
of HepG2 cells was markedly inhibited by transfection with
pSi-ADAM10, pGRIM-19 and pSi-ADAM10-GRIM-19 (P<0.05 compared to
control), and the inhibitory effect of the three plasmids on cell
proliferation was observed beginning on day 2, becoming more
evident on days 4 and 5 (P<0.05, Fig. 2A). Compared to pSi-ADAM10 and
pGRIM-19 treatment, the viability of HepG2 cells was significantly
inhibited in the pSi-ADAM10-GRIM-19 group at different times.
Moreover, BrdU incorporation assays also
demonstrated that the growth rate of HepG2 in the pSi-ADAM10,
pGRIM-19, and pSi-ADAM10-GRIM-19 groups was significantly
diminished compared to the mock and pSi-Scramble groups (P<0.05;
Fig. 2B). Among the HepG2 cell
groups treated with pSi-ADAM10, pGRIM-19 and pSi-ADAM10-GRIM-19,
the lowest incidence of cell proliferation was observed in the
pSi-ADAM10-GRIM-19 treatment group (P<0.05; Fig. 2B). These findings suggest that the
co-expression of GRIM-19 and ADAM10-specific short hairpin RNA
strongly diminished cell proliferative ability in HepG2 cells.
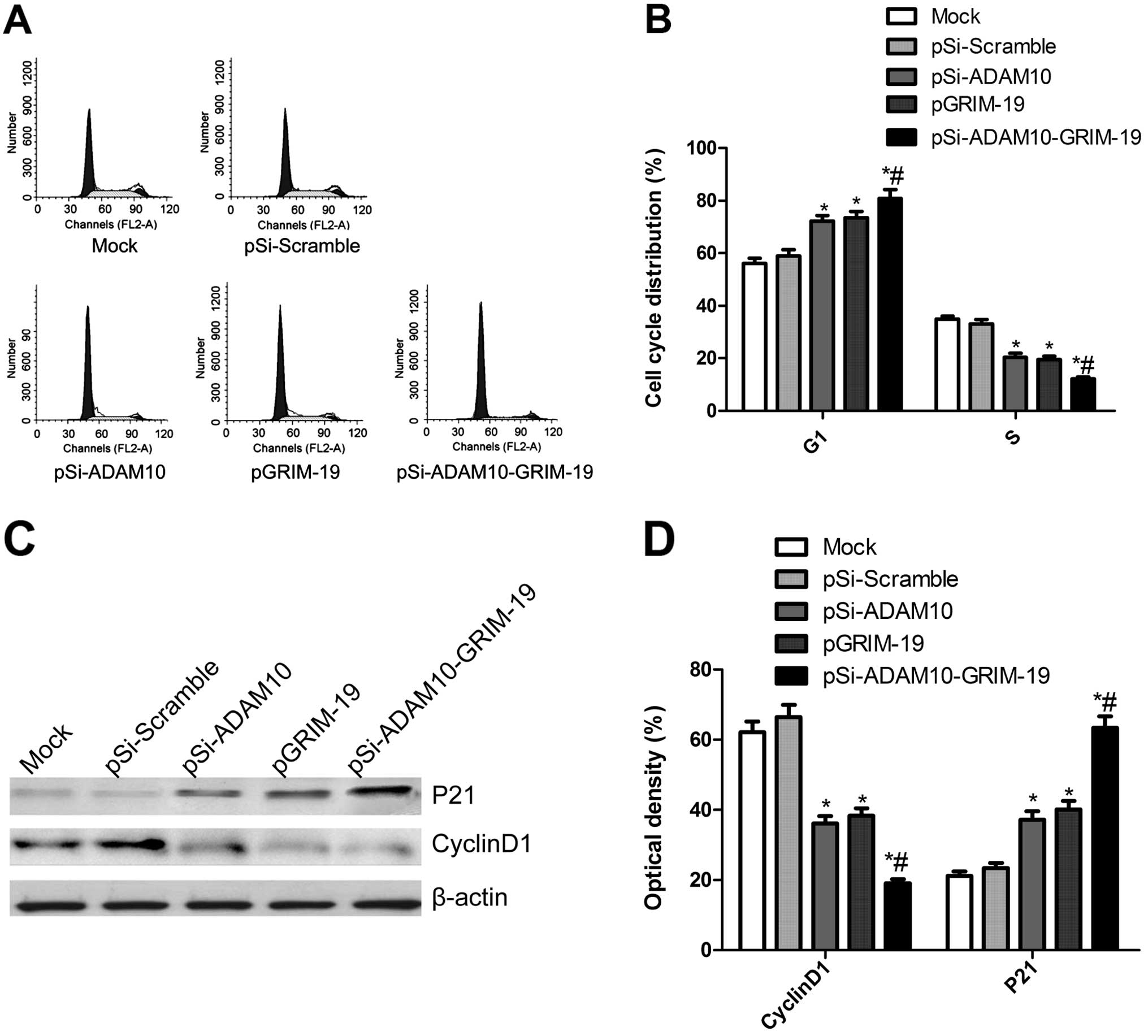
Effect of co-expression plasmid
pSi-ADAM10-GRIM-19 on cell cycle in HepG2 cells
To determine the effects of plasmid pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 on the cell cycle, FACScan flow
cytometry assays were performed. A flow cytometry analysis revealed
