Introduction
Laryngeal cancer is one of the most commonly
occurring malignant cancers of the head and neck region (1). It rates second in incidence among head
and neck cancers, and 11th among all human cancers (2). Advances in chemotherapy, radiation
therapy and surgical techniques have improved the 5-year survival
rate (2), however, recurrence is
prevalent following treatment. Thus, identification of novel
therapeutic targets for laryngeal cancer is crucial.
MicroRNAs (miRNAs) are short, non-coding RNA
molecules that post-transcriptionally regulate the expression of
target genes, and play a role in diverse cell, physiological and
pathophysiological processes (3,4).
miR-221 is coded from chromosome X and functions as an oncogenic
miRNA and is involved in various types of cancer (5). It was reported to be upregulated in
many types of tumor, including glioblastoma, bladder cancer and
papillary tumors of the thyroid (6–8).
miR-221 was shown to affect several cancer pathways by modulating
multiple gene targets such as estrogen receptor-α, p27, p57 and
receptor tyrosine kinase kit 1 (c-kit) (9–12).
To the best of our knowledge, no previous study has
shown the association between miR-221 and laryngeal squamous cell
carcinoma (LSCC). The aim of the present study was to present the
role of miR-221 in laryngeal cancer cell line, Hep-2. Furthermore,
we focused on a target protein of miR-221, apoptotic protease
activating factor-1 (Apaf-1). We also determined the function and
underlying mechanism of miR-221 in Hep-2 cells.
Materials and methods
Cell culture and transfection
The human Hep-2 laryngeal cancer cell line was
purchased from the Shanghai Institutes for Biological Sciences,
Chinese Academy of Sciences. The cells were cultured in PRMI-1640
supplemented with 10% heat-inactivated fetal bovine serum (FBS)
(both from HyClone, Logan, UT, USA) in a humidified cell incubator
with an atmosphere of 5% CO2 at 37°C. Exponentially growing cells
were used for experiments. Hep-2 cells were transfected with
miR-221 antisense and scrambled control oligonucleotides (Thermo
Fisher, Waltham, MA USA) or Apaf-1 siRNA (sc-29201; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
instructions.
Bioinformatics analysis
The analysis of predicted miR-221 targets was
performed using the algorithms TargetScan (http://targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), and miRanda (http://www.microrna.org/microrna/home.do/) website
tools. The minimum free energy predicted for hybridization was
determined by BibiServ analysis (http://bibiserv.techfak.uni-bielefeld.de/genefisher2/).
Luciferase reporter assay
Hep-2 cells were co-transfected using Lipofectamine
2000 reagent (Invitrogen) with 100 ng of firefly luciferase
construct and 300 ng of control-pcDNA3.1 or pcDNA3.1-miR-221
expression vector. A total of 10 ng of pRL-CMV (Promega Biotech
Co., Ltd., Beijing, China) was co-transfected as a normalization
control. Reporter assays were performed 48 h post-transfection
using the Dual-Luciferase Assay system (Promega, Madison, WI, USA),
normalized for transfection efficiency by the co-transfected
Renilla luciferase.
MTT assay
Cells were plated in 96-well plates at a
concentration of 1,500 cells/well and allowed to attach overnight.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT) solution (Sigma-Aldrich, Carlsbad, CA, USA) was then added at
a final concentration of 0.5 mg/ml for 4 h. After 4 h, the cells
were lysed with dimethyl sulfoxide (DMSO; Sigma) and absorbance
rates were measured at 550–560 nm using a microplate reader
(Bio-Rad, Hercules, CA, USA).
Apoptosis analysis
As per the manufacturer’s instructions for the
Apoptosis Assay kit (KeyGen Nanjing, China), the stained cells were
analyzed by flow cytometry (BD Biosciences, Rockville, MD, USA).
Data analysis was performed using CellQuest software (BD
Biosciences).
