Introduction
Colorectal cancer (CRC) is the third most common
malignant neoplasm worldwide and the fourth most common cause of
cancer mortality (1,2). It affects health and life span
(3). Thus, the understanding of the
molecular pathology of CRC is crucial in the identifications of
treatments. Accumulation of multiple genetic alterations transforms
normal colonic epithelial cells to adenocarcinoma cells (4–6).
Genetic alterations are associated with the activation of
proto-oncogenes (KRAS and BRAF) (7–11) and
the inactivation of tumor suppression genes (APC,
DCC/SMAD4, TP53 and PTEN) (12–15).
The APC/β-catenin pathway plays a key role in CRC. Adenomatous
polyposis coli (APC) is the most common initial gene mutated
in CRC and the majority of APC mutations are either frameshift or
nonsense mutations leading to the synthesis of truncated proteins
(16). PTEN is known as a tumor
suppressor through negative regulation of the PI3K/Akt pathway
(17,18). Allele loss close to the PTEN locus
has been reported in 30% of sporadic CRC (19).
MicroRNAs (miRNAs) are small, regulatory, non-coding
RNA molecules that lead to silencing of their genes via mRNA
degradation or preventing mRNA from being translated (20). miRNAs have been involved in the
regulation of a variety of cell processes, such as DNA repair
(21–23), metastasis and metabolism (24–27).
Thus, miRNAs function as oncogenes or tumor suppressors.
Dysregulation of miRNAs has been involved in the progression of CRC
(28,29). Recent studies have examined the
expression patterns of miRNA in CRC and identified a variety of
miRNAs that were dysregulated in CRC (30,31).
miR-135b was reported to function as a key molecule downstream of
the oncogenic pathway involved in cancer progression (32–35).
miR-135b was upregulated in CRCs and its
overexpression was due to APC/β-catenin and PTEN/PI3K pathway
deregulation (36). In the present
study, we showed that miR-135b regulates the stability of PTEN
protein. Deubiquitinating enzyme USP13, which stabilizes the PTEN
protein through direct binding and deubiquitinating of PTEN, was
proven as as a target of miR-135b. Overexpression of miR-135b
downregulates the expression of ubiquitin-specific peptidase 13
(USP13) and reduces the stability of PTEN. miR-135b also promotes
cell proliferation and glycolysis, which could be reversed by the
overexpression of USP13/PTEN. Moreover, knockdown of USP13
upregulates the levels of endogenous miR-135b. These results
revealed positive feedback loops between miR-135b and PTEN
inactivation in CRCs.
Materials and methods
Reagents
Cycloheximide (CHX) was purchased from Sigma-Aldrich
(St. Louis, MO, USA). Anti-PTEN and anti-USP13 were purchased from
Millipore (Bedford, MA, USA). Anti-p-AKT (Ser 473), anti-AKT and
anti-β-actin were purchased from Cell Signaling Technology, Inc.
(Beverly, MA, USA). The real-time PCR Master mix kit was purchased
from Takara (Dalian, China).
Cell cultures and transfections
HCT-116 CRC cell lines were purchased from the
American Type Culture Collection (ATCC, Dallas, TX, USA). Caco2,
CO-115 and 293T cell lines were purchased from the Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China).
HCT-116, Caco2, CO-115 and 293T cells were cultured in DMEM
(Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum (Hyclone, Logan, UT, USA), penicillin (100 U/ml) and
streptomycin (100 μg/ml) in a humidified atmosphere of 5%
CO2 at 37°C. The cells were examined for mycoplasma
contamination periodically and were always found to be negative.
Cell transfections were performed using Lipofectamine 2000
(Invitrogen) following the manufacturer’s instructions.
RT-qPCR for mature miR-135b and PTEN
Total RNA was extracted from cell lines using Trizol
(Invitrogen). RT-qPCR of miR-135b was performed using TaqMan miRNA
reverse transcription kit (Applied Biosystems, Foster City, CA,
USA) and TaqMan human miRNA assay (Assay ID: RNU6B, 001093;
miR-135b, 002261). For detection of the mRNAs of PTEN, total RNA
was extracted by TRIzol (Invitrogen) and subjected to reverse
transcription using the PrimeScript II first-strand cDNA synthesis
kit (Takara) according to the manufacturer’s instructions. The
primers for human PTEN were: forward,
5′-CAAGATGATGTTTGAAACTATTCCAATG-3′ and reverse,
5′-CCTTTAGCTGGCAGACCACAA-3′. RT-qPCR was performed with the 7500
Fast real-time PCR system (Applied Biosystems). miR-135b expression
was normalized to that of RNU6B. PTEN expression was normalized to
GAPDH. Reverse-transcriptase (RT) reactions, including no-template
controls and RT minus controls, were run in a GeneAmp PCR 9700
Thermocycler (Applied Biosystems). After the Ct values of the
target genes were subtracted by that of the internal control
RNU6B/GAPDH, the relative expression level of each target gene was
obtained using the 2−ΔΔCt method.
Metabolism assays
Glucose consumption was measured in the cell lysates
with a Glucose uptake colorimetric assay kit (BioVision, San
Francisco, CA, USA). The extracellular lactate was measured in the
medium using a lactate assay kit (BioVision) according to the
manufacturer’s instructions.
Construction of plasmids
For luciferase reporter experiments, the 3′-UTR
segment of the USP13 gene was amplified by PCR using human
cDNA as templates and inserted into the pGL3-control vector
(Promega, Madison, WI, USA). A mutant construct was generated from
the perfect complementary site using the Quik change site-directed
mutagenesis kit (Stratagene, La Jolla, CA, USA).