that G1 phase cell population was increased in the cells
transfected with pSi-ADAM10, pGRIM-19 and pSi-ADAM10-GRIM-19 as
compared with mock cells, and the cells transfected with plasmid
pSi-Scramble (P<0.05, Fig. 3A and
B). In addition, transfection with pSi-ADAM10, pGRIM-19 and
pSi-ADAM10-GRIM-19 resulted in a considerably lower percentage of
cells in S phase compared with that of mock cells and the cells
transfected with plasmid pSi-Scramble (P<0.05, Fig. 3A and B). Compared to the pSi-ADAM10
and the pGRIM-19 group, pSi-ADAM10-GRIM-19 led to a higher
percentage of cells in G1 phase and lower percentage of cells in S
phase (P<0.05, Fig. 3A and B).
There were no significant differences in cells in G2/M phase among
the groups.
Next, we analyzed the effects of pSi-ADAM10 and
pGRIM-19 group, pSi-ADAM10-GRIM-19 on the expression of cell cycle
relevant proteins, such as cyclin D1 and P21. As shown in Fig. 3C and D, compared to mock cells and
cells transfected with pSi-Scramble, p21 expression was markedly
increased, whereas cyclin D1 expression significantly decreased in
cells transfected with pSi-ADAM10 and pGRIM-19 group,
pSi-ADAM10-GRIM-19 (P<0.05, Fig.
3D). The pSi-ADAM10-GRIM-19 group showed maximally reduced
cyclin D1 expression and increased P21 expression compared to
either the pSi-Stat3 or pGRIM-19 alone (P<0.05, Fig. 3D).
