Introduction
Breast cancer is the most commonly diagnosed cancer
and the second leading cause of cancer-related death in females
worldwide (1). As estrogens acting
via estrogen receptors (ERs) play a critical role in breast cancer
development, growth and progression, and over 70% of breast cancers
are ER-positive (2); ER is
therefore a valuable target for breast cancer therapy (3–5).
Although it is well documented that estrogens such as 17β-estradiol
(E2), usually stimulate the proliferation of ER-positive breast
cancer cells through the regulation of a large number of genes
(6,7), it is a great challenge to determine
which genes or gene networks underlie the estrogen proliferative
response in breast cancer cells.
Recently, long non-coding RNAs (lncRNAs) have been
emerged as an important target in cancer development and
progression. Studies over the years have shown that lncRNAs, which
are non-coding RNAs with a length larger than 200 bp, play a major
role in embryogenesis and cell differentiation, and in the
development and progression of various diseases including cancers
(8–11). lncRNAs may function as either tumor
suppressors or tumor promoters in such prevalent cancers as breast
and prostate cancer (12). Among
lncRNAs, Xist/Tsix, H19, HOTAIR and AIR are a few examples of
lncRNAs whose functions have been investigated.
H19 is a well-known imprinted oncofetal gene, which
is expressed on maternal alleles without a protein product
(13), accumulates in the human
placenta and several fetal tissues, and probably plays a pivotal
role in embryogenesis and fetal growth and development (14). The H19 gene is localized at human
chromosome 11p15.5 (15), in which
a variety of disorders including cancer predisposition for both
pediatric and adult tumors has been implicated (16). Aberrant expression of H19 is
observed in numerous solid tumors, including hepatoma, bladder and
breast cancer (16–18), and it has been suggested to be
involved in oncogenesis and progression of various types of tumors
(19).
Based on the previous observations that H19
expression is associated with the presence of estrogen and
progesterone receptors (20), and
estrogen regulates H19 expression in breast cancer cells (21), we hypothesized that estrogen-induced
cell growth in ER-positive breast cancer cells is mediated through
the upregulation of H19 gene expression. Our results indicate that
H19 lncRNA is a critical factor in estrogen-stimulated cell growth
in breast cancer cells, and blockade of H19 function is a potential
strategy for the treatment of ER-positive breast cancer.
Materials and methods
Cell culture and hormone treatment
The MCF-7 and MDA-MB-231 breast cancer cells were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA). Cells were grown in RPMI-1640 culture medium (Gibco-BRL,
Grand Island, NY, USA) supplemented with 10% fetal bovine serum
(FBS; HyClone Laboratories, Inc., Logan, UT, USA), 20 mM
L-glutamine, 100 U/ml of penicillin and 100 μg/ml of
streptomycin. All cells were maintained in a humidified incubator
with 5% CO2 at 37°C. Four days before drug treatment,
cells were switched to phenol red-free RPMI-1640 medium
supplemented with 5% charcoal-dextran-treated FBS (BI Technologies,
Fullerton, CA, USA), 20 mM L-glutamine, 100 U/ml of penicillin and
100 μg/ml of streptomycin. Medium was changed every 2 days,
and cells were treated with vehicle control [0.1% dimethyl
sulfoxide (DMSO)], various concentrations of E2 and ICI 182780
alone or in combination for the times as indicated in each
experiment. All chemicals and hormones were obtained from
Sigma-Aldrich (St. Louis, MO, USA) except those indicated.
Breast cancer tissue samples
Forty-five patients with breast cancer who underwent
surgery at Xiangya Hospital were enrolled in the present study. The
resected tumor specimens were immediately frozen in liquid nitrogen
and kept at −80°C until RNA extraction. The present study was
approved by the Ethics Committee of Xiangya Hospital, China
(project no. CTXY-140001-5) and a written informed consent was
obtained from each subject.
