Introduction
Colorectal cancer is the third leading cause of
cancer-related deaths worldwide, and the 5-year survival rate is
mostly dependent on the cancer stage, resulting in a survival rate
ranging from 10 to 95% (1). If
patients are not diagnosed in the early stage, this malignancy
seriously threatens patient survival. 5-Fluorouracil (5-FU) is a
widely used therapeutic agent for treating advanced colorectal
cancer (2). However, some patients
develop 5-FU resistance which is a major obstacle to effective
therapy. Therefore, there is urgent need to elucidate the molecular
mechanism underlying the 5-FU resistance in colorectal cancer cells
and develop novel diagnostic biomarkers for 5-FU resistance that
will facilitate the development of therapeutic strategies for
colorectal cancer patients.
Thymidylate synthase (TYMS) is a key therapeutic
target of 5-FU and serves as a predictive biomarker of the cellular
response to 5-FU treatment. The antitumor effect of 5-FU was found
to be promoted by rosemary extract via the downregulation of TYMS
in 5-FU-resistant colorectal cancer cells (3). One study demonstrated that metastatic
colorectal cancer that did not response to 5-FU treatment had a
high TYMS level (4). High TYMS
expression induces 5-FU resistance in human colon cancer cells that
are continuously exposed to 5-FU (5). In addition, TYMS is associated with
5-FU resistance of colon and gastric cancers (6,7).
Emerging evidence demonstrates that ~60% of
protein-coding genes are controlled by microRNAs (miRNAs) (8), which are a conserved class of small
(~22 nt) non-coding RNA molecules and that target mRNA cleavage or
translational repression via binding to the sequences on the 3′
untranslated region (3′UTR) of target mRNAs with complete or
incomplete complementarity (9).
miRNAs play important roles in diverse biological processes, such
as cell proliferation, cell death and developmental timing
(10). Evidence indicates that
aberrant miRNA expression is linked to the initiation and
development of diverse cancers, as well as cancer cell resistance
to therapeutic agents (11–13). For example, miR-129 increases cell
sensitivity to 5-FU by targeting Bcl-2 in colorectal cancer
(14). miR-433 enhances HeLa
cellular sensitivity to 5-FU by downregulating TYMS (15), a first report concerning the
regulation of TYMS by miRNAs in the cell response to 5-FU. However,
the regulation of cell resistance to 5-FU by miR-203 in colorectal
cancer remains known.
In the present study, we found that miR-203 was
downregulated in 5-FU-resistant cell line LoVo/5-Fu compared with
LoVo cells using miRNA microarray. miR-203 expression was inversely
correlated with the extent of 5-FU resistance. miR-203 inhibition
enhanced cell resistance to 5-FU, while miR-203 overexpression
increased 5-FU sensitivity. TYMS was validated as a direct target
of miR-203 and TYMS knockdown resulted in similar effects on the
cellular response to 5-FU as that of miR-203 overexpression.
Finally, we found that the inhibitory effect of 5-FU on colorectal
cancer growth was enhanced by miR-203 overexpression.
Materials and methods
Cell culture
Human normal colorectal mucosa FHC cells and
colorectal cancer cell lines HCT-116, Caco2 and SW480 were cultured
in DMEM. HT29 and DLD1 cells were cultured in RPMI-1640, and LoVo
cells were cultured in F12K medium (Gibco, Life Technologies, USA).
All medium was supplemented with 10% fetal bovine serum (FBS)
(Gibco, Life Technologies) and 1% antibiotic solution (100 U/ml
penicillin and 100 μg/ml streptomycin) (Sigma, USA). The cells were
maintained in a humidified incubator (Thermo Fisher, USA) at 37°C
with 5% CO2. The 5-FU highly resistant cell line
LoVo/5-Fu (HR) was acquired by treating LoVo cells with 5-FU
(Sigma) ranging from a concentration of 0.1 to 2 μg/l for ~11
months. The middle resistant cell line LoVo/5-Fu (MR) was acquired
after ~6 months of treatment.
RNA isolation and qRT-PCR
Total RNAs (including miRNAs) were extracted from
cells or tumor tissues using TRIzol reagent (Invitrogen, USA)
according to the manufacturer’s instructions. After the RNA
concentration was measured, 500 ng of RNA was reverse transcribed
using a specific miR-203 primer by M-MLV reverse transcriptase
(Takara, Japan). qRT-PCR was performed using SYBR-Green PCR Master
Mix and specific miR-203 primers on an ABI 7300 real-time PCR
system. Small nuclear RNA U6 (U6 snRNA) was used as an internal
control to normalize miR-203 expression. The primers for reverse
transcription and PCR were as follows: miR-203 reverse transcript
primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTAGTGGT-3′; miR-203
forward primer, 5′-ACACTCCAGCTGGGGTGAAATGTTTA-3′ and miR-203
reverse primer, 5′-TGGTGTCGTGGAGTCG-3′; U6 forward,
CTCGCTTCGGCAGCACA3′ and U6 reverse, 5′AACGCTTCACGAATTTGCGT-3′.
