Introduction
Colorectal cancer (CRC) is the third most common
cause of cancer-related mortality worldwide (1). Despite the fact that recent advances
in chemotherapeutic regimens in combination with molecular-targeted
compounds have improved the survival rates of CRC patients, the
risk of metastasis and recurrence still remain (2). To improve prognosis, adjuvant
chemotherapy such as 5-fluorouracil (5-FU) plus leucovorin (LV)
(5-FU/LV) and tegafur plus uracil (UFT)/LV has been established as
a generalized regimen. For the treatment of advanced-stage CRC
patients, 5-FU/LV plus irrinotecan (FOLFIRI) and 5-FU/LV plus
oxaliplatin (FOLFOX), combined with molecular-targeted compounds
such as bevacizumab, cetuximab and panitumumab have been approved
(3). Although these combination
therapies of chemotherapy and molecular-targeted agents have
demonstrated survival benefits, the substantial financial cost
involved in these treatments remains an issue (4). Furthermore, chemotherapy without
certain selection tends to lead to overtreatment of patients with
toxic agents that exert severe side-effects. To facilitate
individually tailored treatment for CRC, useful biomarkers for the
selection of patients at high risk of recurrence are required.
Particularly, markers for the selection of high-risk patients in
Duke’s stage B are desirable, since the role of adjuvant
chemotherapy in these patients remains controversial (5).
MicroRNAs (miRNAs) are small 21–25 nucleotide
non-coding RNAs that participate in the regulation of cell
differentiation, cell cycle progression, apoptosis and
tumorigenesis (6–8). They target protein-coding mRNAs at the
post-transcriptional level by direct cleavage of the mRNAs or by
inhibition of protein synthesis (9). Recent evidence indicates that some
miRNAs function as oncogenes (10,11) or
tumor-suppressor genes (12,13)
and play a central role in carcinogenesis. Accumulating evidence
shows that microRNA-21 (miR-21) is an oncogenic miRNA, and
overexpression of this miRNA has been reported in several types of
human malignant solid tumors, including those of the breast, lung
and colon (14). It was
demonstrated that miR-21 promotes carcinogenesis through inhibition
of apoptosis, proliferation, invasion, migration and metastasis
(15). Additionally, high
expression of miR-21 in CRC tissues may be associated with poor
prognosis (16). However, no other
studies have investigated the prognostic value of miR-21 at each
tumor stage.
In the present study, we investigated the
clinicopathological characteristics and the prognostic value of
miR-21 mRNA levels in CRC patients of each tumor stage.
Patients and methods
Patients
A total of 306 CRC patients were studied from
September 2000 to April 2012 at Teikyo University Hospital (Tokyo,
Japan). The median follow-up period was 48 months (range, 24–90
months). All samples were derived from patients who did not receive
any chemotherapy or radiotherapy prior to surgery. Immediately
following surgical resection, primary CRC tissues and normal
adjacent tumor tissues (normal tissue) were mounted by Tissue-Tek
O.C.T. Compound (Sakura Finetechnical Co., Tokyo, Japan) and were
frozen in liquid nitrogen. After surgery, patients with Dukes’
stage A and B were not treated with chemotherapy. Dukes’ stage C
patients were treated with a standard regimen based on 5-FU/LV.
Dukes’ stage D patients were treated with a standard regimen based
on FOLFIRI and FOLFOX, combined with molecular-targeted compounds
(bevacizumab, cetuximab). The study protocol conformed to the
guidelines of the Ethics Committee and was approved by the Review
Board of Teikyo University. Written informed consent was obtained
from all patients.
Follow-up of patients
Postoperative follow-up was performed along the
guidelines published by the Japanese Society for Cancer of the
Colon and Rectum. Confirmation of recurrence in all patients was
required by evaluating imaging or pathological diagnosis. Physical
examination and tumor marker (CEA and CA19-9) testing was conducted
every 3 months for 3 years, and then every 6 months for 5 years.
Computed tomography (CT) or magnetic resonance imaging (MRI) scans
were repeated every 3 months for 3 years, and then 6 months for up
to 5 years after surgery. Colon evaluation, including colonoscopy
or colon radiography was performed every 2 years or annually for 3
years.
LCM and RNA isolation
Frozen tissues were used for laser capture
microdissection (LCM). Ten micrometer-thick frozen sections of CRC
and normal tissues were prepared using a Leica CM 1900 cryostat
(Leica, Germany). The sections were placed on Membrane slides
(Leica), fixed in 75% alcohol for 30 sec and stained with 0.5%
violet-free methyl green (Sigma, USA). After staining, the sections
were air-dried and microdissected using a Leica AS LMD system
(Leica). Total RNAs were extracted by miRNeasy Mini kit (Qiagen
Inc., Valencia, CA, USA) and treated with DNase I according to the
manufacturer’s instructions (Qiagen Inc.). Then, total RNA was
reverse transcribed to complementary DNA (cDNA) using SuperScript
II reverse transcriptase system with random hexamer primers
according to the manufacturer’s protocol (Invitrogen Corporation,
Carlsbad, CA, USA).