RNA extraction and lncRNA screening
Total RNAs from harvested cells or tumor tissues
were prepared using TRIzol reagent (Omega Bio-Tek, Inc., Norcross,
GA, USA) according to the manufacturer’s instructions, and the
concentration was determined using a NanoDrop 2000. The quality of
the RNA samples was in assurance with an A260/A230 ratio >1.7
and an A260/A280 ratio between 1.8 and 2.0. One microgram of total
RNA was subjected to RT-PCR and used for the screening of lncRNAs
by the Disease-Related Human lncRNA Profiler kit following the
manufacturer’s protocol (SBI, Mountain View, CA, USA). A change of
log2(n) >2n was considered a meaningful change.
Quantitative real-time RT-PCR
Total RNA (500 ng) was reversely transcribed in a
total volume of 20 μl with 200 units of reverse
transcriptase, 50 pmol random hexamer and 1 mM deoxynucleotide
triphosphates. The reaction products were then diluted to a total
volume of 100 μl with distilled water. The real-time PCR
reaction consisted of 2 μl of diluted reverse transcription
product, 1X SYBR-Green Master Mix (Applied Biosystems, Foster City,
CA, USA) and 50 nM forward and reverse primers. The reaction was
carried out in a Roche LightCycler 480 Sequence Detection System
for 40 cycles (95°C for 15 sec, 60°C for 1 min) after an initial
10-min incubation at 95°C. Glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) was used as an internal control. The primers used were:
H19, 5′-GTCCGGCCTTCCTGAACACCTT-3′ and 5′-GCTTCACCTTCCAGAGCCGAT-3′;
ERα, 5′-CCACCAA CCAGTGCACCATT-3′ and 5′-GGTCTTTTCGTATCCCACCTTTC-3′;
and GAPDH, 5′-TTGATTTTGGAGGGATCTCGCTC-3′ and
5′-GAGTCAACGGATTTGGTCGTATTG-3′. The level of RNA was expressed as a
fold of the control calculated using the ΔΔCt method.
Knockdown of H19 lncRNA using siRNA and
shRNA
Synthetic RNA oligonucleotides targeting ERα and H19
were obtained from RiboBio Co., Ltd. (Guangzhou, China). The siRNA
sequences used for ERα were: 5′-CCAGUGCACCAUUGAUAAAdTdT-3′. H19
lncRNA was knocked down using a specific H19 siRNA and a hairpin
siRNA vector (shRNAH19) as described by Tsang and Kwok (22). The sequences of H19 siRNA were:
5′-CCTGTAACCAAAAGTGACCG-3′; and the H19 hairpin siRNA sequences
were: 5′-CATCAAAGACACCATCGGA-3′ that were subcloned into pSilencer
2.1-U6 neovector (shRNAH19) (RiboBio Co., Ltd.). A negative control
siRNA and shRNA were purchased from RiboBio.
H19 expression vector construction
Total cellular RNAs from MCF-7 cells were subjected
to reverse-transcription as described above. To construct the H19
expression vector, a 2-step strategy was used. First, the entire
H19 RNA was amplified as two fragments by PCR using the following
pairs of primers: 5′-AAAAGGATCCAGGGCCCTGCTCTGATTGG-3′ and
5′-AAAAAAGCTTCCTCGTCTCCAGCCCGAAC-3′ for the upstream fragment
(1,430 bp); and 5′-GGGCGGG GCGGAGTGAATGAGC-3′ and
5′-AAAAAAGCTTTTGCT GTAACAGTGTTTATTGATGA-3′ for the downstream
fragment (1,170 bp). These two PCR fragments were inserted into the
pGEM-T easy vector and amplified following transformation to E.
coli. Subsequently, the upstream and downstream H19 fragments
in the pGEM-T easy vectors were digested with BamHI plus
HindIII and BbvCI plus HindIII, respectively,
ligated and inserted into the pcDNA3.1 vector as a full-length H19
cDNA. The sequences of the H19 expression vector were confirmed by
DNA sequencing.
Transfection study
Cells were plated in phenol red-free medium
containing 5% stripped FBS in 12-well plates at a density of ~70%.