Viral production and infection
MiRNASelect™ pEP- miR-Null control lentiviral vector
and miRNASelect™ pEP-hsa-mir-135b lentiviral expression vector were
obtained from Cell Biolabs (San Diego, CA, USA). Human miRNA
inhibitors against human miR-135b in the lentiviral vector were
obtained from OriGene (Rockville, MD, USA). Expression lentiviral
vector for shRNA against USP13 was constructed into a pLKO.1
vector. The target sequences were: 5′-CGATTTAAATAGCGACGATTA-3′
(sh-13-1) and 5′-GCC AGTATCTAAATATGCCAA-3′ (sh-13-2). Negative
control vector containing scrambled shRNA was obtained from Addgene
(Cambridge, MA, USA). pLenti-C-Myc-DDK tagged USP13 or PTEN
lentiviral expression vector was purchased from OriGene. The
plasmids were prepared with plasmid maxi kit and transfected in
293T cells (Invitrogen) with the Lipofectamine™ 2000 to produce
lentiviral particles. The cells were infected with lentiviral
particles for 2 h. The cells were then selected with 2.5 mg/ml
puromycin for 72 h.
Western blot analysis
Cells were lysed in a RIPA buffer containing
complete protease/phosphatase inhibitor cocktail (Cell Signaling
Technology, Inc.). The lysate was centrifuged at 12,000 rpm for 15
min at 4°C and the supernatant was collected. Protein concentration
was determined by the Bradford protein assay. The proteins were
separated by SDS-PAGE gel electrophoresis and then transferred onto
PVDF membranes (Millipore). The membranes were blocked with 10% BSA
in TBST for 1 h and incubated with antibodies overnight at 4°C.
After washing with TBST three times, the membranes were probed with
HRP-conjugated secondary antibodies in TBST for 1 h at room
temperature, then washed with PBST three times. The immunobinding
signals were detected by a chemiluminescence method (ECL,
Millipore).
Luciferase activity assay
To directly evaluate the effect of miR-135b on
USP13, the luciferase assay was used. Stable CRC cell lines that
constitutively express human miR-135b were generated. The cells
were transfected with firefly luciferase reporter vector and the