Real-time PCR-based miRNA array
The miRNA expression profiling was performed using
Cancer microRNA qPCR array with the QuantiMir system (System
Biosciences, Mountain View, CA, USA). This system is a real-time
PCR-based array containing a panel of 95 cancer-related mature
miRNA assays and the U6 transcript as a normalization signal. In
brief, total RNA containing small RNA extracted from tissue samples
was first polyadenylated by poly(A) polymerase and then reverse
transcribed to cDNA using a mixture of oligo(dT) adaptor. The cDNA
then served as the template for SYBR real-time PCR using Power SYBR
Master Mix (Life Technologies, Carlsbad, CA, USA), using
miRNA-specific primers provided by the manufacturer. SYBR PCR was
performed using a LightCycler 480 real-time PCR system (Roche
Applied Science, Indianapolis, IN, USA). ΔCt was calculated by
subtracting the Ct values of U6 from the Ct values of the gene of
interest. ΔΔCt was then calculated by subtracting ΔCt of the
control from ΔCt of the sample. The fold change of the gene was
calculated by the equation: 2−ΔΔCt.
TaqMan microRNA assay for miR-21
Total RNAs, including the miRNAs were extracted from
306 primary CRC tissues and normal tissues by miRNeasy Mini kit
(Qiagen Inc.). The miR-21 and RNU6B-specific cDNA were synthesized
using gene-specific primer sets according to the TaqMan microRNA
assays protocol (Life Technologies). RNU6B was used as the internal
control. In brief, reverse transcriptase reactions contained 10 ng
of total RNAs, 50 nmol/stem-loop RT primer, 1X RT buffer, 0.25
mmol/l each of deoxynucleotide triphosphate (dNTP), 3.33 U/μl
MultiScribe reverse transcriptase, and 0.25 U/μl RNase inhibitor.
The 7.5 μl reaction samples were incubated in a GeneAmp PCR System
9700 (Life Technologies) for 30 min at 16°C, 30 min at 42°C, 5 min
at 85°C and then folded at 4°C. Quantitative real-time PCR was
performed using Step One (Life Technologies). The 20 μl PCR samples
included 1.33 μl of RT products, 10 μl of TaqMan Universal PCR
Master Mix, 1 μl of primers and probe mix, and 7.67 μl of
nuclease-free water and were incubated in optical plates. The PCR
conditions were 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 6 sec. Each sample was analyzed in duplicate.
Relative quantification of miRNA expression was calculated using
the 2−ΔΔCt method, where ΔΔCt = ΔCt (Ct miR21
− Ct RNU6B)tumor tissues − ΔCt (Ct
miR21 − Ct RNU6B)normal
tissues.
Statistical analysis
Relationships between miR-21 expression and
clinicopathological factors were analyzed using the Student’s
t-test, the Chi-square test and ANOVA. The cut-off level of miR-21
was determined as the mean level of relative quantity according to
a previously published study (11).
Overall survival (OS) and disease-free survival (DFS) curves were
analyzed using the Kaplan-Meier survival curve method, and the
differences were examined using log-rank tests. Cox
proportional-hazards regression analysis was used to estimate the
univariate and multivariate hazard ratios for OS and DFS. All
p-values are two-sided, and p<0.05 was considered to indicate a
statistically significant result. Statistical analyses were
performed using JMP 9.0 software (SAS Institute, Inc., Cary, NC,
USA).
Results
miRNA profile of CRC
First, we examined the profiling of miRNA expression
in the CRC tissues using the real-time PCR-based miRNA arrays
containing a panel of 95 cancer-related mature miRNAs. In this
assay, we used pooled RNA samples of primary CRC tissues and
matched normal tissues adjacent to the tumor tissues from 5
patients. Table I shows the top
five miRNAs which were upregulated in this assay. miR-21, miR-224,
miR-96, miR-31 and miR-155 were included in this list. Our results
showed the highest upregulation of miR-21 as compared with other
miRNAs. On the basis of these profiling results and other
information from published studies, we selected miR-21 as a marker
for CRC and for use in the following study.
 | Table IFive most highly upregulated miRNAs in
the CRC samples based on the miRNA profiling array. |
Table I
Five most highly upregulated miRNAs in
the CRC samples based on the miRNA profiling array.
| microRNA | MirBase no. | Fold-change |
|---|
| 1 | miR-21 | MIMAT0000076 | 5.10 |
| 2 | miR-224 | MIMAT0000281 | 4.82 |
| 3 | miR-96 | MIMAT0000095 | 4.76 |
| 4 | miR-31 | MIMAT0000089 | 4.53 |
| 5 | miR-155 | MIMAT0000646 | 3.16 |
Expression of miR-21 in each stage of CRC
tissues
The miR-21 expression levels of the primary CRC
tissues were compared for each Dukes’ stage (Fig. 1). These samples were collected from
306 CRC patients. miR-21 levels were normalized by RNU6B levels.
The means (± SE) expression level of miR-21 showed a significant
increase according to the level of advancement of the tumor stage
(p=0.041, Dukes A vs. C; p=0.029, Dukes A vs. D).