Western blotting
After transfection with miR-203 ASO or miR-203
mimics or siRNA against TYMS or scrambled controls for ~48 h,
colorectal cancer cells were harvested and lysed using RIPA buffer
(50 mM Tris-HCl, pH 8.8, 150 mM NaCl, 1% NP-40, 1% sodium
deoxycholate, 0.1% SDS). Proteins were also extracted from the
tumor tissues using RIPA according to the manufacturer’s protocols.
Proteins (30 μg) were separated on a SDS-PAGE gel and then
transferred to a polyvinylidene difluoride (PVDF) membrane,
followed by incubation with 5% milk. The membrane was incubated in
the mouse monoclonal to TYMS antibody (used as a primary antibody)
(1:1,000; Abcam, USA) overnight at 4°C, and subsequently in the
HRP-conjugated goat anti-mouse antibody (used as a secondary
antibody) for 1.5 h. Finally, the protein signals were detected
using enhanced chemiluminescence (ECL) according to the
manufacturer’s instructions. GAPDH was used as a loading control to
normalize the expression of TYMS.
Luciferase assay
The 3′UTR of TYMS containing the binding site for
miR-203 was amplified and cloned in the downstream region of a
luciferase coding gene. Several base mutations within the binding
site were generated using a QuikChange® site-directed
mutagenesis kit (Stratagene, USA). Colorectal cancer cells were
co-transfected with either 40 nM miR-203 ASO or miR-203 mimics and
200 ng of wild-type or mutant TYMS 3′UTR. After transfection for 48
h, cells were harvested to analyze the luciferase intensity using a
Dual-Glo luciferase assay (Promega, USA) according to the
manufacturer’s instructions. Renilla luciferase intensity
was used to normalize the firefly luciferase intensity.
5-FU treatment and cytotoxicity
assay
Colorectal cancer cells transfected with miR-203 ASO
or miR-203 mimics or scrambled controls were seeded in 96-well
plates with 5,000 cells/well. Twenty-four hours after cell
adhesion, an additional 100 μl of 5-FU solution was added for 72 h.
The 5-FU concentration varied from 0.2 to 3.2 μM in the LoVo cells,
from 2 to 32 μM in the SW480, HT29 and LoVo/5-Fu (HR) cells, and
from 1 to 16 μM in the Caco2 cells. The cytotoxicity was measured
using a MTT assay according to the manufacturer’s information.
Xenograft assay
The mouse experiments were approved by the
Institutional Animal Care and Use Committee of Tianjin Union
Medical Center, and performed under specific pathogen-free
conditions. LoVo (1×106) cells were suspended in 100 μl
of phosphate-buffered saline and subcutaneously injected into the
flank of 4–5 week NOD/SCID mice. On day 12 when the tumor size
reached ~100 mm3, the mice were randomly separated into
four groups and subsequently injected with the miR-203-negative
control or miR-203 or 5-FU alone or 5-FU and miR-203 together on
day 12, 15, 18, 21 and 25. Tumor volume was examined by measuring
the length (L) and width (W) with calipers and was calculated with
the formula (LxW2)/2.
Statistical analysis
All experiments were confirmed at least three times.
The data are shown as means ± standard deviation (SD) and one
representative data value is shown in the present study. The
