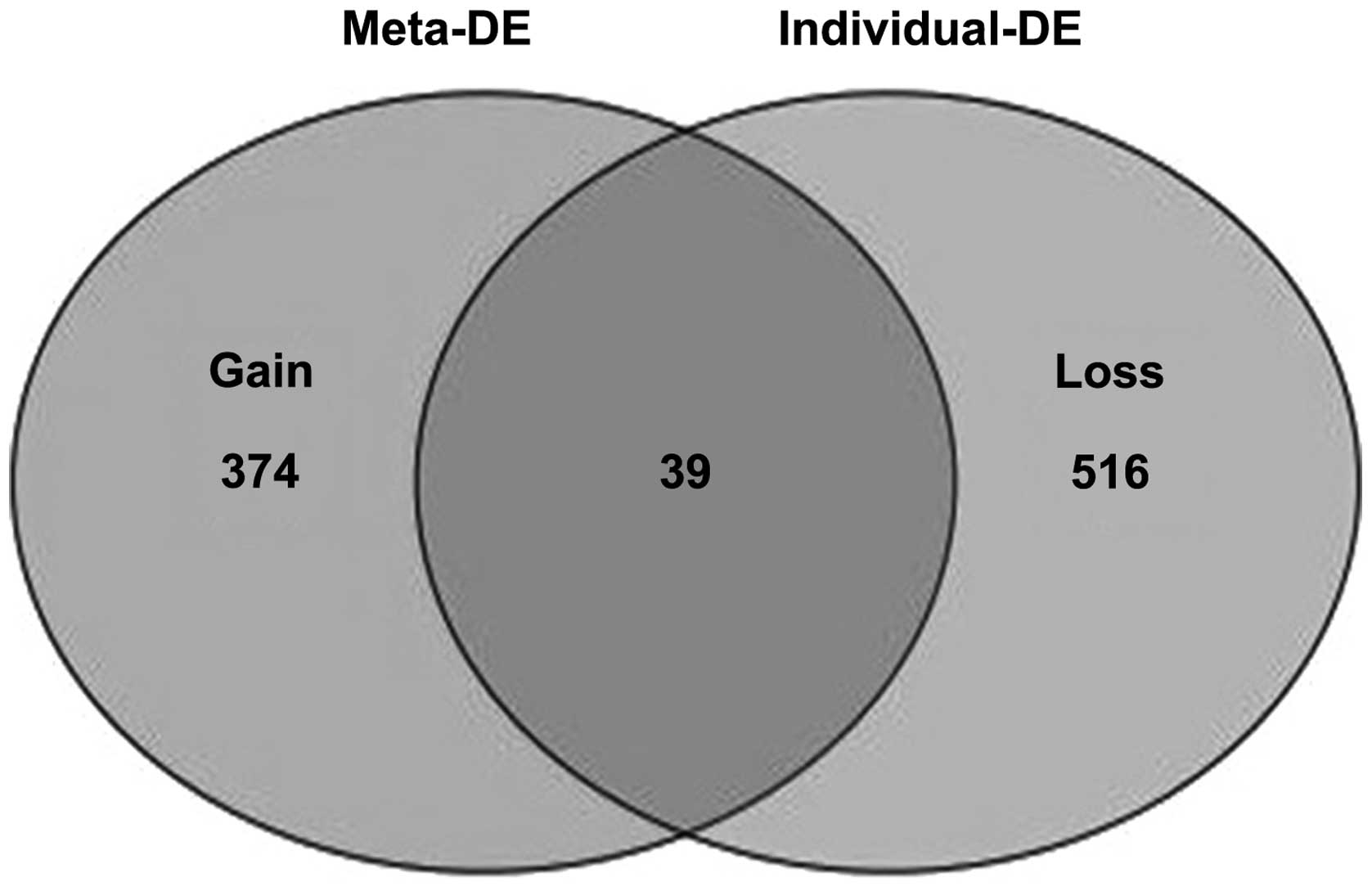
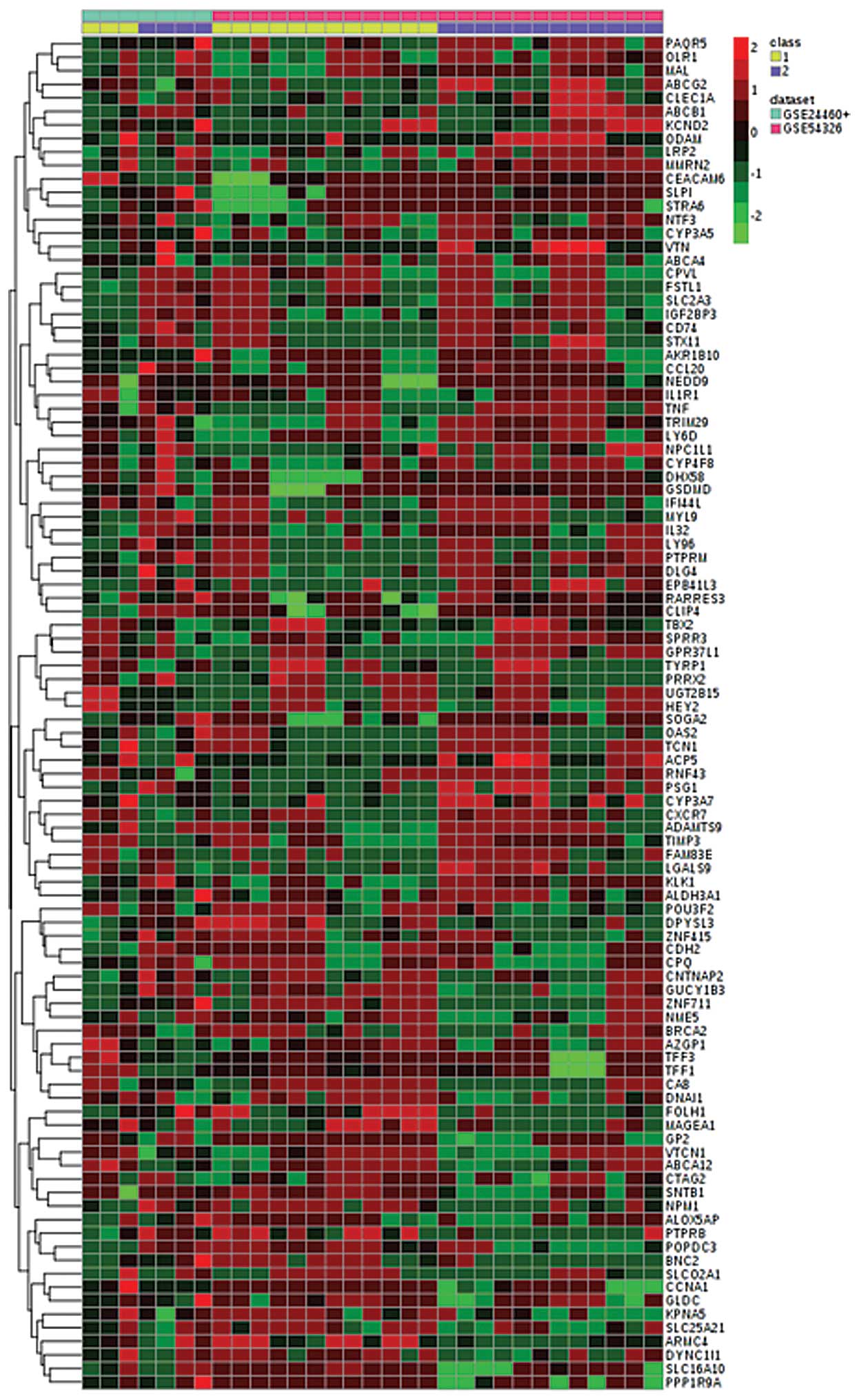
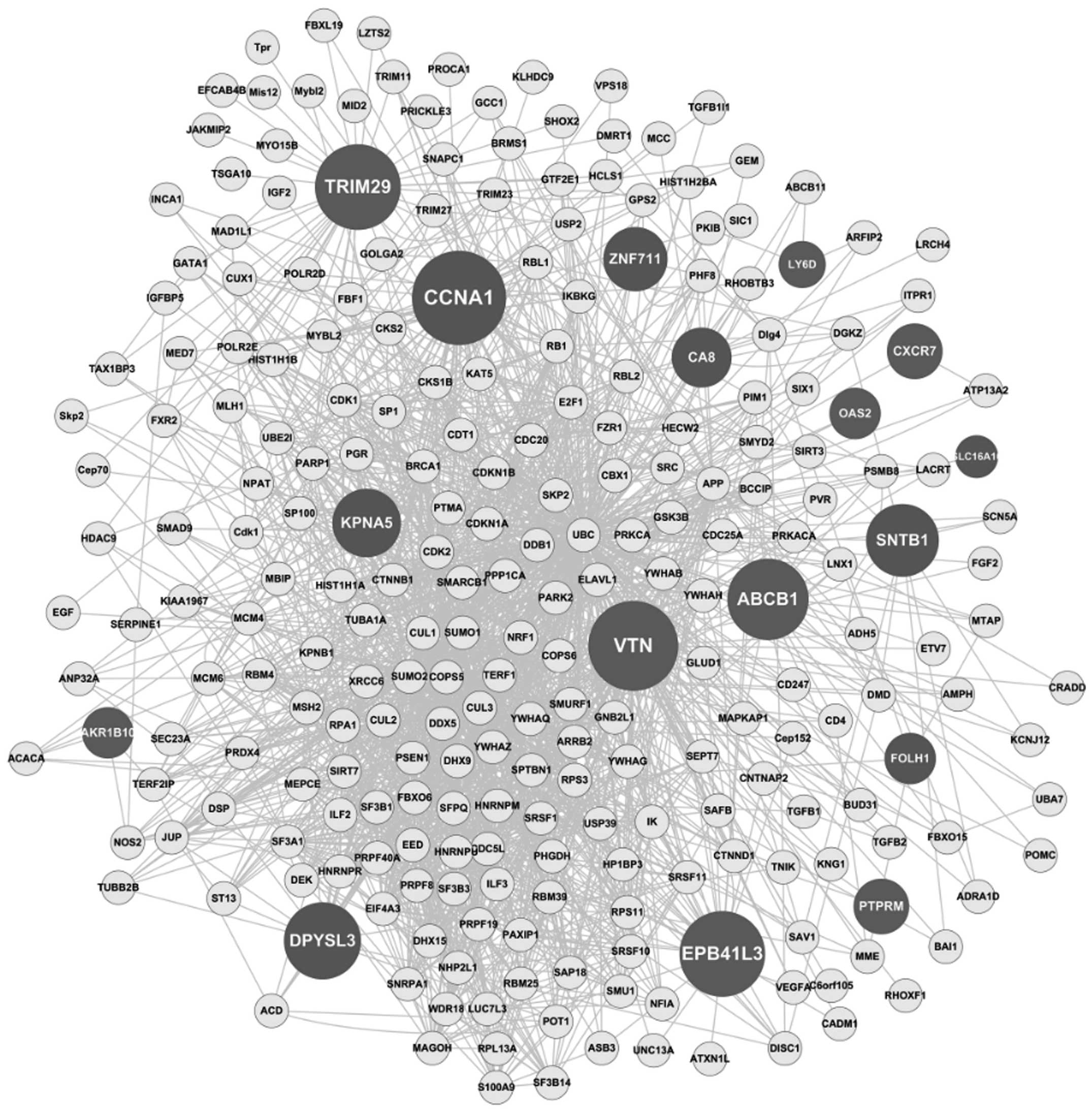
|
1
|
Langer SW, Sehested M and Jensen PB:
Anthracycline extravasation: a comprehensive review of experimental
and clinical treatments. Tumori. 95:273–282. 2009.PubMed/NCBI
|
|
2
|
Gluck S: Adjuvant chemotherapy for early
breast cancer: optimal use of epirubicin. Oncologist. 10:780–791.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreno-Aspitia A and Perez EA:
Anthracycline- and/or taxane-resistant breast cancer: results of a
literature review to determine the clinical challenges and current
treatment trends. Clin Ther. 31:1619–1640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu BH: Strategy in the treatment of
anthracycline-resistant breast cancer. Zhonghua Zhong Liu Za Zhi.
29:241–244. 2007.(In Chinese). PubMed/NCBI
|
|
5
|
Stavrovskaya AA: Cellular mechanisms of
multidrug resistance of tumor cells. Biochemistry (Mosc).
65:95–106. 2000.
|
|
6
|
Banerjee D, Mayer-Kuckuk P, Capiaux G,
Budak-Alpdogan T, Gorlick R and Bertino JR: Novel aspects of
resistance to drugs targeted to dihydrofolate reductase and
thymidylate synthase. Biochim Biophys Acta. 1587:164–173. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Faneyte IF, Kristel PM and van de Vijver
MJ: Multidrug resistance associated genes MRP1, MRP2 and MRP3 in
primary and anthracycline exposed breast cancer. Anticancer Res.