Relationship between clinicopathological
factors and miR-21 expression of the tumor tissues
This study included 306 CRC patients (190 men and
116 women) with a mean age of 65 years (range, 25–86). To evaluate
the correlation between the miR-21 expression levels and the
clinicopathological characteristics, patients were divided into two
groups (high and low expression). The cut-off level (15 as miR-21)
was determined as the mean level of the relative quantity. As shown
in Table II, a statistically
significant association was observed between high miR-21 expression
and depth of invasion, lymphatic and venous invasion, liver
metastasis and Dukes’ stage.
 | Table IIRelationship between
clinicopathological factors and high miR-21 expression in the tumor
tissues. |
Table II
Relationship between
clinicopathological factors and high miR-21 expression in the tumor
tissues.
| Variables | No. of patients
(n=306) | miR-21 high
(n=140) | Positive rate
(%) | P-value |
|---|
| Tumor size (cm) | | | | 0.606 |
| <5 | 171 | 76 | 44.4 | |
| ≥5 | 135 | 64 | 47.4 | |
| Histological
type | | | | 0.502 |
| Well | 210 | 93 | 44.3 | |
| Not well | 95 | 46 | 48.4 | |
| Depth of
invasion | | | | 0.015a |
| pT1 | 26 | 6 | 23.1 | |
| >pT2 | 280 | 134 | 47.9 | |
| Lymphatic
invasion | | | | 0.034a |
| Ly (−) | 188 | 77 | 41.0 | |
| Ly (+) | 118 | 63 | 53.4 | |
| Venous
invasion | | | | 0.010a |
| V (−) | 118 | 43 | 36.4 | |
| V (+) | 188 | 97 | 51.6 | |
| Lymph node
metastasis | | | | 0.067 |
| N (−) | 166 | 68 | 41.0 | |
| N (+) | 140 | 72 | 51.4 | |
| Liver
metastasis | | | | 0.033a |
| H (−) | 269 | 117 | 43.5 | |
| H (+) | 37 | 23 | 62.2 | |
| Peritoneal
dissemination | | | | 0.107 |
| P (−) | 289 | 129 | 44.6 | |
| P (+) | 17 | 11 | 64.7 | |
| Dukes’ stage | | | | 0.036a |