control vector pRL-CMV (coding for Renilla luciferase,
Promega) or plus miR-135b inhibitor lentiviral particles.
Forty-eight hours after transfection, firefly and Renilla
luciferase activities were measured using the Dual-Luciferase
reporter assay system (Promega), and the relative reporter activity
was normalized to Renilla luciferase activity. Each assay
was repeated in six independent experiments.
Cell proliferation analysis
Cell proliferation was examined by using a Cell
Counting kit-8 (Dojindo, Kumamoto, Japan). The cells were plated in
96-well plates at 2×103 cells/well and cultured in
growth medium for 24 h. After 24 h, CCK-8 (10 μl) was added to each
well containing 100 μl of DMEM medium. The plate was incubated for
2 h at 37°C. Absorbance was measured at 450 nm using a microplate
reader (Tecan, Switzerland).
Statistical analysis
Data were analysed using the Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-135b upregulates the stability of
PTEN protein
miR-135b was reported to be upregulated in CRCs and
its overexpression was due to loss of APC and PTEN. APC was proven
to be a target of miR-135b and formed a feedback loop with miR-135b
(36). We determined whether
miR-135b affected the levels of PTEN. To examine this potential
effect, we generated stable Caco2 cell lines that constitutively
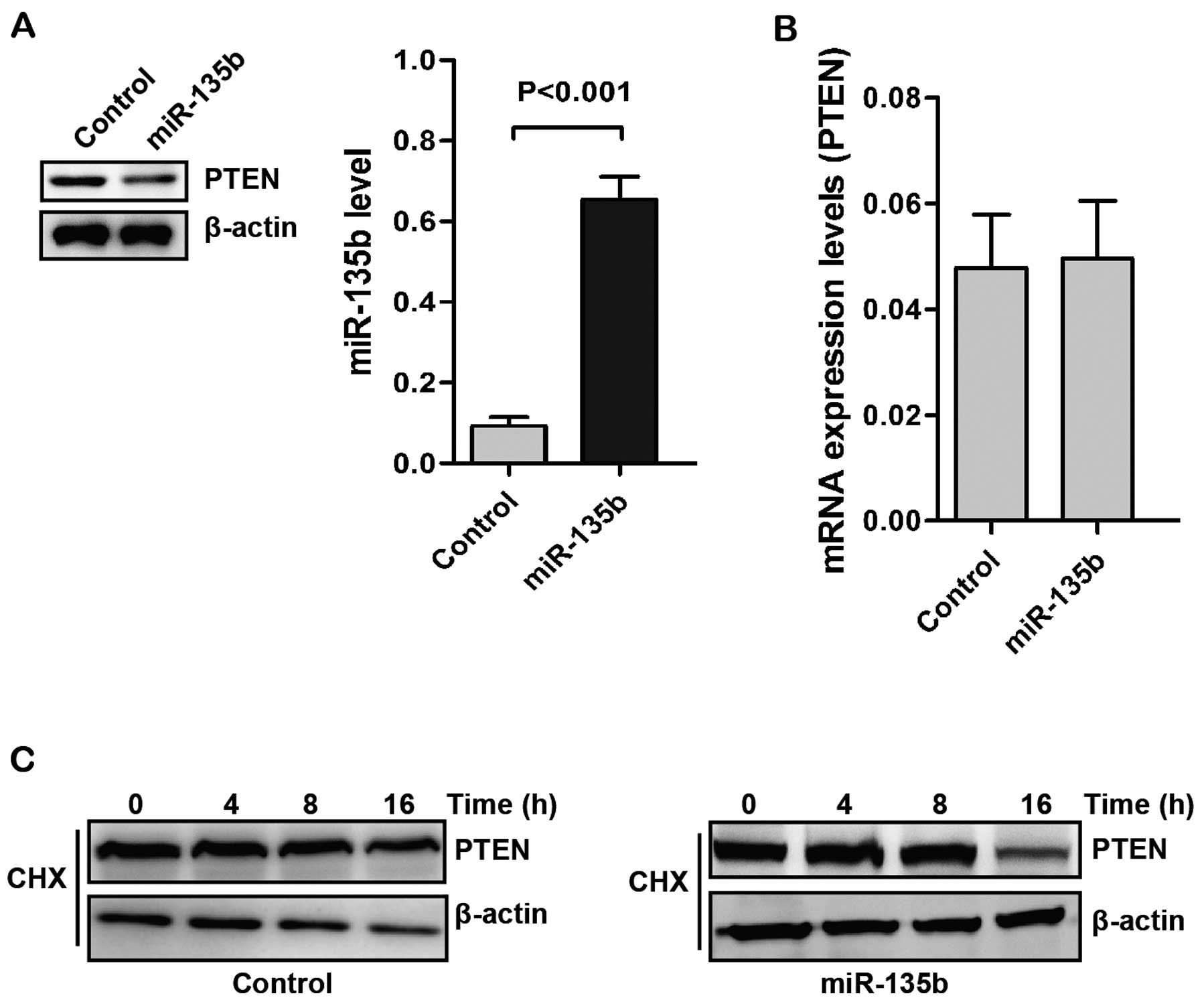
express human miR-135b (Fig. 1A).
Overexpression of miR-135b decreased the levels of PTEN protein. We
then examined the expression of PTEN mRNA by RT-qPCR. In RT-qPCR
analysis, no statistically significant change in PTEN mRNA
expression was observed between the control and miR-135b
overexpressing groups (Fig.
1B).
The role of ubiquitination in modulating PTEN
protein has been identified (37,38).
The regulation of PTEN levels by miR-135b may be the results of
protein stability regulation. We determined the PTEN stability in
Caco2 cells with treatment of cycloheximide (CHX). As shown in
Fig. 1C, upregulation of miR-135b
decreased the stability of PTEN. These results suggested that