statistical significance was evaluated using two-tailed Student’s
t-test between two groups and analyzed using GraphPad Prism
software. A difference was considered statistically significant at
a value of p<0.05.
Results
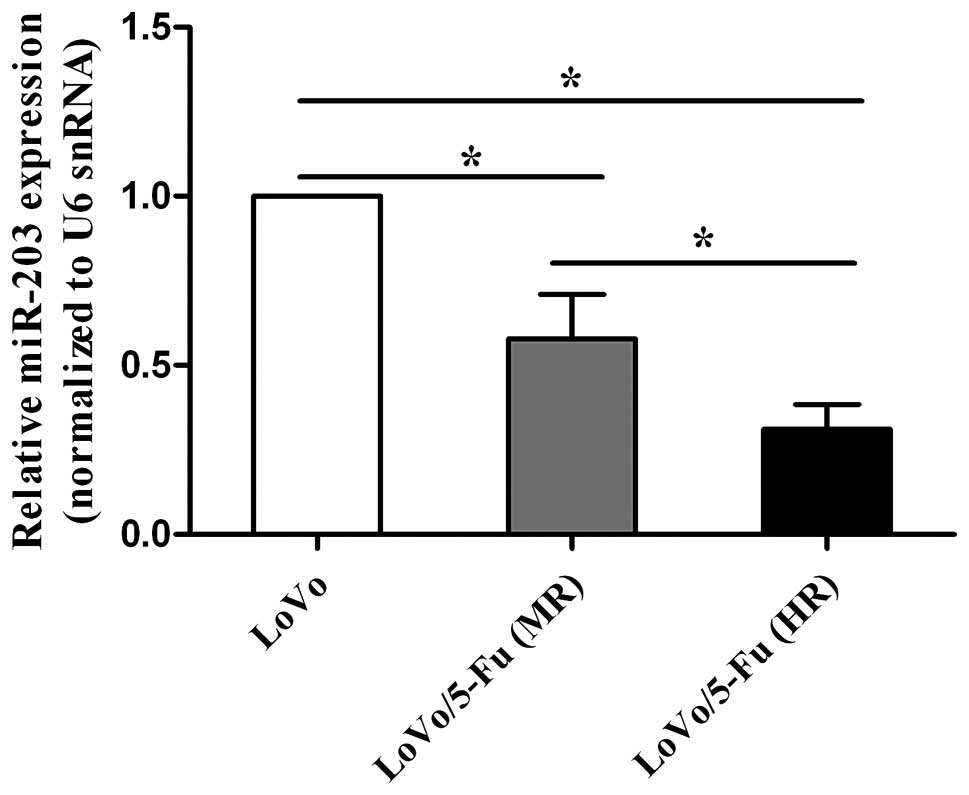
miR-203 is downregulated in 5-FU
resistant cells
To investigate the differentially expressed miRNAs
in 5-FU-resistant cells, we performed miRNA microarray between the
LoVo and LoVo/5-Fu cells and found that miR-203 was expressed to
the lesser degree in the LoVo/5-Fu cells. Cytotoxicity assay
indicated that LoVo/5-Fu (MR) and LoVo/5-Fu (HR) cells had a higher
IC50 value for 5-FU than that of the LoVo cells
(Table I), while there was no
significant difference in the IC50 value for CDDP among
these cell lines. Based on the miRNA microarray data, miR-203
expression was confirmed using qRT-PCR in the LoVo, LoVo/5-Fu (MR)
and LoVo/5-Fu (HR) cell lines (Fig.
1). Taken together, these data indicate that miR-203 expression
may be negatively correlated with the IC50 value of
5-FU, suggesting that miR-203 is closely related to 5-FU
chemosensitivity.
 | Table IThe IC50 values of 5-FU and
CDDP in LoVo and 5-Fu-resistant LoVo cell lines. |
Table I
The IC50 values of 5-FU and
CDDP in LoVo and 5-Fu-resistant LoVo cell lines.
| Cell lines | IC50
(μmol/l)
5-FU | IC50
(μmol/l)
CDDP |
|---|
| LoVo | 1.62±0.14 | 4.13±0.62 |
| LoVo/5-FU (MR) | 4.28±0.47a | 43.91±0.51ns |
| LoVo/5-FU (HR) | 15.45±1.18b,c | 4.28±0.69ns |
miR-203 inhibition enhances 5-FU
chemoresistance in colorectal cancer cells
To determine the effect of miR-203 on the cell
sensitivity to 5-FU in colorectal cancer, we first detected miR-203
expression in a series of colorectal cancer cell lines. qRT-PCR
assay showed that miR-203 was upregulated in most of the colorectal
cancer cell lines, except Caco2 cells (Fig. 2A). Second, we suppressed endogenous
miR-203 expression in LoVo and SW480 cells by transient
transfection with miR-203 ASO or scrambled control. We found that
miR-203 was significantly reduced in the miR-203 ASO-transfected
cells (Fig. 2B). Finally,
cytotoxicity assay indicated that miR-203 inhibition resulted in an
increase in the survival rate of LoVo cells, which implies that
miR-203 inhibition enhances the chemoresistance to 5-FU (Fig. 2C). Similar results were observed in
the SW480 cells (Fig. 2D).
miR-203 overexpression increases 5-FU
chemosensitivity in colorectal cancer cells
To further validate the role of miR-203 in the
cellular response to 5-FU, we overexpressed miR-203 in LoVo/5-Fu
(HR), Caco2 and HT29 cells by transient transfection with miR-203
mimics or mimic control, and miR-203 expression was confirmed by
qRT-PCR assay (Fig. 3A). From the
cytotoxicity assay, we found that miR-203 overexpression reduced
the survival rate of LoVo/5-Fu (HR) cells compared with the
controls (Fig. 3B). In line with
the roles of miR-203 in LoVo/5-Fu (HR) cells, miR-203
overexpression resulted in a decrease in the survival rate of Caco2
cells (Fig. 3C) and HT29 cells
(Fig. 3D). Taken together, these
data suggest that miR-203 overexpression increases the cell
sensitivity to 5-FU.