The transfections of various doses of siRNA or shRNA were performed
using Lipofectamine RNAiMAX or Lipofectamine 2000 reagent
(Invitrogen, Life Technologies, USA) following the manufacturer’s
instructions. Twenty-four hours after transfection, the cells were
treated with various hormones for different times and various
analyses were performed as indicated in each experiment.
Western blot analysis
Western blotting was performed as previously
described with minor modifications (23). Briefly, total cellular proteins were
extracted from the harvested cells using a lysis buffer [62.5 mM
Tris-HCl pH 6.8, 100 mM dithiothreitol (DTT), 2% SDS and 10%
glycerol]. The protein concentrations were determined using the
Bradford method with the Bio-Rad protein assay following the
manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). Cellular
proteins were separated on sodium dodecyl sulfate polyacrylamide
gels and transferred to nitrocellulose membranes. Blots were
incubated in blocking buffer (5% non-fat dry milk in Tris-buffered
saline with 0.5% Tween, TBS-T) at room temperature for 2 h. After
washing with TBS-T, the nitrocellulose was incubated with a
specific antibody against ERα (Santa Cruz Biotechnology, Santa
Cruz, CA, USA) or β-actin (AC-15; Sigma Chemical Co., St. Louis,
MO, USA) overnight at 4°C. Following the incubation with a
horseradish peroxidase-conjugated secondary antibody, the signal
was detected using an ECL Western Blotting system (Promega, Madison
WI, USA) and visualized using the Bio-Rad ChemiDoc MP system.
Cell proliferation assay
For the determination of cell proliferation, MCF-7
cells were plated in 96-well plates at ~10,000 cells/well in phenol
red-free RPMI-1640 media containing 5% charcoal-dextran-stripped
FBS. Twenty-four hours after plating, the cells were treated with
either vehicle control or various hormones as indicated in each
experiment. For H19 knockdown and overexpression study, the cells
were transfected with siRNA, shRNA or H19 expression vector for 24
h, and then treated with various hormones for 48 h. Cell
proliferation was assayed using CellTiter 96 AQueous One Solution
Cell Proliferation Assay as instructed by the manufacturer
(Promega).
Statistical analysis
Each experiment was performed at least three times,
and the data are presented as mean ± SEM. For parametric data, the
Student’s t-test was used to determine the statistical significance
between two groups, and one-way ANOVA following post-hoc
Student-Newman-Keuls test was used to compare the difference among
multiple groups using the SPSS software. A p-value <0.05 was
considered to indicate a statistically significant result.
Results
Screening lncRNAs associated with
estrogen action in breast cancer cells
To identify lncRNAs that are potentially associated
with ER-positive breast cancer cells and with estrogen action, we
employed the Disease-Related Human lncRNA Profiler kit to screen
the expression levels of 83 disease-related lncRNAs in the breast
cancer cells. As shown in Table I,
out of the 83 lncRNAs screened, 18 had a higher expression and 7
had a lower expression as defined by a log2(n) >2n in
the ER-positive MCF-7 cells compared to the ER-negative MDA-MB-231
cells. One of the highest differentially expressed lncRNAs was H19
lncRNA with ~3,300-fold difference between the MCF-7 and MDA-MB-231
cells, which was confirmed using quantitative RT-PCR analysis
(1.00±0.04 in MDA-MB-231 cells vs. 3,335.48±965.37 in MCF-7 cells).
Moreover, out of the 83 lncRNAs screened, only H19 was upregulated
while 3 lncRNAs were downregulated when MCF-7 cells were treated
with 100 nM E2 for 48 h as shown in Table II.
 | Table IThe differentially expressed lncRNAs
between the MCF-7 and MDA-MB-231 cells. |
Table I
The differentially expressed lncRNAs
between the MCF-7 and MDA-MB-231 cells.