miR-135b may regulate the stability of PTEN in post-translational
levels.
Targeting of USP13 by miR-135b
miR-135b is predicted not to target PTEN mRNA. To
determine the function of miR-135b in the regulation of PTEN
stability we used the computational algorithms of TargetScan (ver.
5.2) for prediction analysis to identify target genes of miR-135b.
USP13 is a deubiquitinase that mediates deubiquitination of the
target proteins. USP13 was reported to stabilize the PTEN protein
by direct binding and deubiquitination of PTEN (39). Along with these observations,
reassessment of computational prediction suggested that miR-135b
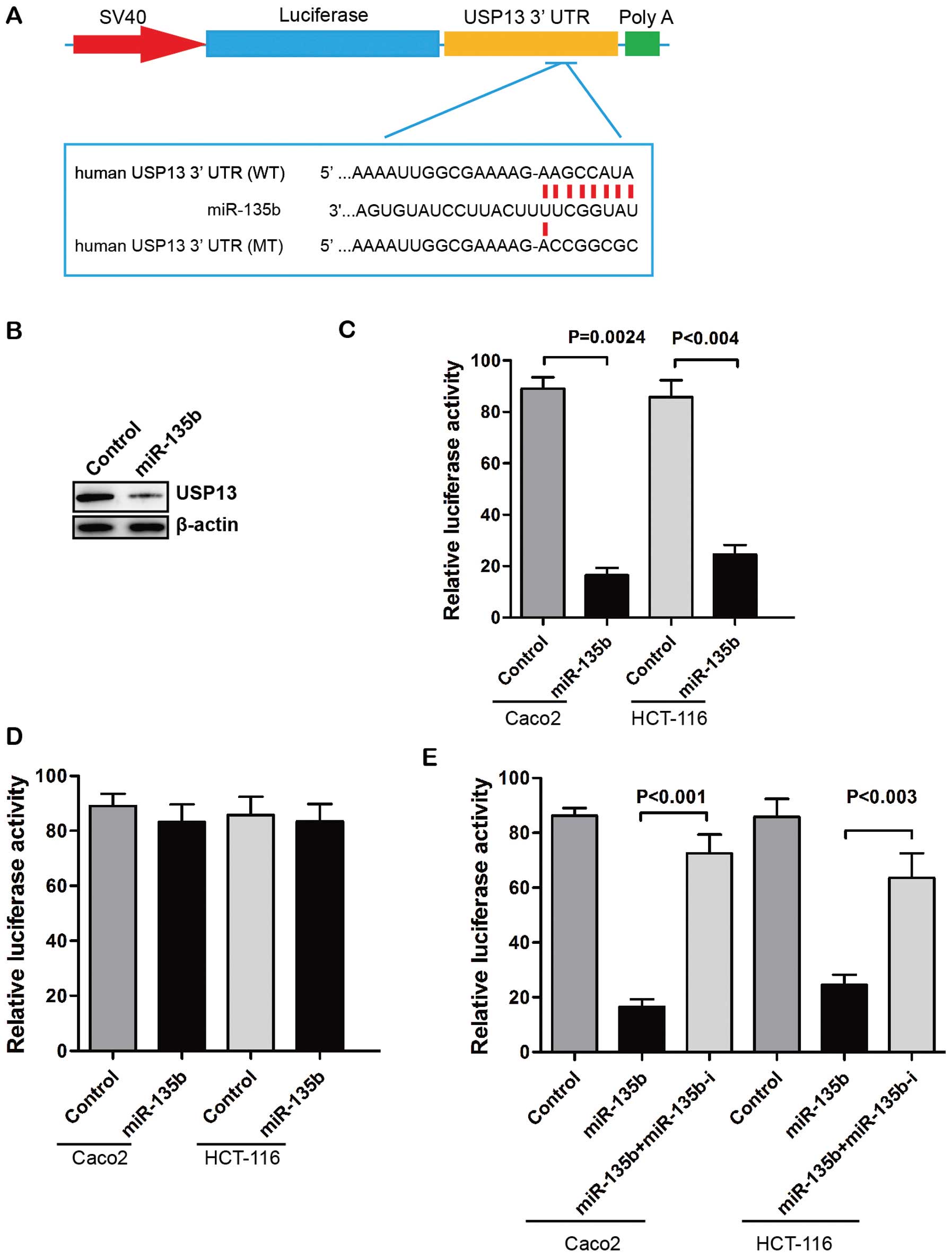
potentially targets USP13 gene (Fig. 2A). Overexpression of miR-135b
suppressed USP13 protein expression in Caco2 cells (Fig. 2B). We also found that
over-expression of miR-135b significantly reduced the luciferase
activity of plasmids containing the 3′-UTR of USP13 (Fig. 2C). The putative mutated target sites
of USP13 3′-UTR were introduced to further investigate the direct
regulatory effect. The reporter assay showed that the effects of
miR-135b repression were eliminated when the target sites of USP13
3′-UTR were mutated (Fig. 2D).
Moreover, the suppressive effects of miR-135b on the USP13
3′-UTR-carrying luciferase were significantly reduced by the
expression of miRNA inhibitor against human miR-135b (Fig. 2E). These results indicated that
miR-135b regulated the expression of USP13 through a direct seed
sequence interaction.
miR-135b promotes glycolysis
The Warburg effect is defined by an increased
glucose uptake via glycolysis as a cellular resource, and is a
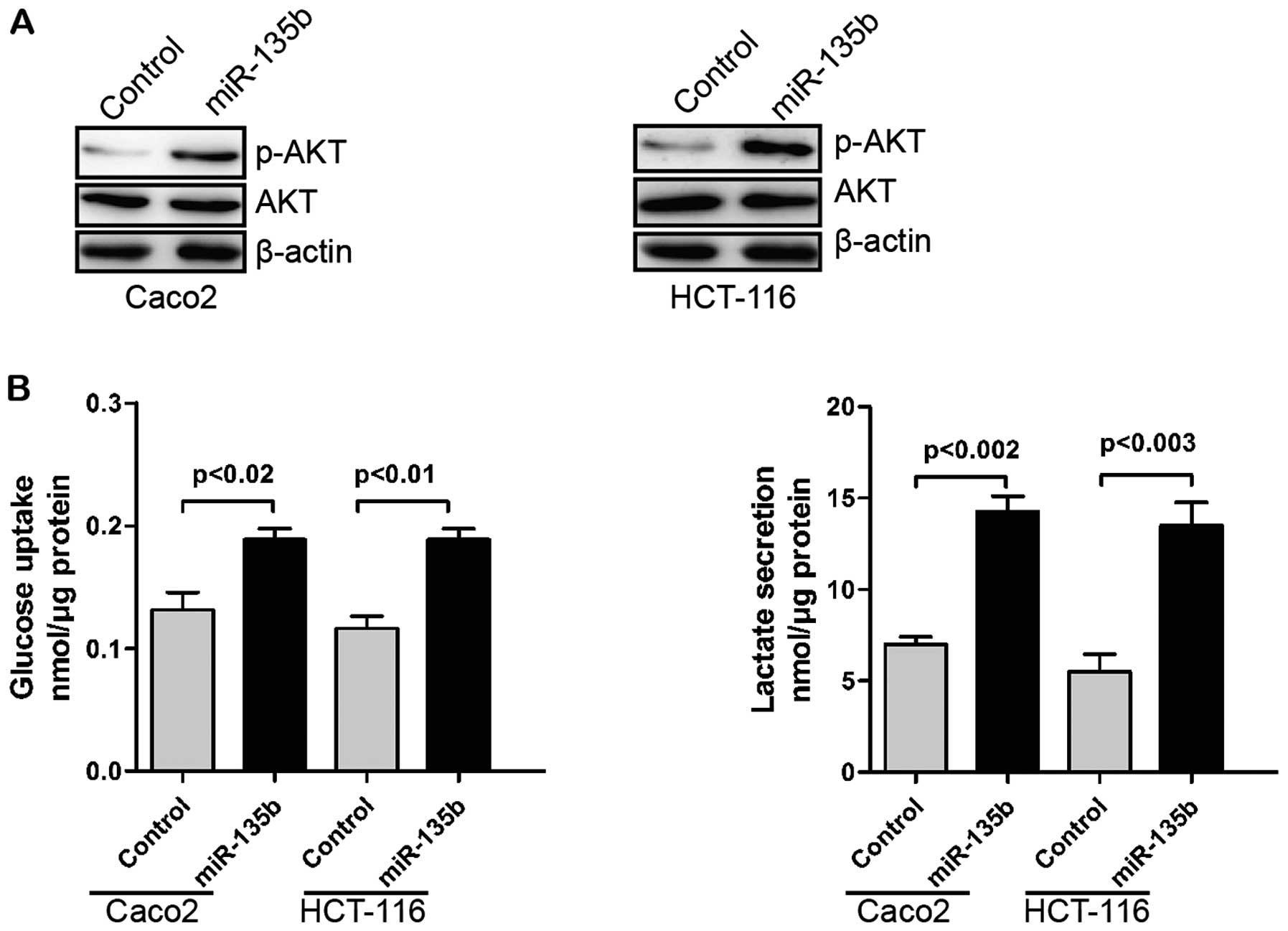
common phenotype of cancer cells (40). AKT was reported to play a critical
role in regulating the Warburg effect (41). USP13 can regulate AKT
phosphorylation by stabilizing PTEN protein. Thus, USP13 can
suppress glycolysis and cell proliferation (39). We hypothesized that miR-135b might
regulate glycolysis through the downregulation of USP13/PTEN.