24:2931–2939. 2004.PubMed/NCBI
|
|
8
|
Liscovitch M and Lavie Y: Cancer multidrug
resistance: a review of recent drug discovery research. IDrugs.
5:349–355. 2002.
|
|
9
|
van Zuylen L, Nooter K, Sparreboom A and
Verweij J: Development of multidrug-resistance convertors: sense or
nonsense? Invest New Drugs. 18:205–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ravdin PM: Anthracycline resistance in
breast cancer: clinical applications of current knowledge. Eur J
Cancer. 31A(Suppl 7): S11–S14. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chien AJ and Moasser MM: Cellular
mechanisms of resistance to anthracyclines and taxanes in cancer:
intrinsic and acquired. Semin Oncol. 35(Suppl 2): S1–S14; quiz S39.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang HC, Kim IJ, Park JH, et al:
Identification of genes with differential expression in acquired
drug-resistant gastric cancer cells using high-density
oligonucleotide microarrays. Clin Cancer Res. 10:272–284. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calcagno AM, Salcido CD, Gillet JP, et al:
Prolonged drug selection of breast cancer cells and enrichment of
cancer stem cell characteristics. J Natl Cancer Inst.
102:1637–1652. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Munkacsy G, Abdul-Ghani R, Mihaly Z, et
al: PSMB7 is associated with anthracycline resistance and is a
prognostic biomarker in breast cancer. Br J Cancer. 102:361–368.
2010. View Article : Google Scholar :
|
|
16
|
Siddiqui AS, Delaney AD, Schnerch A,
Griffith OL, Jones SJ and Marra MA: Sequence biases in large scale
gene expression profiling data. Nucleic Acids Res. 34:e832006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moreau Y, Aerts S, De Moor B, De Strooper
B and Dabrowski M: Comparison and meta-analysis of microarray data:
from the bench to the computer desk. Trends Genet. 19:570–577.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cohn LD and Becker BJ: How meta-analysis
increases statistical power. Psychol Methods. 8:243–253. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosaka Y, Inoue H, Ohmachi T, et al:
Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph
node metastasis in gastric cancer. Ann Surg Oncol. 14:2543–2549.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Yang H, Palmbos PL, et al:
ATDC/TRIM29 phosphorylation by ATM/MAPKAP kinase 2 mediates
radioresistance in pancreatic cancer cells. Cancer Res.
74:1778–1788. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanno Y, Watanabe M, Kimura T, Nonomura K,
Tanaka S and Hatakeyama S: TRIM29 as a novel prostate basal cell
marker for diagnosis of prostate cancer. Acta Histochem.
116:708–712. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dezi M, Fribourg PF, Di Cicco A, et al:
The multidrug resistance half-transporter ABCG2 is purified as a
tetramer upon selective extraction from membranes. Biochim Biophys
Acta. 1798:2094–2101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang G, Wang Z, Luo W, Jiao H, Wu J and
Jiang C: Expression of potential cancer stem cell marker ABCG2 is
associated with malignant behaviors of hepatocellular carcinoma.
Gastroenterol Res Pract. 2013:7825812013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Featherstone JM, Speers AG, Lwaleed BA,
Hayes MC, Cooper AJ and Birch BR: The nuclear membrane in multidrug
resistance: microinjection of epirubicin into bladder cancer cell
lines. BJU Int. 95:1091–1098. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kiyomiya K, Matsuo S and Kurebe M:
Mechanism of specific nuclear transport of adriamycin: the mode of
nuclear translocation of adriamycin-proteasome complex. Cancer Res.
61:2467–2471. 2001.PubMed/NCBI
|
|
26
|
Fujise H, Annoura T, Sasawatari S, Ikeda T
and Ueda K: Trans-epithelial transport and cellular accumulation of
steroid hormones and polychlorobiphenyl in porcine kidney cells
expressed with human P-glycoprotein. Chemosphere. 46:1505–1511.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Doehmer J, Goeptar AR and Vermeulen NP:
Cytochromes P450 and drug resistance. Cytotechnology. 12:357–366.
1993. View Article : Google Scholar : PubMed/NCBI
|