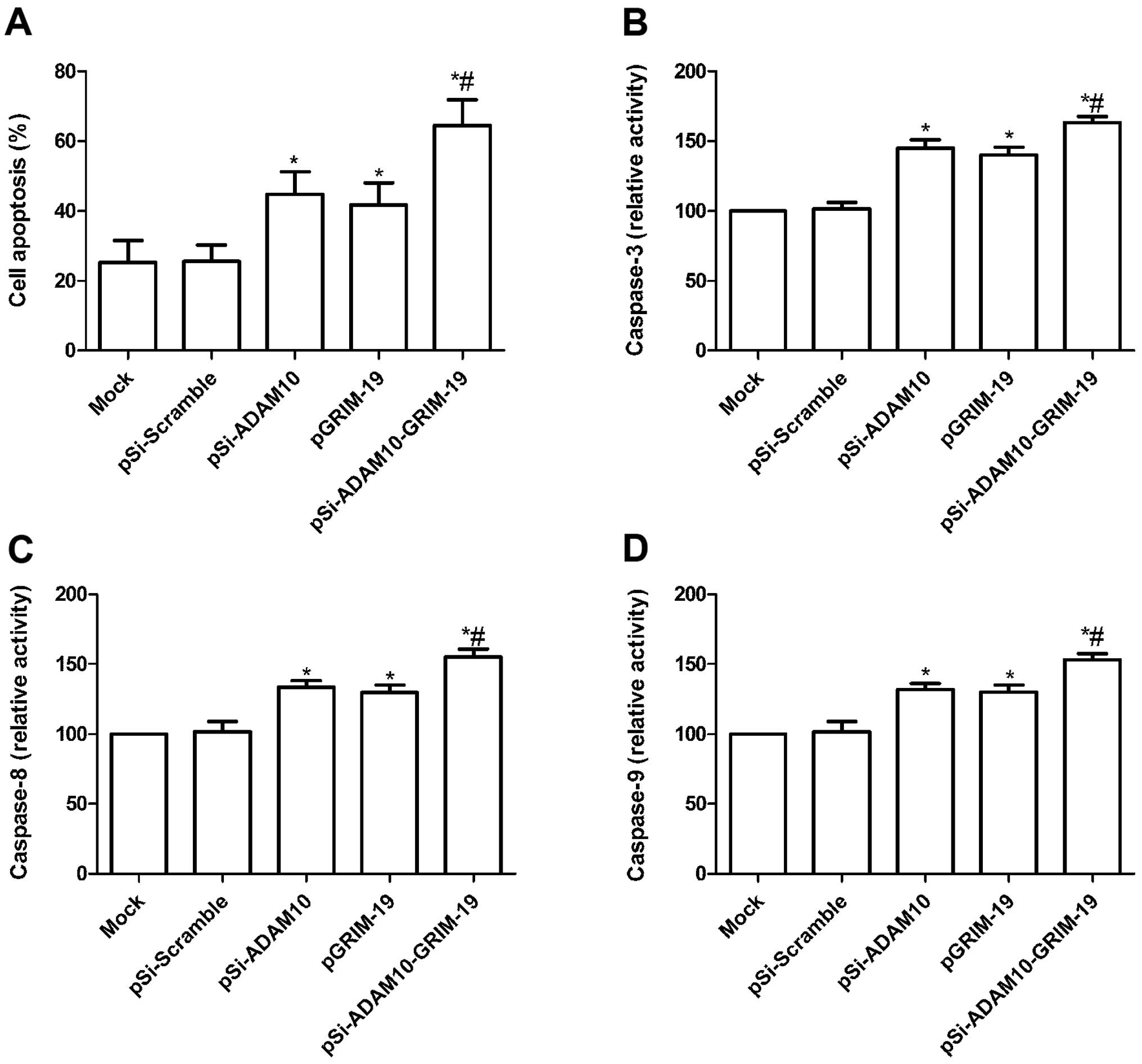
Effect of co-expression plasmid
pSi-ADAM10-GRIM-19 on cell apoptosis in HepG2 cells
To investigate whether plasmid pSi-ADAM10, pGRIM-19
and pSi-ADAM10-GRIM-19 induce apoptosis, TUNEL assay was performed.
Our results showed that HepG2 cells treated with pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 significantly induced cell
apoptosis compared to the mock and pSi-Scramble groups (P<0.05;
Fig. 4A). Treatment with
pSi-ADAM10-GRIM-19 led to a marked increase in apoptotic cells
compared to cells transfected with pSi-ADAM10 and the pGRIM-19
group (P<0.05; Fig. 4A).
To determine the potential mechanism of cell growth
inhibition in vitro, the activities of caspase-3, -8 and -9
were detected using ELISA. The activities of caspase-3, -8 and -9
were found to be significantly increased in the pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 treatment groups, compared to the
mock and pSi-Scramble groups (P<0.05; Fig. 4B–D). Furthermore, the group
transfected with pSi-ADAM10-GRIM-19 showed the highest increase in
activity (P<0.05; Fig.