| A | 44 | 14 | 31.8 | |
| B | 106 | 46 | 43.4 | |
| C | 94 | 43 | 45.7 | |
| D | 62 | 37 | 59.7 | |
Correlation between miR-21 levels and
overall survival and disease-free survival
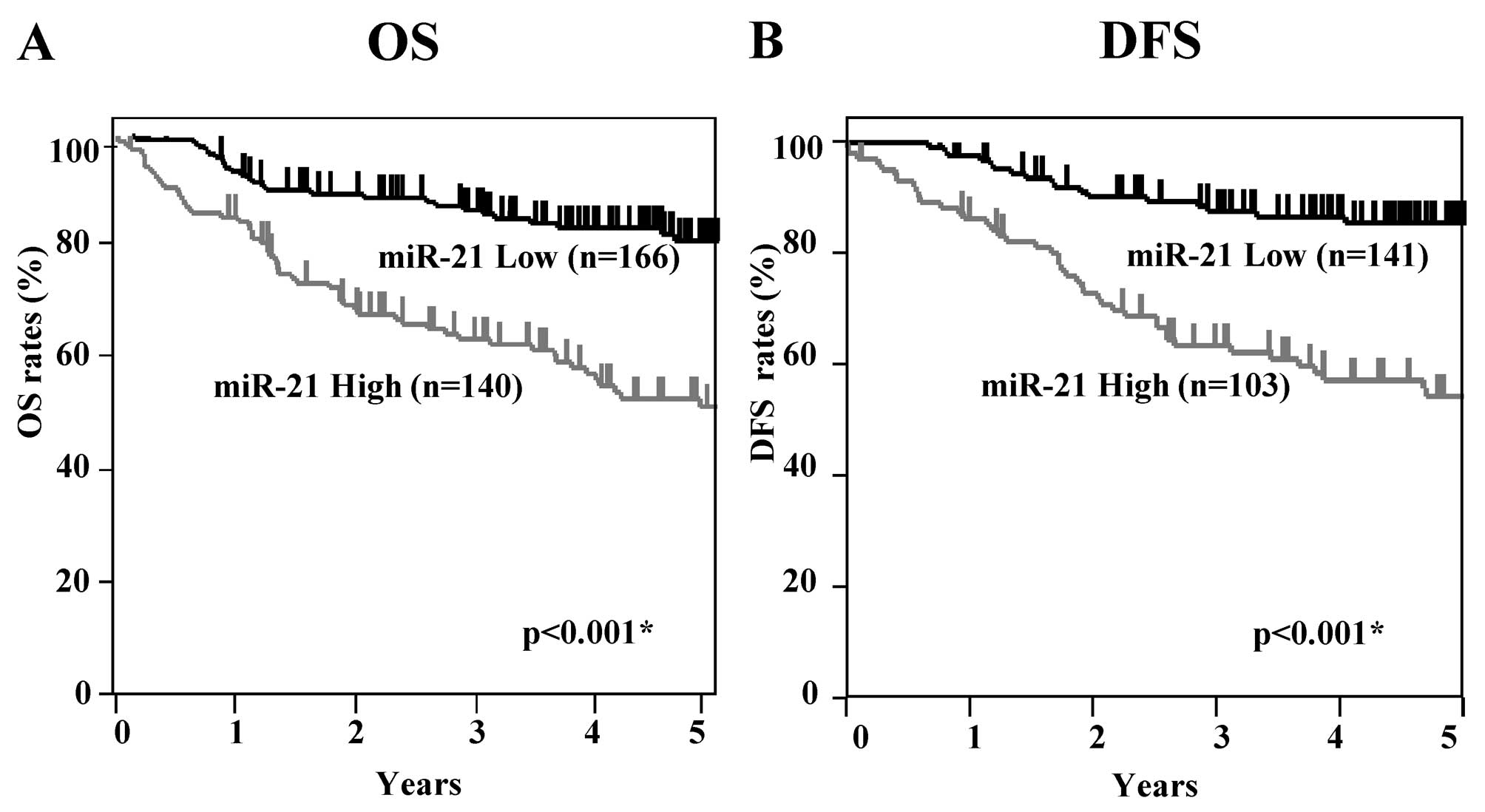
Kaplan-Meier OS and DFS curves of the CRC patients
according to the levels of miR-21 expression were examined
(Fig. 2). All patients were
included for OS analysis (n=306), and patients who had undergone
curative surgery were included for DFS analysis (n=244). The
patients in the high miR-21 expression group showed significantly
worse OS rates than patients in the low miR-21 group (log-rank
p<0.001). DFS analysis revealed that patients in the high miR-21
group had significantly worse survival rates than those in the low
expression group (log-rank p<0.001). These results suggest that
high expression of miR-21 is associated with poorer prognosis in
CRC patients.
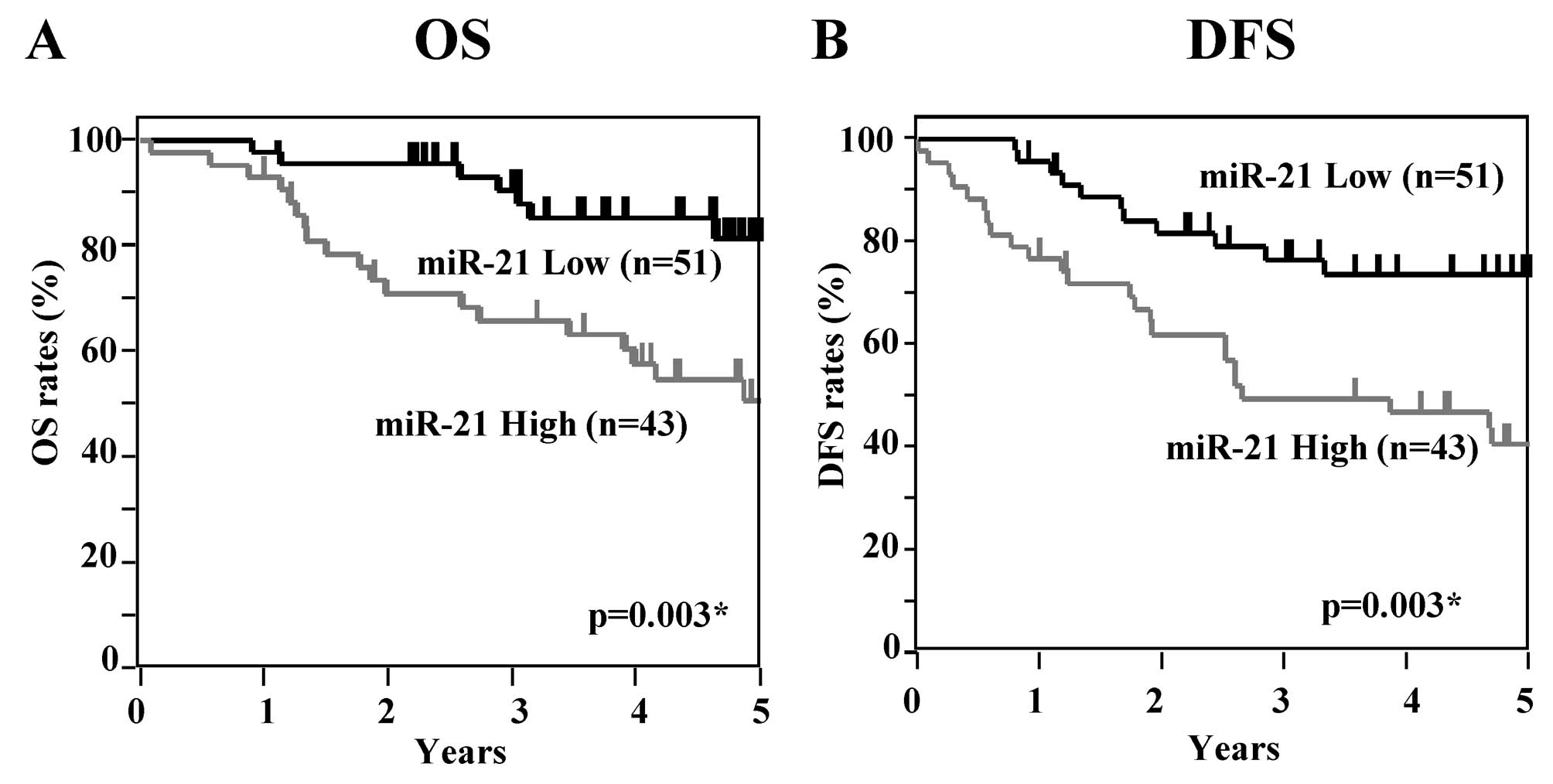
Next, OS and DFS curves at each tumor stage were
examined according to the levels of miR-21. In patients with Dukes’
stage A, significant differences between the high and low miR-21
expression groups were apparent in regards to DFS but not OS (OS
p=0.082, DFS p=0.038) (Fig. 3). In
Dukes’ stage B, patients in the high miR-21 expression group showed
significantly worse OS and DFS rates than those in the low
expression group (OS p=0.003, DFS p=0.002) (Fig. 4). In Dukes’ stage C, patients in the
high miR-21 expression group showed significantly worse OS and DFS
rates than patients with low miR-21 expression (OS, DFS p=0.003)
(Fig. 5). In Dukes’ stage D, OS
analysis revealed that patients with high miR-21 expression had
significantly worse survival rates than those with low miR-21
expression (p=0.015) (Fig. 6).