Cell cycle
The cells were collected and fixed in 70% ethanol at
4°C for 16 h. Fixed cells were then washed once with
phosphate-buffered saline (PBS), resuspended in 500 ml PBS
containing 10 μg/ml propidium iodide (KeyGen) and 50 mg/ml RNase
and incubated for 30 min at room temperature. The cells were then
centrifuged at 1,200 rpm for 5 min, resuspended in 500 ml of PBS
and analyzed with FCM.
Xenograft assays
Animal studies were performed according to the
institutional guidelines. Hep-2 (3×107 in 200 μl) cells
were injected subcutaneously into the right flanks of 6–8 weeks
male nude mice (Charles River, Wilmington, MA, USA). Treatment was
initiated after the tumor diameters reached 3–5 mm by daily
intratumoral injections of PBS, miR-221 antisense or scrambled
control oligonucleotides or Apaf-1 siRNA for two cycles of 3 days.
Tumor growth was then monitored for 30 days. Every five days until
the end of the experiment, one mouse from each group was randomly
selected to be anesthetized, photographed and sacrificed. The tumor
volumes were determined by measuring the length (l) and the width
(w) and calculating the volume (V = lw2/2). Tumor
samples were analyzed by western blotting. Additional mice (n=60)
were used to establish xenografts to obtain survival curves. Mice
with xenografted tumors (as described above) that reached 3–5 mm in
diameter were divided into three treatment groups (n=20 for each).
Survival was monitored until the experiments were terminated due to
the heavy tumor burden.
In situ hybridization of miR-221
A tissue array containing 36 murine tissue samples
from the groups described above was created using a tissue
microarrayer MIA-I (Beecher Instruments, Silver Spring, MD, USA).
In situ hybridization (ISH) was carried out with
double-digoxigenin (DIG)-labeled miRCURY LNA™ miR detection probe
against has-miR-221 (Exiqon, Copenhagen, Denmark) according to the
supplier’s instructions.
Quantitative PCR analysis of miR-221
After thawing at 4°C, 400 μl of cyst fluid was
subjected to small RNA extraction using the mirVana™ miRNA
isolation kit (Ambion, Applied Biosystems, Austin, TX, USA)
according to the manufacturer’s instructions. Quantitative PCR
(qPCR) was analyzed using the Bulge-Loop™ miRNA RT-qPCR Detection
kit (Ribobio Co., Guangzhou, China) and TransStart™ Green qPCR
SuperMix (TransGen Biotech, Beijing, China) according to the
manufacturer’s instructions with the Rotor-gene 6000 system
(Qiagen, Hilden, Germany). The reactions were incubated at 95°C for
30 sec, followed by 40 cycles of 95°C for 30 sec, 60°C for 20 sec,
and 70°C for 1 sec. The relative expression level for miRNA-221 was
computed using the comparative Ct method. miRNA expression was
normalized to small nucleolar RNA U6.
Western blotting
Tissues were rinsed twice with cold PBS buffer and
lysed in an ice-cold lysis buffer containing 150 mM NaCl, 50 mM
Tris-HCl (pH 7.6), 0.1% SDS, 1% Nonidet P-40, and a protease
inhibitor cocktail (Roche, Basel, Switzerland). The extracts were
incubated on ice for 20 min, centrifuged at 12,000 × g for 20 min
at 4°C, and the supernatants were collected. Protein concentrations
were determined using Bradford assay (Bio-Rad), and proteins were
resolved by 10% Bis-Tris gel electrophoresis, transferred to a
nitrocellulose membrane, and western blot analysis was performed.
Rabbit polyclonal IgG anti-Apaf-1 (sc-8339), mouse monoclonal IgG
anti-caspase-3 (sc-7272), mouse monoclonal IgG anti-caspase-8
(sc-56070), mouse monoclonal IgG anti-caspase-9 (sc-73548) and
mouse monoclonal IgG anti-β-actin (sc-47778) were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz).