4B–D).
Effects of co-expression plasmid
pSi-ADAM10-GRIM-19 on cell migration and invasion in HepG2
cells
To ascertain the inhibitory effect of pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 on HepG2 cell migration and
invasion in vitro, we developed a Transwell assay in HepG2
cells. Our results showed that cell migration and invasion were
significantly reduced in the pSi-ADAM10 group, the pGRIM-19 and the
pSi-ADAM10-GRIM-19 treatment group compared to the mock and
pSi-Scramble groups (P<0.05; Fig.
5). Compared to the pSi-ADAM10 or the pGRIM-19 groups, the
pSi-ADAM10-GRIM-19 treatment group strongly inhibited HepG2 cell
migration and invasion (P<0.05; Fig.
5).
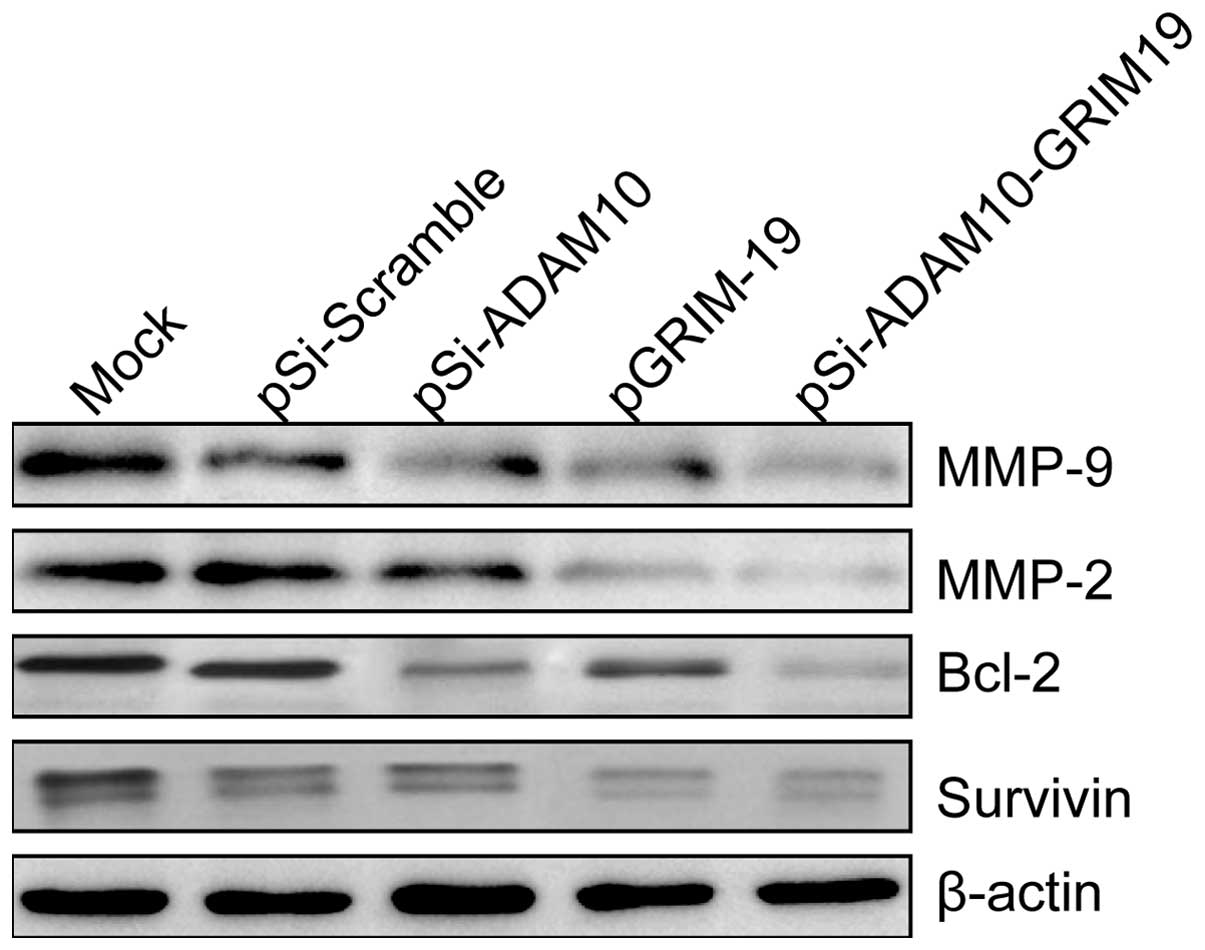
Effects of co-expression plasmid
pSi-ADAM10-GRIM-19 on apoptosis-related proteins and the
invasion-related proteins in HepG2 cells
To determine the potential mechanism of pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 on cell apoptosis and invasion
in vitro, apoptosis-related protein (survivin and Bcl-2) and
invasion-associated protein (MMP-2 and MMP-9) expression levels
were examined using western blot analyses. Western blot analysis
displayed a significant decrease in survivin, Bcl-2, MMP-2 and
MMP-9 proteins in the pSi-ADAM10 group, the pGRIM-19 and the
pSi-ADAM10-GRIM-19 group compared to the mock and pSi-Scramble
groups (Fig. 6). The
pSi-ADAM10-GRIM-19 group showed maximally reduced expression
compared to either the pSi-ADAM10 group or pGRIM-19 (Fig. 6).
Effects of co-expression plasmid
pSi-ADAM10-GRIM-19 on tumor growth in vivo
Finally, we determined whether the co-expressed