| lncRNA | Fold-difference
(log2(n))
n-value |
|---|
| Upregulated | |
| ANRIL | 13.20 |
| H19 | 10.33 |
| NCRMS | 9.81 |
| HOTAIR | 9.61 |
| HOXA1AS
AA489505 | 9.55 |
| HOTAIRM1 | 9.52 |
| HAR1B | 7.11 |
| LOC285194 | 5.97 |
| PCGEM1 | 5.51 |
| HOXA3AS
BI823151 | 3.68 |
| HOXA6AS
AK092154 | 3.32 |
| PCAT-29 | 3.22 |
| BC017743 | 3.05 |
| ST7OT3 | 2.91 |
| BC200 | 2.80 |
| H19-AS | 2.40 |
| HOXA11AS | 2.19 |
| BC043430 | 2.14 |
| Downregulated | |
| HOTTIP | −2.03 |
| UCA1 | −2.05 |
| DGCR5 | −2.06 |
| AK023948 | −2.15 |
| PCAT-32 | −2.20 |
|
LincRNA-SFMBT2 | −3.24 |
| ZEB2NAT | −4.78 |
 | Table IIThe lncRNAs with a change in
expression in MCF-7 cells following treatment with
17β-estradiol. |
Table II
The lncRNAs with a change in
expression in MCF-7 cells following treatment with
17β-estradiol.
| lncRNA | Fold-difference
(log2(n))
n-value |
|---|
| Upregulated | |
| H19 | 2.89 |
| Downregulated | |
| HOXA1AS
AA489505 | −6.93 |
| PCAT-29 | −3.73 |
| TU_0017629 | −4.60 |
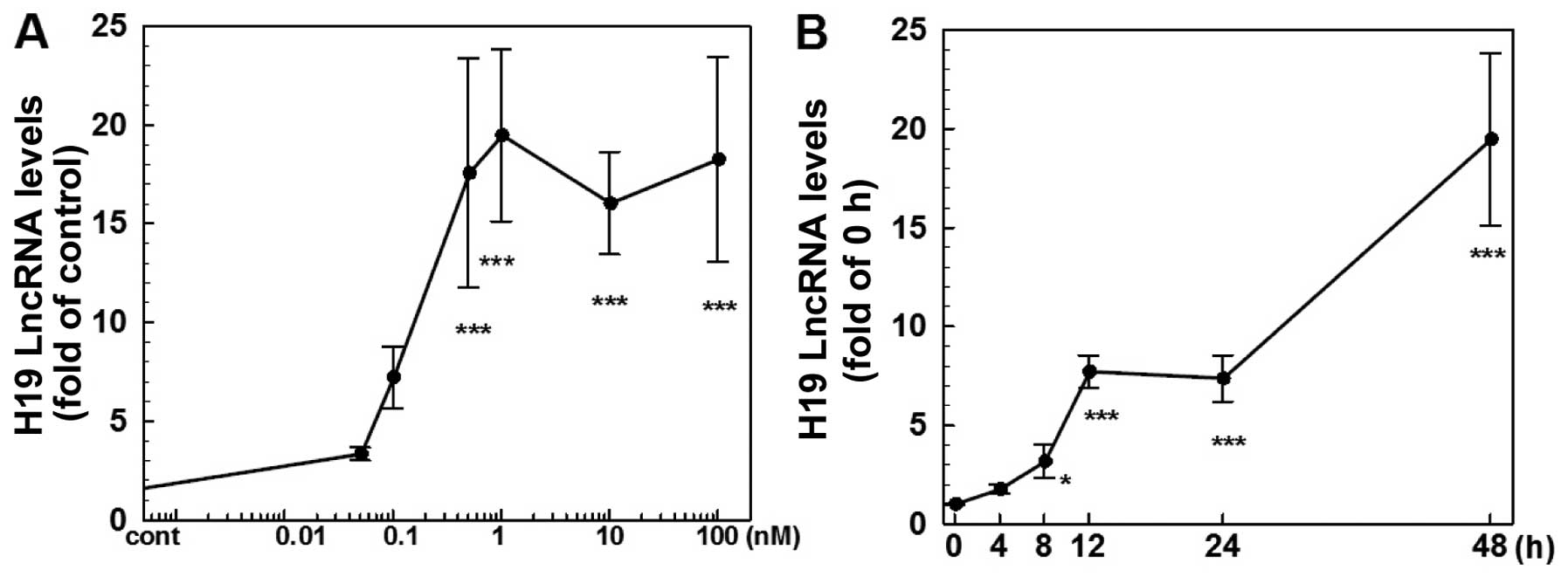
H19 is upregulated by E2 in ER-positive
MCF-7 cells
To further analyze the E2 regulation of H19
expression in breast cancer cells, MCF-7 cells were treated with
various doses of E2 for 4–48 h. As shown in Fig. 1, E2 produced a dose (Fig. 1A) and time-dependent (Fig. 1B) induction of H19 expression. At 48
h of 1 nM E2 treatment, the level of H19 RNA was increased ~19-fold
while there was no significant change observed at 4 h of treatment.