Immunohistochemical staining
Immunohistochemical (IHC) staining was performed on
4-μm sections obtained from formalin-fixed, paraffin-embedded
blocks. Endogenous peroxidase activity was blocked with 3% hydrogen
peroxide for 30 min. Antigen retrieval was carried out in citrate
buffer (10 mM, pH 6.0) for 30 min at 95°C in a microwave oven.
The sections were incubated with primary antibody as described in
western blotting at 4°C overnight. The sections were then incubated
with a biotinylated secondary antibody and then exposed to a
streptavidin complex (HRP; Sigma). Positive reactions were
visualized with 3,3′-diaminobenzidine (DAB) tetrahydrochloride,
followed by counterstaining with hematoxylin (both from Sigma).
Statistical analysis
Statistical analyses were carried out using GraphPad
Prism version 5.00 for Windows (GraphPad Software, San Diego, CA,
USA). Numerical data were presented as means ± SD. The Student’s
t-test and one-way ANOVA analysis was used to determine
significance. Kaplan-Meier survival plots were generated and
comparisons between survival curves were made with the log-rank
statistic. P<0.05 was considered to indicate a statistically
significant result. The experiments were conducted in
triplicate.
Results
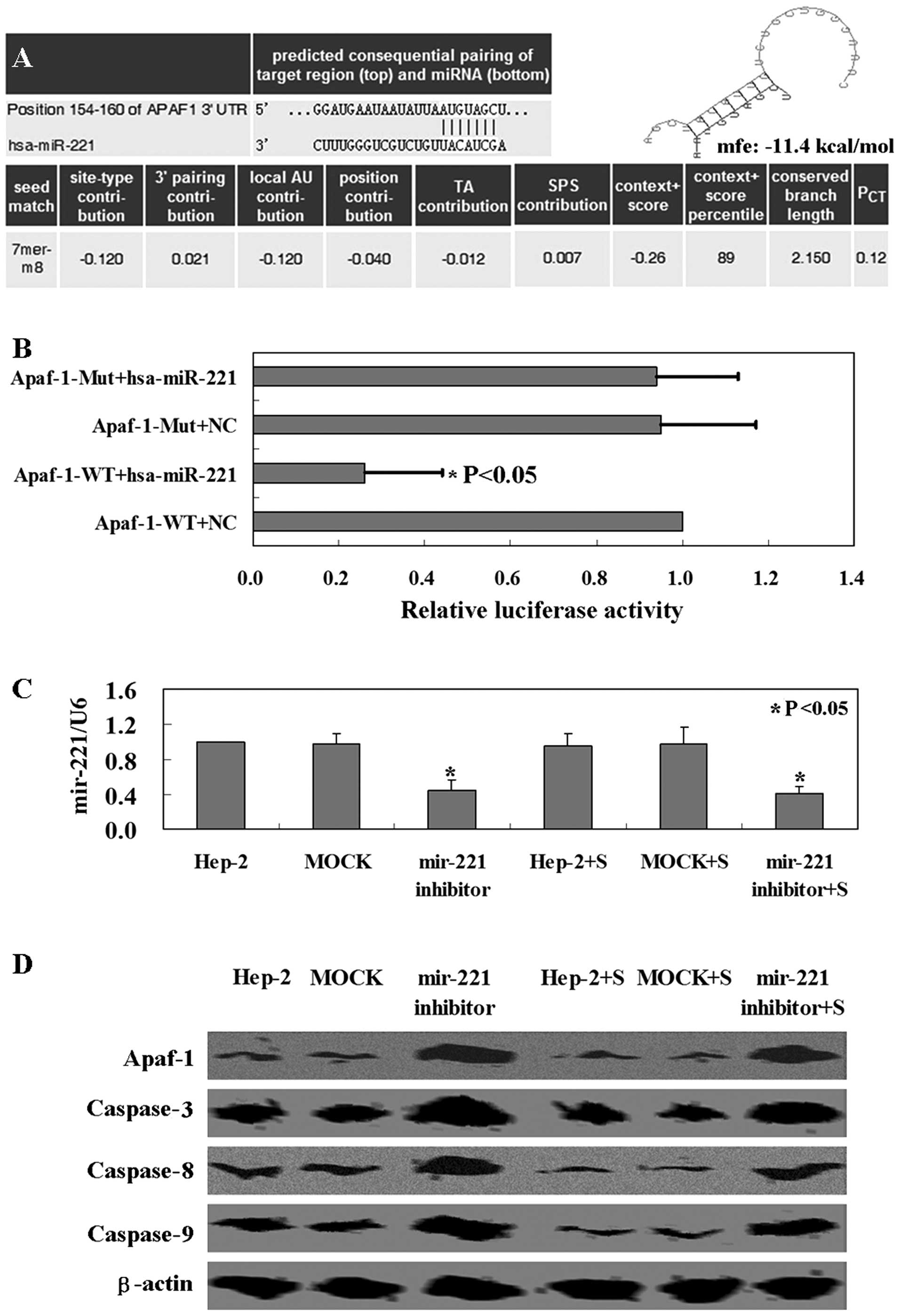
miR-221 interacts specifically with the
3′UTR region of Apaf-1
First of all, we identified Apaf-1 as the
potential target of miR-221 using TargetScan and miRanda online
search programs. A 100% matched sequence was found at the 154–160
nucleotide region of Apaf-1 mRNA 3′UTR (Fig. 1A). The free energy was calculated as
~−11.4 kcal/mol for the hybrid of the Apaf-1 3′UTR region