Overexpression of miR-135b increased AKT phosphorylation (Fig. 3A). Additionally, glucose uptake and
glycolysis of CRC cells were promoted by miR-135b (Fig. 3B).
miR-135b promotes glycolysis and cell
proliferation via downregulation of USP13 or PTEN
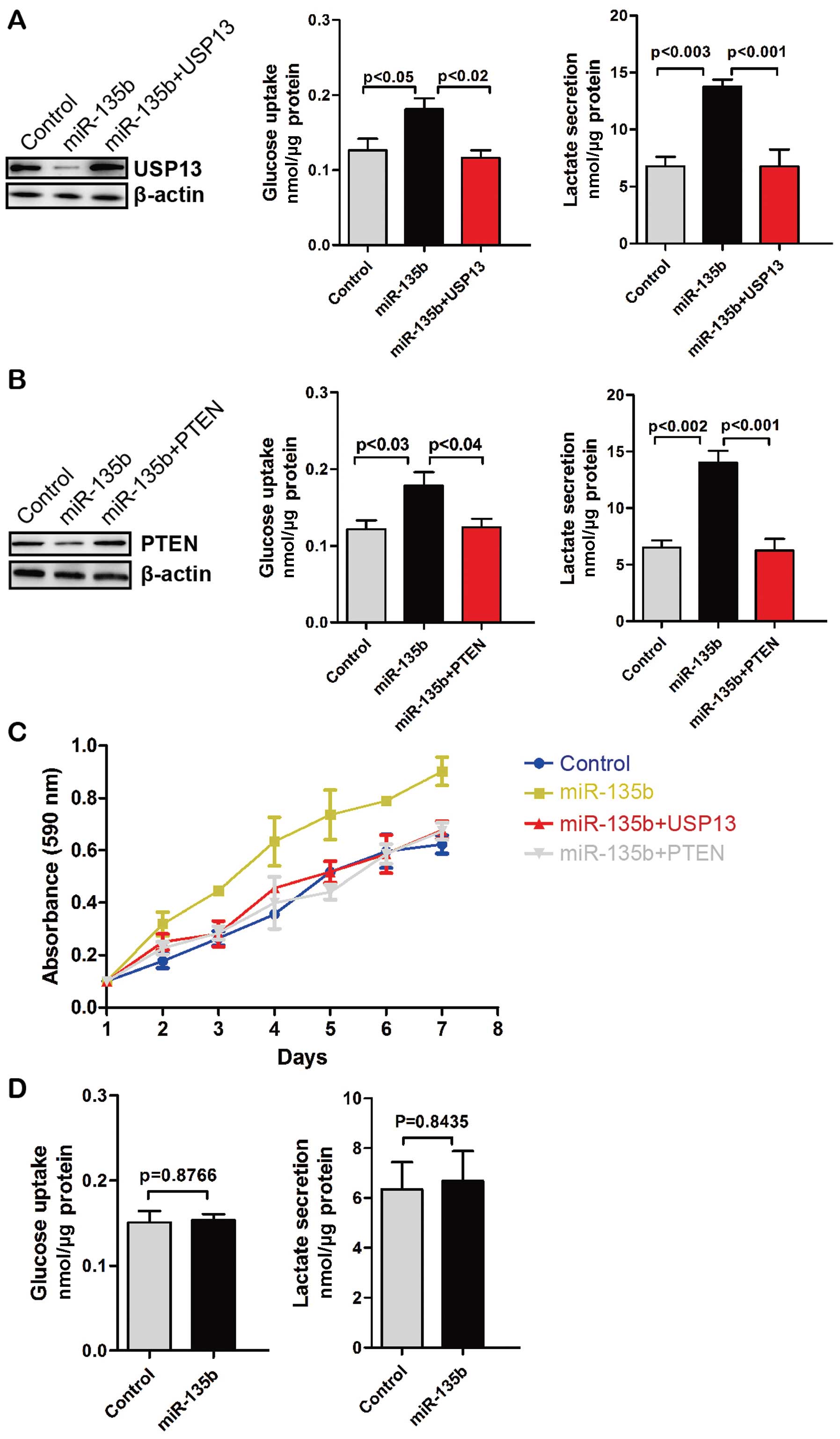
To determine whether miR-135b-mediated upregulation
of glycolysis depends on USP13, we restored the expression of USP13
in miR-135b stably expressing CRC cells. Glucose uptake and
glycolysis in miR-135b-overexpressing CRC cells were decreased with
the restoration of USP13 (Fig. 4A).