These results suggest that high expression of miR-21
is associated with poor prognosis in CRC patients with Dukes’ stage
A, B, C and D.
Univariate and multivariate Cox analyses
for overall survival and disease-free survival
Table III shows
the results of the univariate and multivariate Cox proportional
hazard regression analyses for OS in all of the CRC patients
(n=306). Multivariate analysis was performed for factors that
showed significance in the univariate analysis. In the univariate
analysis of all patients, depth of invasion, lymph node metastasis,
lymphatic and venous invasion, histological type, liver metastasis,
peritoneum dissemination, serum CEA, serum CA19-9, Dukes’ stage and
miR-21 showed a significant association with OS. In the
multivariate analysis, venous invasion, liver metastasis, Dukes’
stage and miR-21 level showed a significant association with OS.
Table IV shows the results of the
univariate and multivariate Cox analyses for DFS in CRC patients
who underwent curative surgery (n=244). In the univariate analysis,
depth of invasion, lymph node metastasis, lymphatic and venous
invasion, serum CEA, serum CA19-9, Dukes’ stage and miR-21 showed a
significant association with OS. In the multivariate analysis,
Dukes’ stage and miR-21 showed a significant association for DFS.
Next, we examined the univariate and multivariate Cox analyses in
patients with Dukes’ stage A, B, C and D, respectively. In the
univariate analysis of Dukes’ stage A patients, no factor showed a
significant association with OS and DFS (data not shown). In the
univariate analysis of Dukes’ stage B patients, venous invasion,
serum CEA and miR-21 showed a significant association with OS
(Table V). In the univariate
analysis for DFS, serum CEA, serum CA19-9 and miR-21 showed
significance. In the multivariate analysis of Dukes’ stage B
patients, only miR-21 showed significance for OS and DFS. In the
univariate analysis of Dukes’ stage C patients, lymphatic invasion,
serum CEA and miR-21 showed a significant association with OS
(Table VI). In the univariate
analysis for DFS, serum CEA, serum CA19-9 and miR-21 showed
significance. In the multivariate analysis of Dukes’ stage C
patients, miR-21 showed a significant association with OS and DFS.
Table VII shows the Cox analysis
in patients with Dukes’ stage D. In the univariate and multivariate
analyses, liver metastasis and miR-21 showed a significant
association with OS.
 | Table IIIUnivariate and multivariate analyses
of the prognostic factors for OS of all patients. |
Table III
Univariate and multivariate analyses
of the prognostic factors for OS of all patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Tumor size | 0.411 | 1.508
(0.984–2.312) | 0.059 | | - | |
| Depth of
invasion | 2.273 | 9.711
(2.161–171.168) | 0.001a | 1.104 | 3.017
(0.579–55.791) | 0.223 |
| Lymph node
metastasis | 1.369 | 3.932
(2.482–6.434) | <0.001a | 0.134 | 1.143
(0.445–3.268) | 0.789 |
| Lymphatic
invasion | 1.056 | 2.875
(1.871–4.467) | <0.001a | 0.288 | 1.334
(0.781–2.291) | 0.292 |
| Venous
invasion | 1.128 | 3.088
(1.860–5.429) | <0.001a | 0.629 | 1.875
(1.005–3.735) | 0.048a |
| Histological
type | 0.767 | 2.153
(1.394–3.307) | 0.001a | 0.158 | 1.171
(0.675–2.007) | 0.571 |
| Liver
metastasis | 2.122 | 8.351
(4.334–14.942) | <0.001a | 1.082 | 2.950
(1.177–6.879) | 0.022a |
| Peritoneum
dissemination | 1.648 | 5.194
(3.103–8.426) | <0.001a | 0.279 | 1.321
(0.624–2.68) | 0.456 |
| Serum CEA | 1.841 | 6.303
(3.710–11.453) | <0.001a | 0.496 | 1.643
(0.475–5.41) | 0.425 |
| Serum CA19-9 | 0.851 | 2.343
(1.658–3.337) | <0.001a | −0.081 | 0.922
(0.503–1.662) | 0.789 |
| Dukes’ stage | 1.140 | 3.127
(2.190–4.587) | <0.001a | 0.716 | 2.046
(1.121–3.765) | 0.020a |
| miR-21 | 1.125 | 3.080
(1.970–4.937) | <0.001a | 1.058 | 2.880
(1.696–5.084) | <0.001a |
 | Table IVUnivariate and multivariate analyses
of prognostic factors for DFS of all patients. |
Table IV
Univariate and multivariate analyses
of prognostic factors for DFS of all patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Tumor size | −0.110 | 0.896
(0.519–1.503) | 0.683 | | - | |