and miR-221 by the Bibiserv analysis (Fig. 1A). Secondly, we confirmed the
hypothesis that miR-221 targets the 3′UTR region of Apaf-1
using a luciferase reporter assay. Co-transfection of miR-221
expression vector along with the full-length 3′UTR of Apaf-1
caused a significant decrease in luciferase units compared to the
controls (p<0.05, Fig. 1B).
Furthermore, we decreased miR-221 expression in Hep-2 cells using
miR-221 antisense (p<0.05, Fig.
1C). Compared with mock-transfected or untransfected ones,
Apaf-1 protein in Hep-2 cells was markedly increased after miR-221
antisense transfection (p<0.05, Fig.
1D). The results clearly demonstrated that miR-221 targeted
specifically the 3′UTR region of Apaf-1.
Effect of miR-221 and Apaf-1 on Hep-2
cells in vitro and in vivo
The MTT assay showed that the proliferation rate of
Hep-2 cells with miR-221 antisense transfection was decreased
compared to the untransfected or mock-transfected cells (p<0.05,
Fig. 2A). The cells with Apaf-1
knockdown showed a higher proliferation rate than the untransfected
or mock-transfected ones (p<0.05, Fig. 2A). As shown in Fig. 2B, the ratio of apoptotic Hep-2 cells
with miR-221 antisense transfection was 5- to 6-fold higher than
that in the untransfected or mock-transfected ones (p<0.05).
Apaf-1 knockdown cells exhibited a lower apoptotic ratio than other
cells (p<0.05, Fig. 2B). As
evidenced by PI staining, miR-221 antisense caused a significant
increase in the fraction of S-phase cells (Fig. 2C). Apaf-1 knockdown partly repaired
miR-221-induced S-phase arrest and promoted cell cycle restoration
(Fig. 2C).