GRIM-19 and siRNA-ADAM10 (pSi-ADAM10-GRIM-19) could synergistically
inhibit tumor growth in a xenograft tumor model. Tumor growth was
monitored for 35 days. On day 35, animals were sacrificed and final
tumor weights and tumor volume were determined. Tumor weight and
volume of mice treated with pSi-ADAM10, pGRIM-19 and
pSi-ADAM10-GRIM-19 were significantly diminished when compared to
those that received PBS (mock group) and pSi-Scramble treatment
(P<0.05; Fig. 7A–C). Moreover,
the inhibition of cancer growth in the pSi-ADAM10-GRIM-19 group was
more evident than in the former two groups (P<0.05; Fig. 7A–C). In the present study, we also
examined the expression of ADAM10 and GRIM-19 in grafted tumor
tissues by western blot analysis. Our results showed that GRIM-19
expression levels were clearly increased in the groups treated with
the pGRIM-19 and pSi-ADAM10-GRIM19 plasmids (P<0.05, Fig. 7D and E), while ADAM10 expression
markedly decreased in the groups treated with pGRIM-19 or
pSi-ADAM10-GRIM-19 (P<0.05; Fig. 7D
and E). We also assessed the efficacy of co-expressed GRIM-19
and siRNA-ADAM10 (pSi-ADAM10-GRIM-19) in inducing cell apoptosis
in vivo using TUNEL assay. As shown in Fig. 7F, cells treated with pSi-ADAM10,
pGRIM-19 and pSi-ADAM10-GRIM-19 significantly induced apoptosis
compared to the mock and the pSi-Scramble group (P<0.05).
Treatment with pSi-ADAM10-GRIM-19 resulted in a marked increase in
apoptotic cells compared to cells treated with pSi-ADAM10 and
pGRIM-19 (P<0.05, Fig. 7F).
These results suggest that co-expressed GRIM-19 and siRNA-ADAM10
synergistically inhibit tumor growth in vivo.