| Depth of
invasion | 1.342 | 3.825
(1.196–23.331) | 0.020a | 0.449 | 1.566
(0.261–12.422) | 0.630 |
| Lymph node
metastasis | 0.943 | 2.567
(1.550–4.307) | 0.001a | −1.015 | 0.363
(0.039–2.828) | 0.339 |
| Lymphatic
invasion | 0.544 | 1.723
(1.027–2.854) | 0.040a | −0.239 | 0.788
(0.440–1.382) | 0.408 |
| Venous
invasion | 1.250 | 3.490
(1.434–5.131) | 0.004a | 0.762 | 2.143
(0.532–3.142) | 0.303 |
| Histological
type | 0.337 | 1.401
(0.804–2.360) | 0.227 | | -
(0.675–2.007) | |
| Serum CEA | 0.595 | 1.814
(1.246–2.660) | 0.002a | 0.522 | 1.686
(0.937–3.041) | 0.081 |
| Serum CA19-9 | 0.703 | 2.020
(1.361–2.993) | 0.001a | 0.521 | 1.684
(0.902–3.094) | 0.101 |
| Dukes’ stage | 0.782 | 2.186
(1.283–3.885) | 0.004a | 0.706 | 2.026
(1.110–3.877) | 0.021a |
| miR-21 | 1.275 | 3.578
(2.098–6.359) | <0.001a | 1.078 | 2.940
(1.682–5.360) | <0.001a |
 | Table VUnivariate and multivariate analyses
of prognostic factors for OS and DFS in Dukes’ stage B
patients. |
Table V
Univariate and multivariate analyses
of prognostic factors for OS and DFS in Dukes’ stage B
patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| For overall
survival (OS) |
| Tumor size | −0.436 | 0.647
(0.173–2.053) | 0.467 | | - | |
| Lymphatic
invasion | 0.094 | 0.910
(0.202–3.051) | 0.887 | | - | |
| Venous
invasion | 1.893 | 6.641
(1.291–121.369) | 0.020a | 1.625 | 5.078
(0.968–93.408) | 0.056 |
| Histological
type | 0.001 | 1.001
(0.222–3.355) | 0.999 | | - | |
| Serum CEA | 1.296 | 3.655
(1.149–13.720) | 0.028a | 0.964 | 2.621
(0.818–9.906) | 0.106 |
| Serum CA19-9 | 0.337 | 1.401
(0.311–4.698) | 0.623 | | - | |
| miR-21 | 0.982 | 7.125
(1.876–46.376) | 0.003a | 1.813 | 6.130
(1.607–39.991) | 0.006a |
| For disease-free
survival (DFS) |
| Tumor size | −0.726 | 0.484
(0.174–1.177) | 0.112 | | - | |
| Lymphatic
invasion | −0.248 | 0.780
(0.256–1.972) | 0.618 | | - | |
| Venous
invasion | 0.503 | 1.653
(0.679–4.610) | 0.278 | | - | |
| Histological
type | −0.141 | 0.868
(0.285–2.195) | 0.778 | | - | |
| Serum CEA | 0.971 | 2.641
(1.137–6.413) | 0.024a | 0.723 | 2.060
(0.852–5.164) | 0.108 |
| Serum CA19-9 | 0.941 | 2.562
(1.023–5.985) | 0.045a | 0.671 | 1.957
(0.757–4.752) | 0.159 |
| miR-21 | 0.659 | 3.734
(1.532–10.422) | 0.003a | 1.215 | 3.369
(1.373–9.448) | 0.007a |
 | Table VIUnivariate and multivariate analyses
of prognostic factors for OS and DFS in Dukes’ stage C
patients. |
Table VI
Univariate and multivariate analyses
of prognostic factors for OS and DFS in Dukes’ stage C
patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| For overall
survival (OS) |
| Tumor size | −0.432 | 0.649
(0.277–1.425) | 0.286 | | - | |
| Lymphatic
invasion | 0.828 | 2.288
(1.039–5.390) | 0.040a | | - | |
| Venous
invasion | 0.388 | 1.475
(0.662–3.595) | 0.351 | | - | |
| Histological
type | 0.010 | 1.010
(0.413–2.257) | 0.981 | | - | |
| Serum CEA | 0.803 | 2.233
(1.025–5.106) | 0.043a | 0.727 | 2.069
(0.949–4.736) | 0.068 |
| Serum CA19-9 | 0.737 | 2.089
(0.949–4.531) | 0.067 | | - | |
| miR-21 | 1.228 | 3.413
(1.499–8.743) | 0.003a | 1.179 | 3.251)
(1.425–8.340) | 0.005a |
| For disease-free
survival (DFS) |
| Tumor size | −0.054 | 0.947
(0.477–1.842) | 0.874 | | - | |
| Lymphatic
invasion | 0.521 | 1.683
(0.862–3.390) | 0.128 | | - | |
| Venous
invasion | 0.662 | 1.940
(1.943–4.385) | 0.073 | | - | |
| Histological
type | 0.137 | 1.147
(0.551–2.267) | 0.703 | | - | |
| Serum CEA | 0.817 | 2.264
(1.160–4.557) | 0.017a | 0.434 | 1.544
(0.663–3.571) | 0.312 |
| Serum CA19-9 | 0.940 | 2.560
(1.308–4.989) | 0.007a | 0.562 | 1.754
(0.775–4.067) | 0.179 |
| miR-21 | 1.030 | 2.801
(1.405–5.952) | 0.003a | 0.921 | 2.512
(1.250–5.372) | 0.009a |
 | Table VIIUnivariate and multivariate analyses
of prognostic factors for OS in Dukes’ stage D patients. |
Table VII
Univariate and multivariate analyses
of prognostic factors for OS in Dukes’ stage D patients.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Tumor size | 0.090 | 1.094
(0.590–2.130) | 0.781 | | - | |
| Lymph node
metastasis | 0.642 | 1.900
(0.933–4.307) | 0.079 | | - | |
| Lymphatic
invasion | 0.443 | 1.558
(0.839–3.037) | 0.163 | | - | |