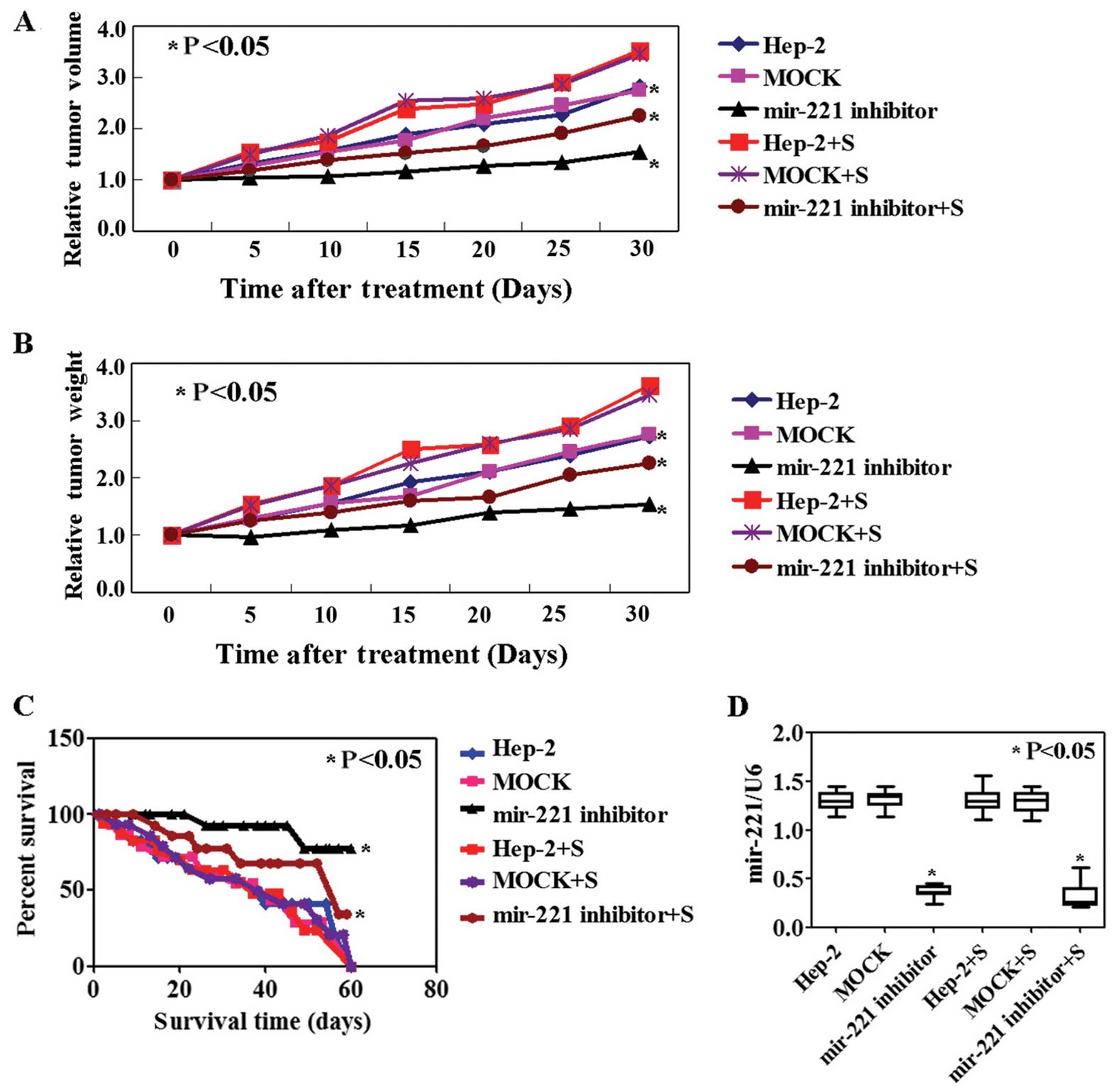
In vivo efficacy of miR-221 and Apaf-1 in
a human Hep-2 xenograft mouse model
The success of intratumoral injection of miR-221
antisense was demonstrated using qPCR and ISH, respectively. The
results of qPCR (p<0.05, Fig.
3D) and ISH (Fig. 3E) showed a
lower level of miR-221 in the tumor tissues of the mice transfected
with miR-221 antisense as compared to other groups. The tumor
volume and weights of the mice transfected with miR-221 antisense
were lower than those of the PBS or mock group. However, the tumor
volume and weights of the Apaf-1 knockdown group was higher than
the other groups (p<0.05, Fig. 3A
and B). Correspondingly, the survival rate of mice with miR-221
antisense was significantly improved, while the Apaf-1 knockdown
group showed a poor survival rate (p<0.05, Fig. 3C).