Discussion
Cancer is caused by multiple factors, making single
gene therapy difficult. Therefore, several studies have focused on
blocking several signaling pathways involved in the proliferation
and survival of cancer cells. Combined gene therapy can kill tumor
cells by synergistically targeting several genes involved in tumor
occurrence and/or development. Numerous studies have shown that
combined gene therapy is more effective in inhibiting cancer growth
and improving the prognosis compared to single gene therapy. For
example, Wen et al reported that co-expressed survivin-shRNA
and GRIM-19 can induce the apoptosis of laryngeal cancer cells and
inhibit their proliferation, and co-expressed survivin-shRNA and
GRIM-19 is more effective than psi-survivin and p-GRIM-19 alone
(24). Recently, Wang et al
reported that co-expressed Stat3-specific shRNA and GRIM-19
synergistically and more effectively suppressed thyroid tumor
growth in vitro and in vivo (25). Li et al demonstrated that a
combined strategy of co-expressed E6-specific siRNA and p53
synergistically and more effectively suppressed cervical tumor
growth when compared with single treatment (26). Zhang et al reported that the
co-expressed Stat3-specific shRNA and GRIM-19 synergistically and
more effectively suppressed prostate tumor growth and metastases
when compared to treatment with either single agent alone (16). In the present study, our data showed
that simultaneous expression of ADAM10-specific shRNA and GRIM-19
in HepG2 cells significantly suppressed the proliferation,
migration and invasion in vitro and tumor growth in
vivo, when compared to the controls, either ADAM10-specific
siRNA or GRIM19 alone. These studies as well as our findings
indicate that combined gene therapy may be a more effective method
compared to single gene therapy, for the treatment of various
cancers.
Apoptosis is closely related to the elimination of
potentially malignant cells, hyperplasia and tumor progression
(27). It has been shown that
apoptosis is an important feature of cancer cells and abnormal
apoptosis plays an important role in the development and
progression of cancer (28).
Therefore, controlling the apoptosis of cancer cells has biological
and clinical significance (29). It
is well known that downregulation of ADAM10 or overexpression of
GRIM-19 alone could cause tumor growth inhibition in hepatocellular
carcinoma (HCC) (12,22). However, this is the first report
showing that simultaneous expression of ADAM10-specific siRNA and
GRIM-19 (pSi-ADAM10-GRIM-19) in HepG2 tumor cells causes additive
effects on cell apoptosis in vitro and in vivo. In
addition, Bcl-2 and caspase pathways have been considered as
classical apoptotic pathways and these two pathways mediate most of
the cell apoptosis. Therefore, we also assessed the effect of
co-expressed ADAM10-specific shRNA and GRIM-19 on these two
pathways. The results demonstrated the anticipated findings:
plasmid co-expressed ADAM10-specific shRNA and GRIM-19
(pSi-ADAM10-GRIM-19) upregulated caspase-3, -8 and -9 activity,
whereas expression of Bcl-2 and survivin was decreased when
compared to the controls, pSi-ADAM10 or pGRIM-19 alone. Similarly,
our results showed that G1 phase cell population was increased in
the cells transfected with pSi-ADAM10-GRIM-19 as compared with
cells transfected with pSi-ADAM10 and pGRIM-19. These results
suggest that the co-expression of siRNA-ADAM10 with GRIM-19 caused
the strongest growth suppression of HCC cells, and was an effective
strategy which could be adopted in clinics and may result in
improved therapeutic outcome.
HCC is often lethal due to its aggressive
metastasis, and migration and invasion are important features of
cancer cells in general (30).
Therefore, prevention of HCC recurrence and metastasis has
biological and clinical significance. In previous studies, ADAM10
was reported to promote cell invasiveness and migration in HepG2
cells (31). A recent study also
showed that downregulation of ADAM10 by siRNA could inhibit HCC
cell migration and invasion (12).