At doses ranging from 0.05 to 1 nM, E2 caused a dose-dependent
induction of H19 expression with a maximal effect observed at 1 nM
(Fig. 1A). On the other hand,
treatment of ER-negative MDA-MB-231 cells with E2 at doses up to
100 nM failed to induce H19 expression (data not shown).
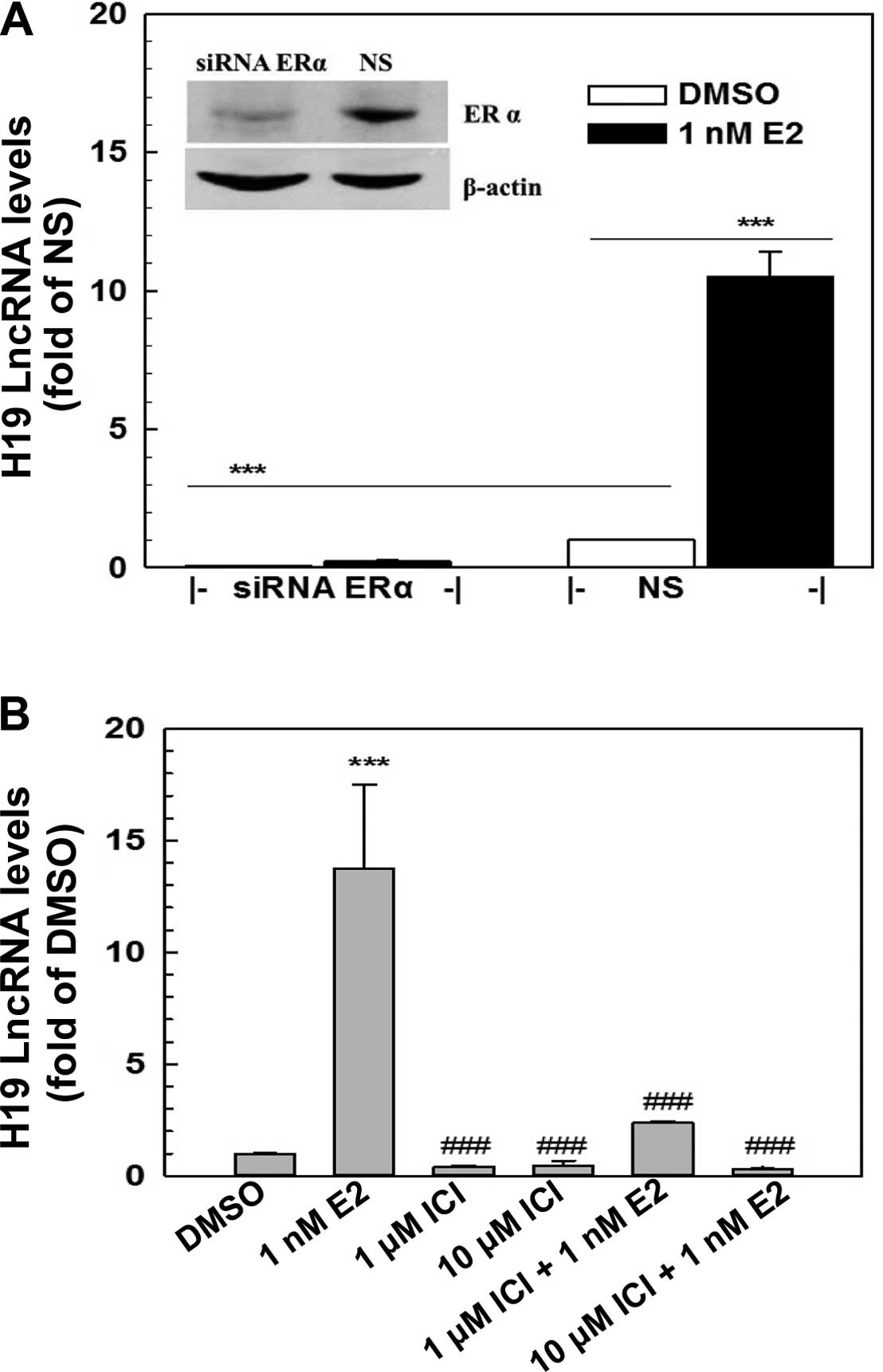
E2-induced H19 expression is mediated
through ERα
The genomic action of estrogens is mainly mediated
through estrogen receptors, ERα and/or ERβ. ERα yet not ERβ, was
expressed in the MCF-7 cells as demonstrated by RT-PCR and western
blot analysis (Fig. 2, and data not
shown). To determine whether E2 induction of H19 expression is
mediated via ERα, a specific ER antagonist, ICI 182780, and a
specific ERα siRNA were employed to block ERα action and to knock