Mechanism of miR-221 in Hep-2 cells
To determine the mechanisms of miR-221, western
blotting and IHC were carried out to measure the changes of
possible proteins. The level of Apaf-1 protein was observed to be
higher in Hep-2 cells with miR-221 antisense transfection compared
with untreated cells (Fig. 1D).
Expression levels of caspase-3, −8 and −9 were also higher and
associated with the Apaf-1 expression (Fig. 1D). The same mechanism of miR-221 was
confirmed using the tissues of Hep-2 xenograft mouse model
(Fig. 3E). The results suggested
that miR-221 inhibits Apaf-1 expression in Hep-2 cells, followed
with the downregulation of caspase-3, −8 and −9.
Discussion
The results of the present study have demonstrated
that inhibition of miR-221 in Hep-2 cells induced apoptosis in
vitro and in vivo. miR-221 is upregulated in multiple
malignancies (6–8). Overexpression of miR-221 promoted
cancer cell proliferation by its ability to inhibit the expression
of the cyclin-dependent kinase inhibitors CDKN1B/p27 (13,14).
We also confirmed that miR-221 inhibition suppressed cell
proliferation and induced apoptosis in Hep-2 cells. Previous
studies have shown that miR-221 promotes tumor cells entering the S
phase from the G1 phase (15,16).
In the present study, the downregulation of miR-221 was identified
to induce S-phase arrest. Park et al (17) found that anti-miR-221 treatment
improved survival of the orthotopic tumor xenograft mice compared
with the scrambled control. Consistent with that study, we found
miR-221 inhibition decreased tumor volume and weights, although the
survival rate of Hep-2 xenograft mouse models was improved.
Furthermore, we found that miR-221 and Apaf-1
mRNA 3′-UTR have complementary binding sites using bioinformatics
prediction software including TargetScan, PicTar and miRanda.
However, whether Apaf-1 is a novel target of miR-221 in LSCC
has yet to be reported. The hypothesis was demonstrated using the
luciferase assay. The results of the luciferase assay showed a
direct interaction between miR-221 and the target site in the
3′-UTR of the Apaf-1 mRNA. In addition, we confirmed that a
decreased miR-221 level was associated with the upregulation of
Apaf-1 expression in vitro and in vivo. The results
collectively confirmed Apaf-1 as a novel target of
miR-221.
Apaf-1 is a component protein of the apoptosome
(18). Apoptosis was induced by
exogenous and endogenous factors (19). In the intrinsic apoptotic pathway,
cytochrome c, Apaf-1 and caspase-9 precursor are interacted,
and then caspase-9 precursor becomes activated (20). Apaf-1 downregulation has been
connection with decreased apoptosis and correlated with adverse
prognosis in colorectal cancer (21). Overexpression of Apaf-1 in the U87
cells has an effect on inhibiting cell proliferation and promoting
apoptosis (22). Consistent with
previous studies, our results also revealed Apaf-1 expression
induced Hep-2 cell apoptosis and cell cycle arrest. Furthermore,
results of the present study showed that downregulation of miR-221
caused elevated expression levels of the Apaf-1 apoptotic pathway
proteins caspase-3, −8 and −9.
In conclusion, the present study reports the
knockdown of endogenous miR-221-inhibited proliferation, induced
apoptosis and cell cycle arrest of laryngeal cancer cell. miR-221
was demonstrated to possess apoptosis resistance function in
laryngeal cancer cell by targeting Apaf-1. Of note is that we
confirmed the antitumor roles of miR-221 in animal models. miR-221
may therefore be employed as a novel therapeutic target of
laryngeal cancer.
Acknowledgements
This study was supported by the Doctoral Scientific
Start-up Funds of Liaoning Province (no. 20141041).