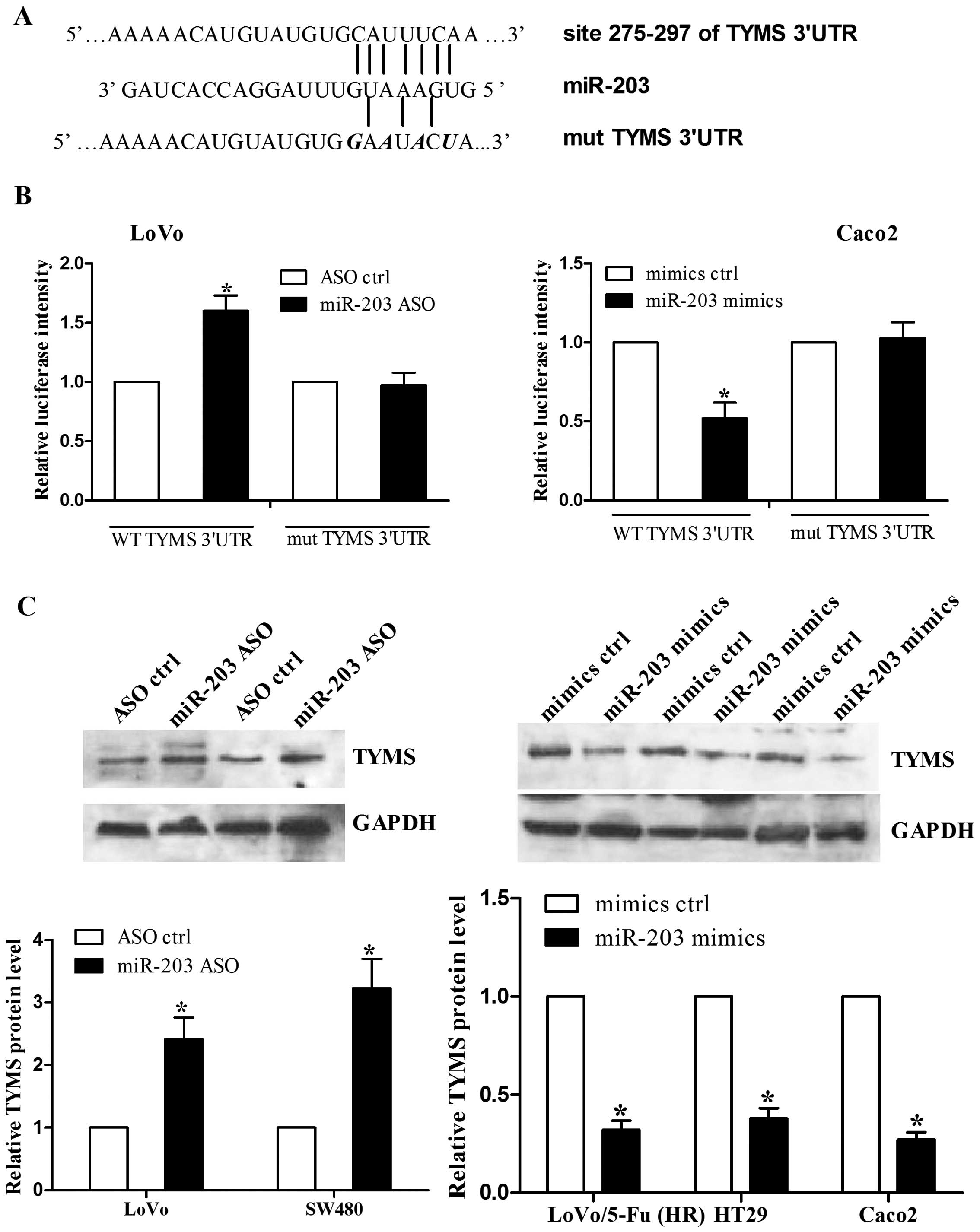
TYMS is a direct target of miR-203
We discovered that there was a putative binding site
for miR-203 in the 3′UTR of TYMS mRNA (Fig. 4A), which has been validated to be
involved in 5-FU chemoresistance. We then constructed a luciferase
reporter that contained the TYMS 3′UTR downstream of the luciferase
coding gene. In addition, a mutant TYMS 3′UTR was also generated as
shown in Fig. 4A. LoVo and Caco2
cells were co-transfected with either miR-203 ASO or miR-203 mimics
and wild-type or mutant TYMS 3′UTR for luciferase assay. As shown
in Fig. 4B, we found that LoVo
cells with miR-203 ASO had a higher level of TYMS 3′UTR intensity
than the control cells, while Caco2 cells with miR-203 mimics had a
lower intensity. However, neither miR-203 ASO nor miR-203 mimics
affected the intensity of mutant TYMS 3′UTR (Fig. 4B). The luciferase assay suggests
that miR-203 suppresses TYMS expression via binding to its 3′UTR.
To further investigate the roles of miR-203 on TYMS protein level,
colorectal cancer cells transfected with either miR-203 ASO or
miR-203 mimics were subjected to western blot assay (Fig. 4C). We found that miR-203 inhibition
increased TYMS protein levels, while miR-203 overexpression reduced
TYMS expression. Overall, these data indicate that TYMS is a direct
target of miR-203 in colorectal cancer cells.