down ERα expression in the MCF-7 cells, respectively. Transfection
of a specific ERα siRNA (100 nM) into MCF-7 cells resulted in a
knockdown of ERα of >70% as demonstrated by western blotting
(Fig. 2A), leading to a complete
elimination of E2-induced H19 expression (Fig. 2A). Furthermore, the E2-induced H19
expression was significantly blocked by ICI 182780, a specific ER
antagonist, in a dose-dependent manner (Fig. 2B). The addition of 10 μM ICI
182780 in the medium completely blocked the E2 induction of H19
expression. These data revealed that the E2 induction of H19
expression in MCF-7 cells was mediated through ERα.
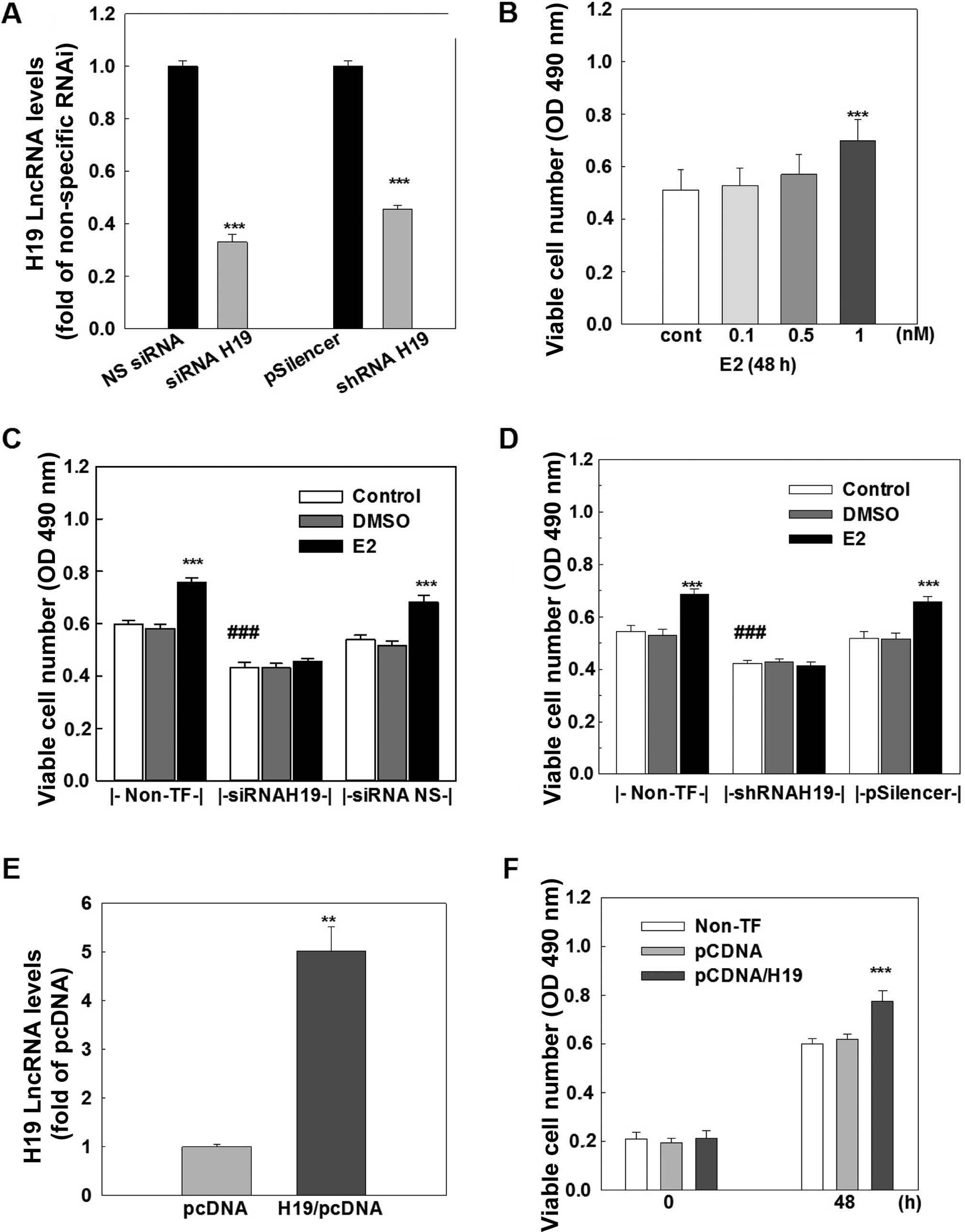
H19 lncRNA promotes cell growth in MCF-7
cells
The functional importance of H19 lncRNA was
investigated by analyzing cell growth and cell cycle distribution
in the MCF-7 cells. Treatment of MCF-7 cells with E2 produced a
dose-dependent increase in viable cell number, and an ~30%
induction of viable cells was observed at 1 nM E2 treatment
(Fig. 3B). This E2-induced cell
growth was completely blocked by transfection of either a specific
siRNA (Fig. 3C) or a specific shRNA
(Fig. 3D), which knocked down H19
lncRNA by 70 and 55%, respectively (Fig. 3A). In contrast, the transfection of
either a non-specific siRNA or a non-specific shRNA that did not
affect H19 expression had no effect on E2-induced cell growth.
Notably, knockdown of H19 lncRNA caused a significant decrease
(~30%) in viable cell number in the MCF-7 cells without E2
treatment (Fig. 3C and D).