References
|
1
|
Chen YF, Luo RZ, Li Y, et al: High
expression levels of COX-2 and P300 are associated with unfavorable
survival in laryngeal squamous cell carcinoma. Eur Arch
Otorhinolaryngol. 270:1009–1017. 2013. View Article : Google Scholar :
|
|
2
|
Chu EA and Kim YJ: Laryngeal cancer:
diagnosis and preoperative work-up. Otolaryngol Clin North Am.
41:673–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang TC and Mendell JT: microRNAs in
vertebrate physiology and human disease. Annu Rev Genomics Hum
Genet. 8:215–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Latronico MV, Catalucci D and Condorelli
G: Emerging role of microRNAs in cardiovascular biology. Circ Res.
101:1225–1236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ergun S, Arman K, Temiz E, et al:
Expression patterns of miR-221 and its target caspase-3 in
different cancer cell lines. Mol Biol Rep. 41:5877–5881. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ciafrè SA, Galardi S, Mangiola A, et al:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gottardo F, Liu CG, Ferracin M, et al:
MicroRNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He H, Jazdzewski K, Li W, et al: The role
of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad
Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Medina R, Zaidi SK, Liu CG, et al:
MicroRNAs 221 and 222 bypass quiescence and compromise cell
survival. Cancer Res. 68:2773–2780. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
le Sage C, Nagel R, Egan DA, et al:
Regulation of the p27Kip1 tumor suppressor by miR-221
and miR-222 promotes cancer cell proliferation. EMBO J.
26:3699–3708. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gramantieri L, Fornari F, Ferracin M, et
al: MicroRNA-221 targets Bmf in hepatocellular carcinoma and
correlates with tumor multifocality. Clin Cancer Res. 15:5073–5081.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu X, Wang Q, Chen J, et al: Clinical
significance of miR-221 and its inverse correlation with
p27Kip1 in hepatocellular carcinoma. Mol Biol Rep.
38:3029–3035. 2011. View Article : Google Scholar
|
|
13
|
Visone R, Russo L, Pallante P, et al:
MicroRNAs (miR)-221 and miR-222, both overexpressed in human
thyroid papillary carcinomas, regulate p27Kip1 protein
levels and cell cycle. Endocr Relat Cancer. 14:791–798. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gillies JK and Lorimer IA: Regulation of
p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle.
6:2005–2009. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahmed FE, Jeffries CD, Vos PW, et al:
Diagnostic microRNA markers for screening sporadic human colon
cancer and active ulcerative colitis in stool and tissue. Cancer
Genomics Proteomics. 6:281–295. 2009.PubMed/NCBI
|
|
16
|
Felli N, Fontana L, Pelosi E, et al:
MicroRNAs 221 and 222 inhibit normal erythropoiesis and
erythroleukemic cell growth via kit receptor down-modulation. Proc
Natl Acad Sci USA. 102:18081–18086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park JK, Kogure T, Nuovo GJ, et al:
miR-221 silencing blocks hepatocellular carcinoma and promotes
survival. Cancer Res. 71:7608–7616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li P, Nijhawan D, Budihardjo I, et al:
Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9
complex initiates an apoptotic protease cascade. Cell. 91:479–489.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lian S, Shi R, Bai T, et al:
Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by
targeting apoptotic protease-activating factor-1. Curr Pharm Des.
19:6382–6389. 2013. View Article : Google Scholar
|
|
20
|
Zang YS, Zhong YF, Fang Z, et al: MiR-155
inhibits the sensitivity of lung cancer cells to cisplatin via
negative regulation of Apaf-1 expression. Cancer Gene Ther.
19:773–778. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paik SS, Jang KS, Song YS, et al: Reduced
expression of Apaf-1 in colorectal adenocarcinoma correlates with
tumor progression and aggressive phenotype. Ann Surg Oncol.
14:3453–3459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L, Zhang J, Han L, et al:
Downregulation of miR-221/222 sensitizes glioma cells to
temozolomide by regulating apoptosis independently of p53 status.
Oncol Rep. 27:854–860. 2012.
|