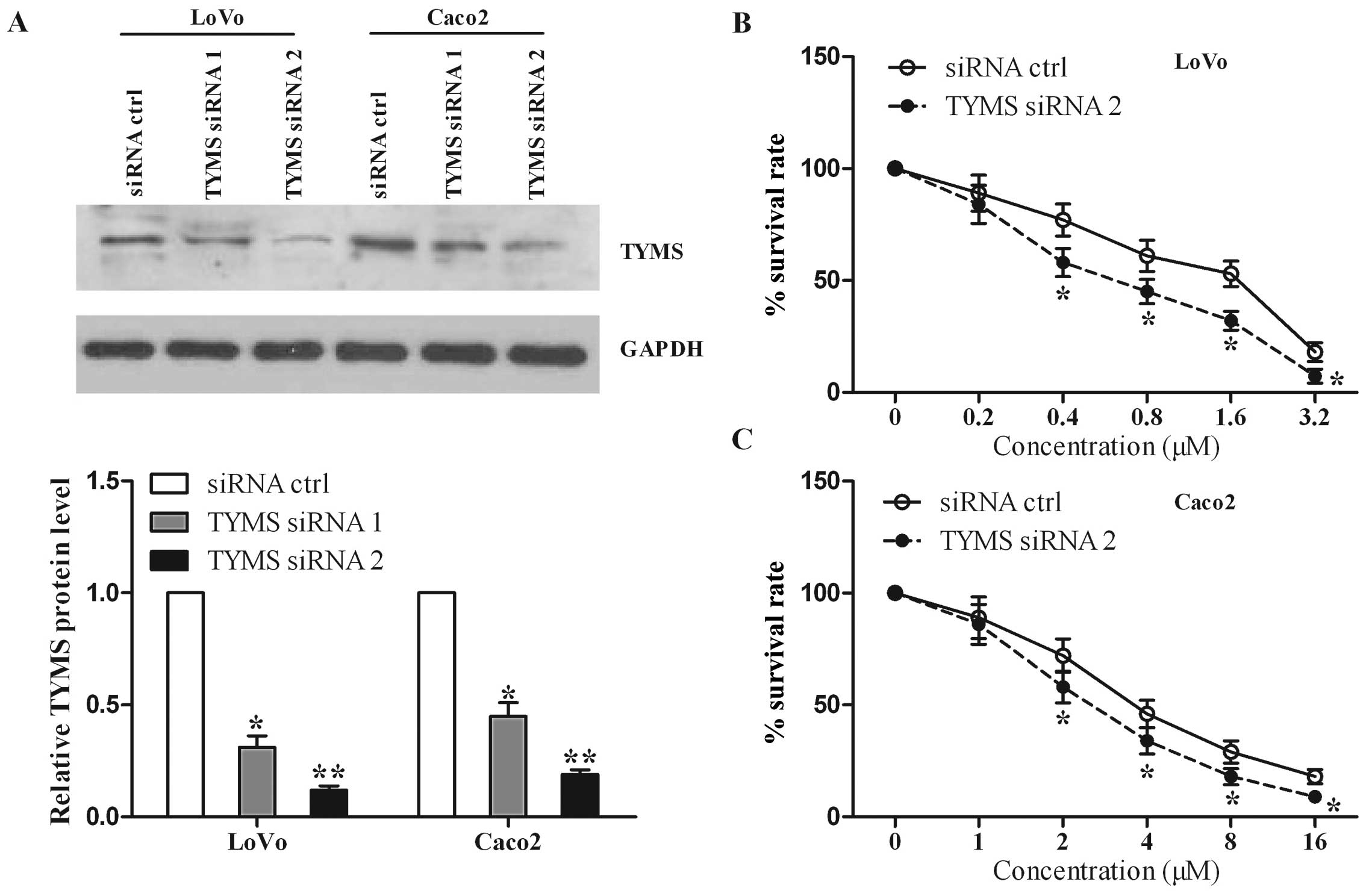
Silencing of TYMS increases 5-FU
chemosensitivity
Considering that TYMS is a target of miR-203, we
next investigate the effects of TYMS on the cell sensitivity to
5-FU in colorectal cancer cells. LoVo and Caco2 cells were
transfected with TYMS-specific siRNAs and scrambled siRNA control.
As shown in Fig. 5A, we found that
cells with siRNA1 and siRNA2 specific for TYMS had a lower TYMS
protein level than that in the control. Since siRNA2 resulted in a
more marked decrease in TYMS level than siRNA1, we chose siRNA2 for
the cytotoxicity assay. We found that TYMS knockdown suppressed the
survival rate of the LoVo cells compared with the control (Fig. 5B). Similar data were obtained for
Caco2 cells (Fig. 5C). These
results indicate that TYMS knockdown increases cell sensitivity to
5-FU, similar to the roles of miR-203 overexpression.