| Venous
invasion | 0.205 | 1.228
(0.558–3.240) | 0.633 | | - | |
| Histological
type | 0.534 | 1.705
(0.932–3.185) | 0.084 | | - | |
| Liver
metastasis | −0.756 | 0.470
(0.258–0.860) | 0.015a | −0.796 | 0.451
(0.246–0.831) | 0.011a |
| Peritoneum
dissemination | 0.422 | 1.525
(0.768–2.864) | 0.218 | | - | |
| Serum CEA | −0.722 | 0.486
(0.215–1.306) | 0.141 | | - | |
| Serum CA19-9 | −0.551 | 0.577
(0.278–1.194) | 0.136 | | - | |
| miR-21 | 0.860 | 2.363
(1.276–4.524) | 0.006a | 0.895 | 2.448
(1.316–4.712) | 0.005a |
These results suggest that miR-21 levels of tumor
tissues have an independent prognostic value for OS and DFS in CRC
patients with Dukes’ stage B, C and D.
Discussion
In the present study, we examined the prognostic
value of miR-21 in CRC patients at each tumor stage. Our results
demonstrated that the high expression of miR-21 in CRC tissues
indicated a significantly poor prognosis for OS and DFS in CRC
patients with Dukes’ stage B and C, and for OS in patients with
Dukes’ stage D.
It has been reported that miR-21 is one of the
prominent miRNAs implicated in the genesis and progression of human
cancer (17). Not only has it been
implicated in the promotion of tumor growth, proliferation and
response to chemotherapy, but studies have also shown that miR-21
is overexpressed in several human malignant solid tumors such as
glioblastoma, hepatocellular and pancreatic cancer and CRC
(9,18–20).
In the present study, we demonstrated a significantly increased
expression of miR-21 in the CRC tissues, along with the progression
of tumor stage. It has been reported that the expression of miR-21
is associated with more advanced stages of CRC (16). Furthermore, we examined the
association of miR-21 expression and clinicopathological factors,
and found that high miR-21 expression had a significant association
with depth of invasion, lymphatic and venous invasion, liver
metastasis and Dukes’ stage. Another study revealed that CRC
patients overexpressing miR-21 are more likely to have lymph node
and distant metastasis (17).
The prognostic value of miR-21 has been reported in
many types of cancer, including CRC. In the present study, we
demonstrated that miR-21 expression in tumor tissues was
significantly associated with the poor prognosis of patients with
CRC. Similar results were reached independently by Schetter et
al (16). However, the
significance of miR-21 expression in each tumor stage has not been
previously reported. In the present study, the prognostic
significance of miR-21 levels was investigated using the
Kaplan-Meier method and Cox multivariate analysis on 306 patients
with CRC at each Dukes’ stage. In these analysis, our data
demonstrated the existence of an independent prognostic impact of
miR-21 for OS and DFS in CRC patients with Dukes’ stage B and C,
and for OS in patients with Dukes’ stage D. In Dukes’ stage A
patients, miR-21 was associated with a significantly worse
prognosis for DFS in the Kaplan-Meier analysis. However, a
significant prognostic impact was not demonstrated in the Cox
analysis of Dukes’ stage A patients. This may be due to the low
number of patients who suffered recurrence and death. To the best
of our knowledge, this is the first study to clarify the
significance of the prognostic value of miR-21 at each stage in CRC
patients. At present, the role of adjuvant chemotherapy for Dukes’
stage B patients remains controversial. Many Dukes’ stage B
patients will benefit from therapy. However, if surgery is
curative, additional therapy may harm the quality of life with
little therapeutic benefit. Therefore, it is important to develop
novel biomarkers to identify high-risk patients who may be suitable
for therapeutic intervention. In the present study, we established
that miR-21 has potential for the selection of high-risk patients
in need of adjuvant chemotherapy among Dukes’ stage B patients.
miRNAs target protein-coding mRNAs at the
post-transcriptional level by direct cleavage of the mRNAs in two
different ways: one is direct cleavage of the target mRNAs and the
other is inhibition of protein synthesis (9,21,22).