Similarly, overexpression of PTEN decreased glucose uptake and
glycolysis (Fig. 4B). Moreover,
miR-135b promoted the proliferation of HCT-116 CRC cells but not
with the restoration of USP13 or overexpression of PTEN (Fig. 4C). CO-115 CRC cells have mutations
leading to the premature stop codon of PTEN. Thus, we overexpressed
miR-135b in CO-115 cells. We found that miR-135b did not affect
glucose uptake and glycolysis in CO-115 cells (Fig. 4D). Therefore, miR-135b promotes cell
proliferation and glycolysis via the downregulation of USP13 or
PTEN.
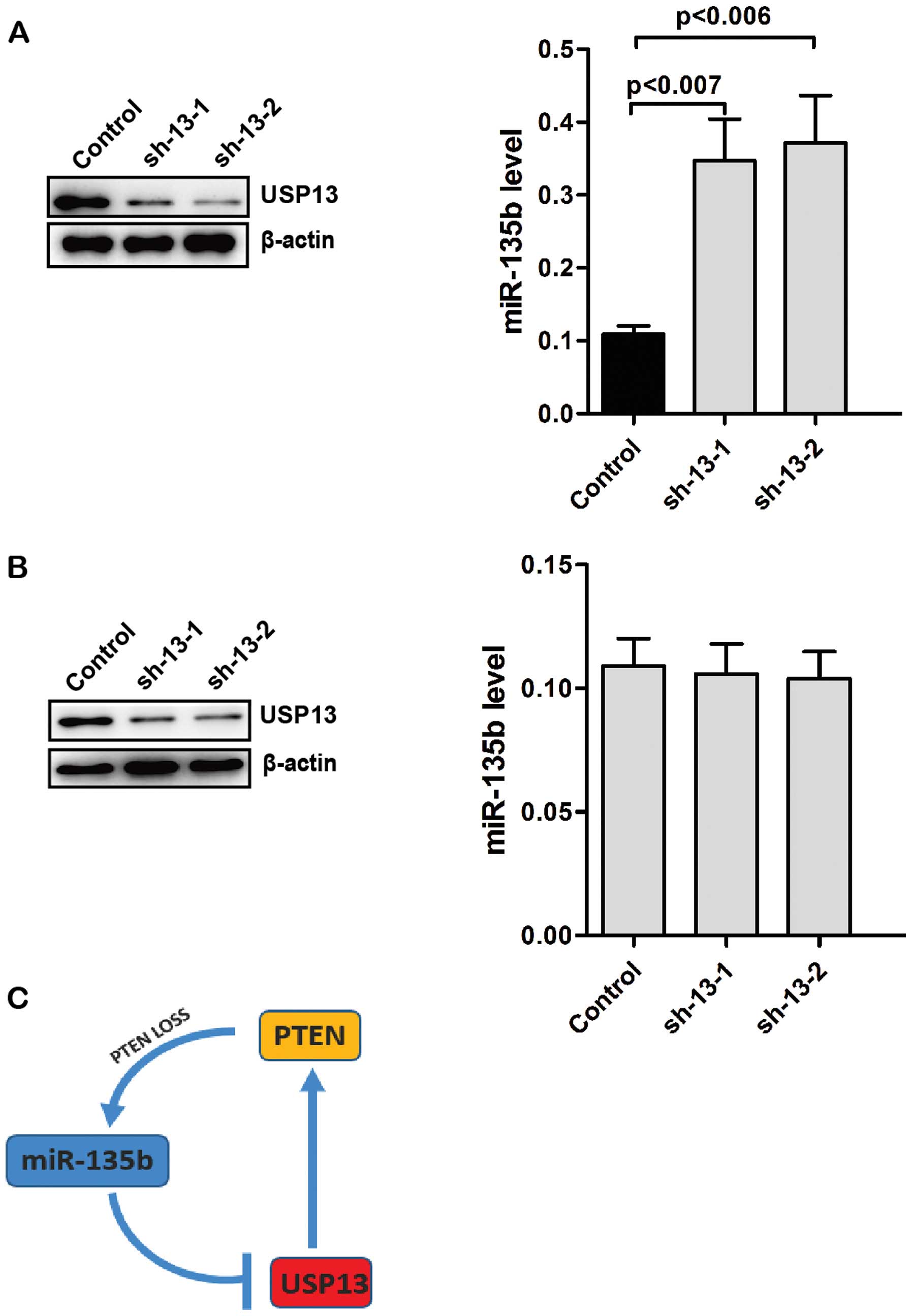
Knockdown of USP13 increases the levels
of endogenous miR-135b
PTEN is a negative regulator of the PI3K pathway and
overexpression of miR-135b partly depends on PI3K pathway. We
determined whether the levels of USP13 affect the expression of
endogenous miR-135b. Knockdown of USP13 in Caco2 cells decreased
the levels of PTEN (Fig. 5A). By
contrast, the levels of endogenous miR-135b were significantly
increased in USP13-knockdwon Caco2 cells (Fig. 5A). We knocked down the expression of
USP13 in CO-115 cells. However, knockdown of USP13 did not affect
the levels of endogenous miR-135b in CO-115 cells (Fig. 5B). These results suggested that loss
of USP13 upregulates the levels of endogenous miR-135b by reducing
the stability of PTEN.