In addition, it has been shown that overexpression of GRIM19
inhibited HCC cell migration and invasion. Thus, in the present
study, we constructed a co-expression plasmid (pSi-ADAM10-GRIM-19)
that was capable of co-expressing a shRNA that targets the ADAM10
and GRIM-19, then transfected it into HepG2 cells. Our results
showed that HepG2 cell migration and invasion were significantly
reduced in the pSi-ADAM10-GRIM-19 treatment group compared to the
pSi-ADAM10 group and pGRIM-19 alone. Furthermore, we also
discovered this plasmid could inhibit the expression of MMP-2 and
MMP-9, which are directly linked to angiogenesis and the
degradation of basement membrane collagen, and their expression and
activity correlate with metastatic ability and the prognosis of
cancer (32). Also, downregulation
of MMP-2 and MMP-9 expression contributes to the inhibition of
cancer cell invasion and metastasis (33,34).
In summary, our results demonstrated that
simultaneous expression of ADAM10-specific siRNA and GRIM-19
(pSi-ADAM10-GRIM-19) in HepG2 tumor cells significantly inhibited
cell proliferation, cell cycle, migration and invasion and induced
cell apoptosis in vitro, and it also suppressed tumor growth
in a mouse model, when compared to the controls, pSi-ADAM10 or
pGRIM-19 alone. These results suggest that a combined strategy of
co-expressed ADAM10-specific siRNA and GRIM19 synergistically and
more effectively suppresses HCC tumor growth, and has therapeutic
potential for the treatment of HCC.
Acknowledgements
The authors acknowledge the financial support
provided by the Development of Science and Technology Plan Projects
of Jilin (no. 209Z0198).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klein T and Bischoff R: Active
metalloproteases of the A disintegrin and metalloprotease (ADAM)
family: biological function and structure. J Proteome Res.
10:17–33. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloproteases: multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee SB, Schramme A, Doberstein K, et al:
ADAM10 is upregulated in melanoma metastasis compared with primary
melanoma. J Invest Dermatol. 130:763–773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gaida MM, Haag N, Günther F, et al:
Expression of A disintegrin and metalloprotease 10 in pancreatic
carcinoma. Int J Mol Med. 26:281–288. 2010.PubMed/NCBI
|
|
7
|
Wang YY, Ye ZY, Li L, Zhao ZS, Shao QS and
Tao HQ: ADAM 10 is associated with gastric cancer progression and
prognosis of patients. J Surg Oncol. 103:116–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW
and Liu TY: Increase of disintergin metalloprotease 10 (ADAM10)
expression in oral squamous cell carcinoma. Cancer Lett. 245:33–43.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Liu S, Liu K, et al: A
disintegrin and metalloprotease (ADAM)10 is highly expressed in
hepatocellular carcinoma and is associated with tumour progression.
J Int Med Res. 42:611–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Endres K and Fahrenholz F: Upregulation of
the α-secretase ADAM10 - risk or reason for hope? FEBS J.
277:1585–1596. 2010.
|
|
11
|
Murai T, Miyazaki Y, Nishinakamura H, et
al: Engagement of CD44 promotes Rac activation and CD44 cleavage
during tumor cell migration. J Biol Chem. 279:4541–4550. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yue Y, Shao Y, Luo Q, Shi L and Wang Z:
Downregulation of ADAM10 expression inhibits metastasis and
invasiveness of human hepatocellular carcinoma HepG2 cells. Biomed
Res Int. 2013:4345612013.PubMed/NCBI
|
|
13
|
Mora LB, Buettner R, Seigne J, et al:
Constitutive activation of Stat3 in human prostate tumors and cell
lines: direct inhibition of Stat3 signaling induces apoptosis of
prostate cancer cells. Cancer Res. 62:6659–6666. 2002.PubMed/NCBI
|
|
14
|
Angell JE, Lindner DJ, Shapiro PS, Hofmann
ER and Kalvakolanu DV: Identification of GRIM-19, a novel cell
death-regulatory gene induced by the interferon-β and retinoic acid
combination, using a genetic approach. J Biol Chem.
275:33416–33426. 2000.PubMed/NCBI
|
|
15
|
Alchanati I, Nallar SC, Sun P, et al: A
proteomic analysis reveals the loss of expression of the cell death
regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene.