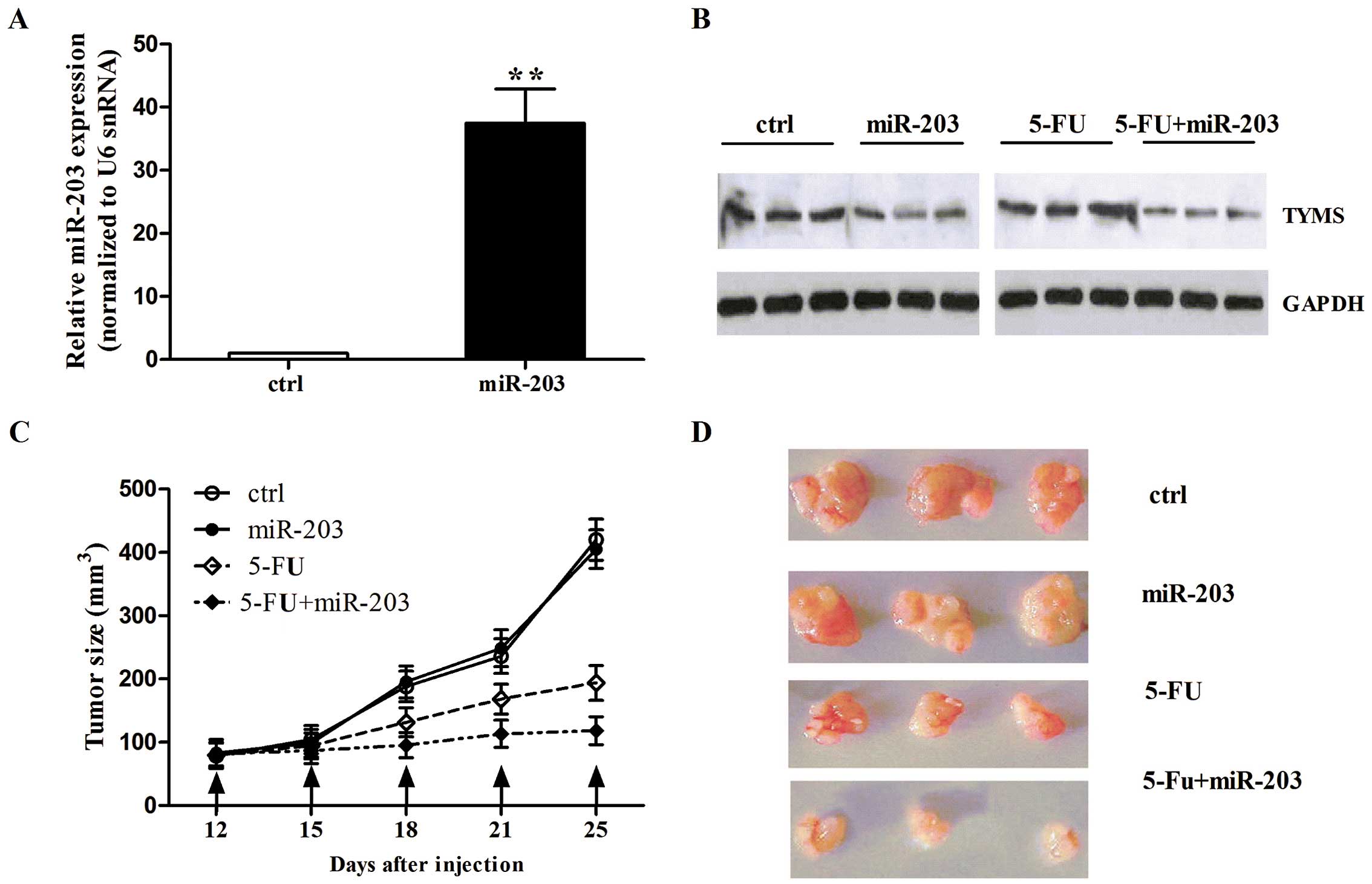
miR-203 enhances 5-FU cytotoxicity in
vivo
To determine whether the delivering of miR-203 in
vivo increases the cytotoxic effect of 5-FU, we established a
mouse colorectal cancer model by subcutaneously injected
106 LoVo cells in NOD/SCID mice. When the volume of the
tumor xenografts reached ~100 mm3 (on day 12), we
randomly separated the mice into four groups and each group was
treated with either miR-203 control or miR-203, or 5-FU alone, or
miR-203 together with 5-FU. The injection was performed every three
days until day 25 when the tumors were collected. miR-203
expression was quantified by qRT-PCR in the tumor xenografts. We
found that miR-203 expression increased in the mice treated with
miR-203 (Fig. 6A), indicating that
the delivery of miR-203 in vivo was successful. We then
detected TYMS expression in the tumor tissues. As shown in Fig. 6B, the mice with miR-203 had a lower
TYMS protein level than this level in the mice treated with miR-203
control or 5-FU alone. We also found that miR-203 alone did not
affect tumor volume compared with the control. However, 5-FU
treatment had a significant inhibitory effect on tumor growth. More
importantly, when mice were treated with miR-203 and 5-FU together,
tumor volume was inhibited more than with 5-FU treatment alone
(Fig. 6C). The images of tumor
xenografts are shown at day 25 in Fig.
6D. Taken together, these data suggest that miR-203 enhances
the inhibitory effect of 5-FU on tumor growth via the
downregulation of TYMS.