Discussion
As described earlier, miR-135b functions as a key
molecule downstream of oncogenic pathways involved in CRC
progression (36). miR-135b and its
networks are potential therapeutic targets in CRC. The levels of
miR-135b in serum were found to be biomarkers for early detection
of CRC (42). Xu et al
showed that miR-135b is correlated with the clinical parameters of
CRC (43). miR-135b was found to be
involved in CRC cell invasion (34). Moreover, the overexpression of
miR-135b was due to APC/β-catenin and PTEN/PI3K pathway
deregulation. APC is also the target of miR-135b (36). In the present study, we have shown
that miR-135b can regulate the stability of PTEN protein.
Deubiquitinating enzyme USP13 can stabilize the PTEN protein by
direct binding and deubiquitination of PTEN (39). We used the computational algorithms
of TargetScan for prediction analysis to identify target genes of
miR-135b and found that USP13 is a potential target of miR-135b
(Fig. 2A). Luciferase assay showed
that miR-135b significantly reduced the USP13 3′-UTR-carrying
luciferase (Fig. 2C). However, the
reporter assay showed that the effects of miR-135b repression were
eliminated when the target sites of USP13 3′-UTR were mutated
(Fig. 2D). Moreover, the
suppressive effects of miR-135b on the USP13 3′-UTR-carrying
luciferase were significantly reduced by miR-135b inhibitor
(Fig. 2E). This result indicates
that USP13 is a target of miR-135b.
The Warburg effect is a common phenotype of cancer
cells by increasing glucose uptake via glycolysis (44). AKT was reported to play a critical
role in regulating the Warburg effect. USP13 can regulate AKT
phosphorylation by stabilizing PTEN protein (39). Thus, USP13 can suppress glycolysis
and cell proliferation. miR-135b could promote AKT phosphorylation
and enhance glycolysis in Caco2 and HCT-116 CRC cells with
wild-type PTEN (45). PTEN
inactivation by mutation or allelic loss also occurs in CRCs.
CO-115 CRC cells have mutations leading to the premature stop codon
of PTEN (45). In CO-115 cells, we
found that miR-135b did not affect glucose uptake and glycolysis.
Therefore, miR-135b promotes glucose uptake and glycolysis of CRC
cells via the downregulation of PTEN. Moreover, miR-135b promoted
the proliferation of HCT-116 CRC cells, but not with the
restoration of USP13 or overexpression of PTEN. APC was proven to
be a target of miR-135b and form a feedback loop with miR-135b
(36). We also found that knockdown
of USP13 upregulated the levels of endogenous miR-135b in the
presence of wild-type PTEN. miR-135b can activate the expression of
miR-135b itself by inhibiting the expression of USP13, thus
attenuating the inhibitory effect of PTEN. These results reveal
positive feedback loops between miR-135b and PTEN inactivation via
USP13 in CRCs. Thus, this study provides support for the
therapeutic potential of miR-135b in the targeted therapy of
CRC.
References
|
1
|
Labianca R, Nordlinger B, Beretta GD, et
al: Early colon cancer: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 24:vi64–vi72. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paquin MC, Leblanc C, Lemieux E, Bian B
and Rivard N: Functional impact of colorectal cancer-associated
mutations in the transcription factor E2F4. Int J Oncol.
43:2015–2022. 2013.PubMed/NCBI
|
|
3
|
Garcia Sanchez J: Colonoscopic polypectomy
and long-term prevention of colorectal cancer deaths. Rev Clin Esp.
212:4082012.(In Spanish).
|
|
4
|
Nystrom M and Mutanen M: Diet and
epigenetics in colon cancer. World J Gastroenterol. 15:257–263.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim YI: Nutritional epigenetics: impact of
folate deficiency on DNA methylation and colon cancer
susceptibility. J Nutr. 135:2703–2709. 2005.PubMed/NCBI
|
|
6
|
Yamashita K, Dai T, Dai Y, Yamamoto F and
Perucho M: Genetics supersedes epigenetics in colon cancer
phenotype. Cancer Cell. 4:121–131. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Markman B, Javier Ramos F, Capdevila J and
Tabernero J: EGFR and KRAS in colorectal cancer. Adv Clin Chem.
51:71–119. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu S, Xiao X, Lu J, Qian X, Liu B and Feng
J: Colorectal cancer patients with low abundance of KRAS mutation
may benefit from EGFR antibody therapy. PLoS One. 8:e680222013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yokota T: Are KRAS/BRAF mutations potent
prognostic and/or predictive biomarkers in colorectal cancers?
Anticancer Agents Med Chem. 12:163–171. 2012. View Article : Google Scholar :
|
|
10
|
Hamilton SR: BRAF mutation and
microsatellite instability status in colonic and rectal carcinoma:
context really does matter. J Natl Cancer Inst. 105:1075–1077.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tonini G, Imperatori M, Vincenzi B, Frezza
AM and Santini D: Rechallenge therapy and treatment holiday:
different strategies in management of metastatic colorectal cancer.
J Exp Clin Cancer Res. 32:922013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fodde R: The APC gene in colorectal
cancer. Eur J Cancer. 38:867–871. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Woodford-Richens KL, Rowan AJ, Gorman P,
et al: SMAD4 mutations in colorectal cancer probably occur before
chromosomal instability, but after divergence of the microsatellite
instability pathway. Proc Natl Acad Sci USA. 98:9719–9723. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de Weger VA, Turksma AW, Voorham QJ, et
al: Clinical effects of adjuvant active specific immunotherapy
differ between patients with microsatellitestable and
microsatelliteinstable colon cancer. Clin Cancer Res. 18:882–889.