25:7138–7147. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang L, Gao L, Li Y, et al: Effects of
plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PC-3M
tumor cell growth. Clin Cancer Res. 14:559–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li F, Ren W, Zhao Y, et al: Downregulation
of GRIM-19 is associated with hyperactivation of p-STAT3 in
hepatocellular carcinoma. Med Oncol. 29:3046–3054. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lufei C, Ma J, Huang G, et al: GRIM-19, a
death-regulatory gene product, suppresses Stat3 activity via
functional interaction. EMBO J. 22:1325–1335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Yang J, Roy SK, et al: The cell
death regulator GRIM-19 is an inhibitor of signal transducer and
activator of transcription 3. Proc Natl Acad Sci USA.
100:9342–9347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang Y, Yang M, Yang H and Zeng Z:
Upregulation of the GRIM-19 gene suppresses invasion and metastasis
of human gastric cancer SGC-7901 cell line. Exp Cell Res.
316:2061–2070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang G, Chen Y, Lu H and Cao X: Coupling
mitochondrial respiratory chain to cell death: an essential role of
mitochondrial complex I in the interferon-β and retinoic
acid-induced cancer cell death. Cell Death Differ. 14:327–337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao H, Liu J, Liu G, et al: Depletion of
GRIM-19 accelerates hepatocellular carcinoma invasion via inducing
EMT and loss of contact inhibition. J Cell Physiol. 227:1212–1219.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Y, Peng S, Yu H, et al: RNAi-mediated
downregulation of NOB1 suppresses the growth and
colony-formation ability of human ovarian cancer cells. Med Oncol.
29:311–317. 2012.
|
|
24
|
Wen LJ, Gao LF, Jin CS, et al: Small
interfering RNA survivin and GRIM-19 co-expression salmonella
plasmid inhibited the growth of laryngeal cancer cells in vitro and
in vivo. Int J Clin Exp Pathol. 6:2071–2081. 2013.PubMed/NCBI
|
|
25
|
Wang GM, Ren ZX, Wang PS, et al:
Plasmid-based Stat3-specific siRNA and GRIM-19 inhibit the growth
of thyroid cancer cells in vitro and in vivo. Oncol
Rep. 32:573–580. 2014.PubMed/NCBI
|
|
26
|
Li X, Li Y, Hu J, et al: Plasmid-based
E6-specific siRNA and co-expression of wild-type p53 suppresses the
growth of cervical cancer in vitro and in vivo. Cancer Lett.
335:242–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
28
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chau BN and Wang JY: Coordinated
regulation of life and death by RB. Nat Rev Cancer. 3:130–138.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yau WL, Lam CS, Ng L, et al:
Over-expression of miR-106b promotes cell migration and metastasis
in hepatocellular carcinoma by activating epithelial-mesenchymal
transition process. PLoS One. 8:e578822013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan S, Lei S and Wu S: ADAM10 is
overexpressed in human hepatocellular carcinoma and contributes to
the proliferation, invasion and migration of HepG2 cells. Oncol
Rep. 30:1715–1722. 2013.PubMed/NCBI
|
|
32
|
Xue YJ, Xiao RH, Long DZ, et al:
Overexpression of FoxM1 is associated with tumor progression in
patients with clear cell renal cell carcinoma. J Transl Med.
10:2002012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Braicu EI, Gasimli K, Richter R, et al:
Role of serum VEGFA, TIMP2, MMP2 and MMP9 in monitoring response to
adjuvant radiochemotherapy in patients with primary cervical cancer
- results of a companion protocol of the randomized NOGGO-AGO phase
III clinical trial. Anticancer Res. 34:385–391. 2014.
|
|
34
|
Lai WW, Hsu SC, Chueh FS, et al: Quercetin
inhibits migration and invasion of SAS human oral cancer cells
through inhibition of NF-κB and matrix metalloproteinase-2/-9
signaling pathways. Anticancer Res. 33:1941–1950. 2013.PubMed/NCBI
|