Bioinformatically predicted targets for miR-21 that have been
experimentally validated include the tumor-suppressor genes:
programmed cell death 4 (PDCD4), phosphatase and tensin homologue
(PTEN), tropomyosin 1 (TPM1), reversion-inducing cysteine-rich
protein (RECK), ras homing gene family member B or maspin (23–28).
PDCD4 has been characterized as a novel tumor-suppressor gene that
acts as a suppressor of transformation, tumorigenesis and
progression, invasion and metalloproteinase activation and as an
inducer of apoptosis. We previously demonstrated a significant
inverse relationship between miR-21 and PDCD4 mRNA in CRC samples
(29). miR-21 targets the 3′-UTR of
the PDCD4 gene at nucleotides 228–249 with perfect complementarity,
thereby post-transcriptionally regulating its expression (15). Through the inhibition of PDCD4
tumor-suppressor gene by miR-21, tumor growth may be promoted,
leading to a poor prognosis. We will await further study to clarify
the tumor regulation mediated by miR-21.
In conclusion, the present study demonstrated that
miR-21 may be a valuable biomarker for identifying high-risk CRC
patients with Dukes’ stage B or C. To clarify the prognostic value
of miR-21 in Dukes’ stage A patients, further study with a large
number of cases is required.
Acknowledgements
We thank Miss J. Tamura for her excellent technical
assistance, and all members of the colorectal group for their
clinical suggestions. This study was supported by a Grant-in-Aid
for Scientific Research (C) (24591984) and (C) (25462070).
Abbreviations:
|
miR-21
|
microRNA-21
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
CRC
|
colorectal cancer
|
References
|
1
|
Gill S, Thomas RR and Goldberg RM: Review
article: colorectal cancer chemotherapy. Aliment Pharmacol Ther.
18:683–692. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aggarwal S and Chu E: Current therapies
for advanced colorectal cancer. Oncology. 19:589–595.
2005.PubMed/NCBI
|
|
3
|
André T, Boni C, Mounedji-Boudiat L, et
al: Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment
for colon cancer. N Engl J Med. 350:2343–2351. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meropol NJ and Schulman KA: Cost of cancer
care: issues and implications. J Clin Oncol. 25:180–186. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chung KY and Kelsen D: Adjuvant therapy
for stage II colorectal cancer; who and with what? Curr Treat
Options Gastroenterol. 9:272–280. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aslam MI, Taylor K, Pringle JH and Jameson
JS: microRNAs are novel biomarkers of colorectal cancer. Br J Surg.
96:702–710. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar
|
|
8
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caldas C and Brenton JD: Sizing up miRNAs
as cancer genes. Nat Med. 11:712–714. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L,
Kipps T, Negrini M, Bullrich F and Croce CM: Frequent deletions and
down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in
chronic lymphocytic leukemia. Proc Natl Acad Sci USA.
99:15524–15529. 2002. View Article : Google Scholar
|
|
11
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M,
Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C,
Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M and
Croce CM: A microRNA signature associated with prognosis and
progression in chronic lymphocytic leukemia. N Engl J Med.
353:1793–1801. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
16
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu
CG, Calin GA, Croce CM and Harris CC: MicroRNA expression profiles
associated with prognosis and therapeutic outcome in colon
adenocarcinoma. JAMA. 299:425–436. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR21, miR-31, miR-143 and miR-145 is related to
clinicopathological features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar
|
|
18
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I,
Calin GA, Querzoli P, Negrini M and Croce CM: MicroRNA gene
expression deregulation in human breast cancer. Cancer Res.
65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang KH, Miler N, Kheirelseid EA, et al:
MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg
Oncol. 37:597–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang ZX, Lu BB, Wang H, et al: MicroRNA-21
modulates chemosensitivity of breast cancer cells to doxorubicin by
targeting PTEN. Arch Med Res. 42:281–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ziyan W, Shuhua Y, Xiufang W and Xianoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar
|
|
27
|
Liu M, Tang Q, Qiu M, et al: miR-21
targets the tumor suppressor RhoB and regulates proliferation,
invasion and apoptosis in colorectal cancer cells. FEBS Lett.
585:2998–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Torres A, Torres K, Paszkowski T, et al:
Highly increased maspin expression corresponds with up-regulation
of miR-21 in colorectal cancer: a preliminary report. Int J Gynecol
Cancer. 21:8–14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Horiuchi A, Iinuma H, Akahane T, Shimada R
and Watanabe T: Prognostic significance of PDCD4 expression and
association with microRNA-21 in each Dukes’ stage of colorectal
cancer patients. Oncol Rep. 27:1384–1392. 2012.PubMed/NCBI
|