Discussion
Resistance to chemotherapy is one of the primary
obstacles to effective cancer treatment (16), yet the mechanism is poorly clear in
numerous types of cancers. In the present study, we demonstrated
the molecular mechanism of 5-FU resistance in colorectal cancer
cells. We found that miR-203 was downregulated in 5-FU-resistant
LoVo/5-Fu cells compared with LoVo cells, and there was an inverse
correlation between miR-203 and 5-FU resistance. miR-203 inhibition
enhanced 5-FU chemoresistance, in contrast, miR-203 overexpression
increased the sensitivity to 5-FU in colorectal cancer cells.
Furthermore, TYMS was identified as a target of miR-203, and
miR-203 suppressed the TYMS protein level. Silencing of TYMS had
similar effects on 5-FU chemoresistance as that of miR-203
overexpression. Importantly, the inhibitory effect of 5-FU on tumor
growth was enhanced by miR-203 in vivo.
miR-203 suppresses colon cancer cell proliferation
and induces cell apoptosis, as well as increases the sensitivity to
paclitaxel by downregulating Akt2 and its downstream genes that are
involved in chemoresistance (17).
miR-203 also reverses the chemoresistance to 5-FU by targeting Bmi1
in breast cancer cells (18). In
addition, miR-203 suppresses cell proliferation, migration and
invasion in prostate cancer (19),
indicating that miR-203 plays tumor-suppressive roles. In line with
the findings in previous studies, our data showed that miR-203
increased colorectal cancer cell sensitivity to 5-FU in
vitro and in vivo via the suppression of TYMS.
Surprisingly, miR-203 was reported to induce resistance to
oxaliplatin by downregulating ATM kinase in colorectal cancer cells
(20). Oxaliplatin causes cell
apoptosis by inducing DNA damage, while paclitaxel plays antitumor
roles by inducing M phase-associated apoptosis (21), and the action of 5-FU mainly depends
on the interference of DNA synthesis. Different targets of miR-203
may be responsible for the cytotoxicity effects of oxaliplatin,
paclitaxel and 5-FU on cancer cells, resulting in the different
roles of miR-203 in chemotherapy. Previous studies indicate that
miR-203 acts as a tumor-suppressor gene (19,22).
However, in a tumor growth study, we found that miR-203
overexpression did not affect tumor growth without 5-FU treatment.
Our experimental concentration of miR-203 (5 nM) was much lower
than the miR-203 used in the previous report (miR-203 lentivirus)
(22). It is possible that miR-203
does not function as a tumor suppressor at a lower
concentration.
To investigate the molecular mechanism of miR-203 in
the cellular response to 5-FU, we validated that TYMS is a direct
target of miR-203. miR-203 inhibition increased the luciferase
intensity of TYMS 3′UTR, while miR-203 overexpression resulted in
opposite effects. However, the inhibitory roles of miR-203 in TYMS
expression was abolished when the binding site for miR-203 on TYMS
3′UTR was mutated. Furthermore, the TYMS protein level was
suppressed by miR-203. TYMS is a cytosolic enzyme that alters the
methylation of dUMP to dTMP and subsequently affects DNA synthesis
and repair (23). The inhibition of
TYMS is considered as a primary mechanism of 5-FU action (24). Accumulating evidence shows that
patients with low TYMS expression have a higher response to 5-FU
and overall survival (6,25,26).
In contrast, overexpression of TYMS results in cell resistance to
5-FU (27,28). Accordingly, our functional
experiment showed that silencing of TYMS increased cell sensitivity
to 5-FU and mediated the inhibitory effects of miR-203 on tumor
growth under 5-FU exposure. Therefore, TYMS can act as a predictive
biomarker of the cellular response to 5-FU and a therapeutic target
for 5-FU-based chemotherapy.
In conclusion, the present study indicates that
miR-203 enhanced the cell resistance to 5-FU in vitro and
in vivo via the downregulation of TYMS in colorectal cancer,
and provides novel insight into the molecular mechanism of 5-FU
resistance. The present study will facilitate the development of a
therapeutic strategy for 5-FU-based chemotherapy.
Acknowledgements
The study was supported by a grant (no.
2012-WSZD068) from the 2012 Medical Research Mentoring Program of
Qingdao City.
References
|
1
|
Coget J, Borrini F, Susman S and Sabourin
JC: Colorectal carcinomas in 2013: the search for powerful
prognostic markers is still on the go! Cancer Biomark. 14:145–150.
2014.PubMed/NCBI
|
|
2
|
No authors listed. Efficacy of adjuvant
fluorouracil and folinic acid in colon cancer. International
Multicentre Pooled Analysis of Colon Cancer Trials (IMPACT)
investigators. Lancet. 345:939–944. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
González-Vallinas M, Molina S, Vicente G,
et al: Antitumor effect of 5-fluorouracil is enhanced by rosemary
extract in both drug sensitive and resistant colon cancer cells.
Pharmacol Res. 72:61–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Etienne MC, Chazal M, Laurent-Puig P, et
al: Prognostic value of tumoral thymidylate synthase and p53 in
metastatic colorectal cancer patients receiving fluorouracil-based
chemotherapy: phenotypic and genotypic analyses. J Clin Oncol.