2012. View Article : Google Scholar
|
|
15
|
Berg M, Danielsen SA, Ahlquist T, et al:
DNA sequence profiles of the colorectal cancer critical gene set
KRAS-BRAF- PIK3CA-PTEN-TP53 related to age at disease onset. PLoS
One. 5:e139782010. View Article : Google Scholar
|
|
16
|
Ilyas M, Tomlinson IP, Rowan A, Pignatelli
M and Bodmer WF: Beta-catenin mutations in cell lines established
from human colorectal cancers. Proc Natl Acad Sci USA.
94:10330–10334. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Georgescu MM: PTEN tumor suppressor
network in PI3K-Akt pathway control. Genes Cancer. 1:1170–1177.
2010. View Article : Google Scholar
|
|
18
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012.PubMed/NCBI
|
|
19
|
Nassif NT, Lobo GP, Wu X, et al: PTEN
mutations are common in sporadic microsatellite stable colorectal
cancer. Oncogene. 23:617–628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin Q, Furong W and Baosheng L: Multiple
functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer
Res. 33:502014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y and Taniguchi T: MicroRNAs and DNA
damage response: implications for cancer therapy. Cell Cycle.
12:32–42. 2013. View
Article : Google Scholar :
|
|
22
|
Krishnan K, Steptoe AL, Martin HC, et al:
MicroRNA-182-5p targets a network of genes involved in DNA repair.
RNA. 19:230–242. 2013. View Article : Google Scholar :
|
|
23
|
He YQ, Sheng JQ, Ling XL, et al: Estradiol
regulates miR-135b and mismatch repair gene expressions via
estrogen receptor-beta in colorectal cells. Exp Mol Med.
44:723–732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dumortier O, Hinault C and Van Obberghen
E: MicroRNAs and metabolism crosstalk in energy homeostasis. Cell
Metab. 18:312–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhai H and Ju J: Implications of microRNAs
in colorectal cancer development, diagnosis, prognosis, and
therapeutics. Front Genet. 2:782011. View Article : Google Scholar :
|
|
29
|
Dong Y, Wu WK, Wu CW, Sung JJ, Yu J and Ng
SS: MicroRNA dysregulation in colorectal cancer: a clinical
perspective. Br J Cancer. 104:893–898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schetter AJ, Okayama H and Harris CC: The
role of microRNAs in colorectal cancer. Cancer J. 18:244–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brunet Vega A, Pericay C, Moya I, et al:
microRNA expression profile in stage III colorectal cancer:
Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep.
30:320–326. 2013.PubMed/NCBI
|
|
32
|
Khatri R and Subramanian S: MicroRNA-135b
and its circuitry networks as potential therapeutic targets in
colon cancer. Front Oncol. 3:2682013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu CW, Ng SC, Dong Y, et al:
Identification of microRNA-135b in stool as a potential noninvasive
biomarker for colorectal cancer and adenoma. Clin Cancer Res.
20:2994–3002. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu W, Wang Z, Yang P, et al: MicroRNA-135b
regulates metastasis suppressor 1 expression and promotes migration
and invasion in colorectal cancer. Mol Cell Biochem. 388:249–259.
2014. View Article : Google Scholar
|
|
35
|
Munding JB, Adai AT, Maghnouj A, et al:
Global microRNA expression profiling of microdissected tissues
identifies miR-135b as a novel biomarker for pancreatic ductal
adenocarcinoma. Int J Cancer. 131:E86–E95. 2012. View Article : Google Scholar
|
|
36
|
Valeri N, Braconi C, Gasparini P, et al:
MicroRNA-135b promotes cancer progression by acting as a downstream
effector of oncogenic pathways in colon cancer. Cancer Cell.
25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JT, Shan J, Zhong J, et al:
RFP-mediated ubiquitination of PTEN modulates its effect on AKT
activation. Cell Res. 23:552–564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Trotman LC, Koppie T, et al:
NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell.
128:129–139. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Zhang P, Wei Y, et al:
Deubiquitination and stabilization of PTEN by USP13. Nat Cell Biol.
15:1486–1494. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Donohoe DR, Collins LB, Wali A, Bigler R,
Sun W and Bultman SJ: The Warburg effect dictates the mechanism of
butyrate-mediated histone acetylation and cell proliferation. Mol
Cell. 48:612–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Najafov A and Alessi DR: Uncoupling the
Warburg effect from cancer. Proc Natl Acad Sci USA.
107:19135–19136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Faltejskova P, Bocanek O, Sachlova M, et
al: Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum:
evidence against their usage as biomarkers in colorectal cancer.
Cancer Biomark. 12:199–204. 2012.PubMed/NCBI
|
|
43
|
Xu XM, Qian JC, Deng ZL, et al: Expression
of miR-21, miR-31, miR-96 and miR-135b is correlated with the
clinical parameters of colorectal cancer. Oncol Lett. 4:339–345.
2012.PubMed/NCBI
|
|
44
|
Zhao Y, Butler EB and Tan M: Targeting
cellular metabolism to improve cancer therapeutics. Cell Death Dis.
4:e5322013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ahmed D, Eide PW, Eilertsen IA, et al:
Epigenetic and genetic features of 24 colon cancer cell lines.
Oncogenesis. 2:e712013. View Article : Google Scholar : PubMed/NCBI
|