20:2832–2843. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Copur S, Aiba K, Drake JC, Allegra CJ and
Chu E: Thymidylate synthase gene amplification in human colon
cancer cell lines resistant to 5-fluorouracil. Biochem Pharmacol.
49:1419–1426. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Johnston PG, Lenz HJ, Leichman CG, et al:
Thymidylate synthase gene and protein expression correlate and are
associated with response to 5-fluorouracil in human colorectal and
gastric tumors. Cancer Res. 55:1407–1412. 1995.PubMed/NCBI
|
|
7
|
Yeh KH, Shun CT, Chen CL, et al: High
expression of thymidylate synthase is associated with the drug
resistance of gastric carcinoma to high dose 5-fluorouracil-based
systemic chemotherapy. Cancer. 82:1626–1631. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang LY, Liu M, Li X and Tang H:
miR-490-3p modulates cell growth and epithelial to mesenchymal
transition of hepatocellular carcinoma cells by targeting
endoplasmic reticulum-Golgi intermediate compartment protein 3
(ERGIC3). J Biol Chem. 288:4035–4047. 2013. View Article : Google Scholar :
|
|
9
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fujita Y, Kojima K, Hamada N, et al:
Effects of miR-34a on cell growth and chemoresistance in prostate
cancer PC3 cells. Biochem Biophys Res Commun. 377:114–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hwang JH, Voortman J, Giovannetti E, et
al: Identification of microRNA-21 as a biomarker for
chemoresistance and clinical outcome following adjuvant therapy in
resectable pancreatic cancer. PLoS One. 5:e106302010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bitarte N, Bandres E, Boni V, et al:
MicroRNA-451 is involved in the self-renewal, tumorigenicity, and
chemoresistance of colorectal cancer stem cells. Stem Cells.
29:1661–1671. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karaayvaz M, Zhai H and Ju J: miR-129
promotes apoptosis and enhances chemosensitivity to 5-fluorouracil
in colorectal cancer. Cell Death Dis. 4:e6592013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gotanda K, Hirota T, Matsumoto N and Ieiri
I: MicroRNA-433 negatively regulates the expression of thymidylate
synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa
cells. BMC Cancer. 13:3692013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Djeu JY and Wei S: Clusterin and
chemoresistance. Adv Cancer Res. 105:77–92. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li JA, Chen YX, Zhao JF, Kong FR and Zhang
YD: miR-203 reverses chemoresistance in p53-mutated colon cancer
cells through downregulation of Akt2 expression. Cancer Lett.
304:52–59. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin J, Zheng G, Jia X, et al: A
Bmi1-miRNAs cross-talk modulates chemotherapy response to
5-fluorouracil in breast cancer cells. PLoS One. 8:e732682013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Viticchie G, Lena AM, Latina A, et al:
MiR-203 controls proliferation, migration and invasive potential of
prostate cancer cell lines. Cell Cycle. 10:1121–1131. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Wan G, Spizzo R, et al: miR-203
induces oxaliplatin resistance in colorectal cancer cells by
negatively regulating ATM kinase. Mol Oncol. 8:83–92. 2014.
View Article : Google Scholar
|
|
21
|
Jordan MA and Wilson L: Microtubules as a
target for anticancer drugs. Nat Rev Cancer. 4:253–265. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian L, Li M, Ge J, et al: MiR-203 is
downregulated in laryngeal squamous cell carcinoma and can suppress
proliferation and induce apoptosis of tumours. Tumour Biol.
35:5953–5963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lehman NL: Future potential of thymidylate
synthase inhibitors in cancer therapy. Expert Opin Investig Drugs.
11:1775–1787. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Houghton JA, Tillman DM and Harwood FG:
Ratio of 2′-deoxyadenosine-
5′-triphosphate/thymidine-5′-triphosphate influences the commitment
of human colon carcinoma cells to thymineless death. Clin Cancer
Res. 1:723–730. 1995.PubMed/NCBI
|
|
25
|
Popat S, Matakidou A and Houlston RS:
Thymidylate synthase expression and prognosis in colorectal cancer:
a systematic review and meta-analysis. J Clin Oncol. 22:529–536.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lenz HJ, Hayashi K, Salonga D, et al: p53
point mutations and thymidylate synthase messenger RNA levels in
disseminated colorectal cancer: an analysis of response and
survival. Clin Cancer Res. 4:1243–1250. 1998.PubMed/NCBI
|
|
27
|
Cho YB, Chung HJ, Lee WY, et al:
Relationship between TYMS and ERCC1 mRNA expression and in vitro
chemosensitivity in colorectal cancer. Anticancer Res.
31:3843–3849. 2011.PubMed/NCBI
|
|
28
|
Galbiatti AL, Caldas HC, Maniglia JV,
Pavarino EC and Goloni-Bertollo EM: Gene expression profile of
5-fluorouracil metabolic enzymes in laryngeal cancer cell line:
predictive parameters for response to 5-fluorouracil-based
chemotherapy. Biomed Pharmacother. 68:515–519. 2014. View Article : Google Scholar : PubMed/NCBI
|