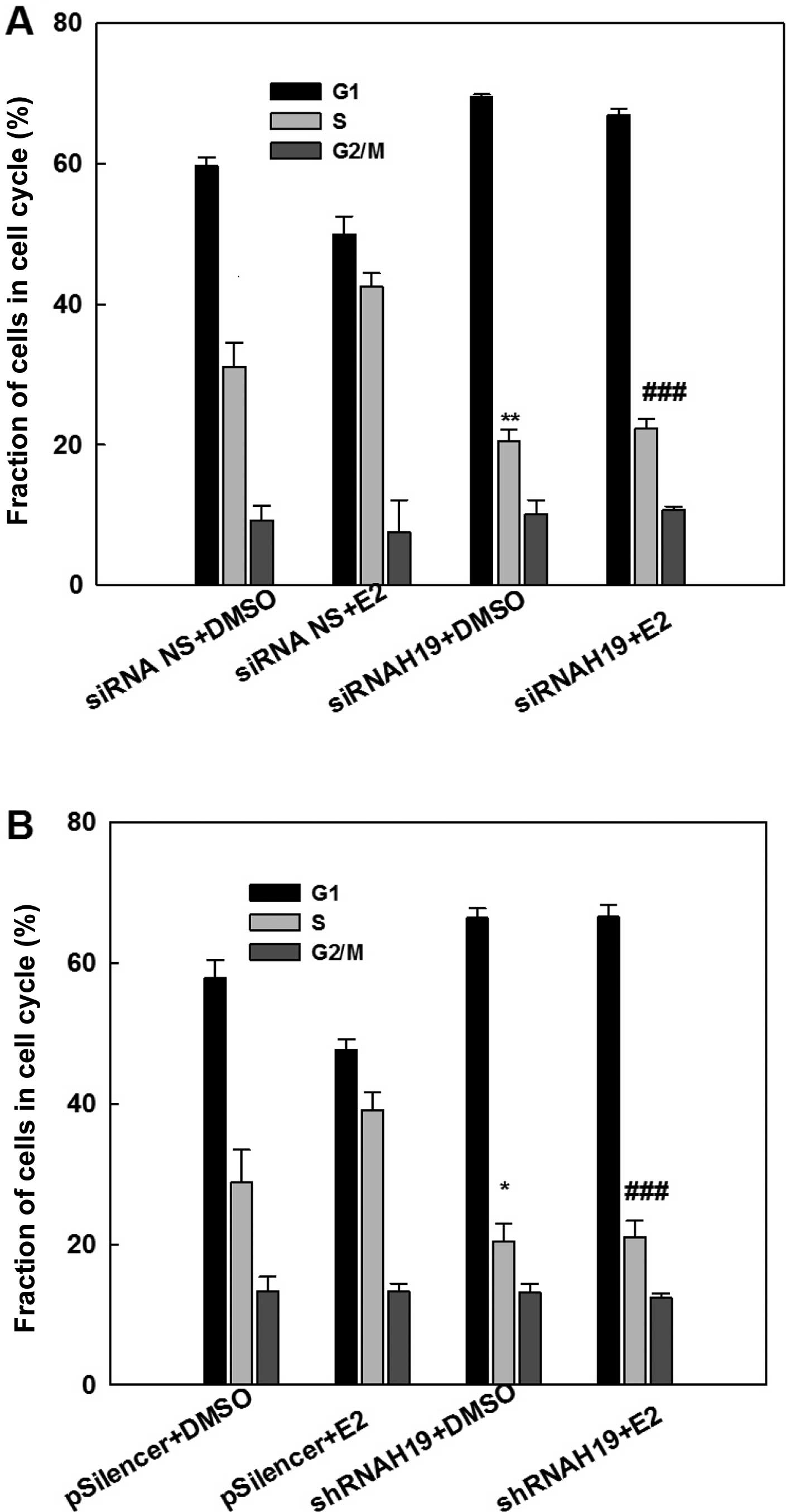
Consistent with the cell growth study, cell cycle
analysis with flow cytometry demonstrated that E2 treatment
increased the fraction of S-phase cells, an indication of an
increase in DNA biosynthesis (Fig.
4). This E2-induced elevation in the S-phase cell fraction was
eliminated by the knockdown of H19 lncRNA using either a specific
siRNA (Fig. 4A) or a specific shRNA
(Fig. 4B). Similarly, knockdown of
H19 lncRNA decreased the S-phase fraction of cells in MCF-7 cells
without E2 treatment. This analysis demonstrated that expression of
H19 is important for the survival and maintenance of MCF-7 breast
cancer cells.
To further determine the role of H19 lncRNA in cell
growth, H19 lncRNA was overexpressed in the MCF-7 cells. The
transfection of an H19 expression vector for 48 h resulted in a
4–5-fold increase in the H19 lncRNA level in the MCF-7 cells
(Fig. 3E) and a significant
increase in cell growth, which was not observed in cells
transfected with a negative control vector (Fig. 3F). Taken together, these data
suggest that H19 lncRNA possesses cell proliferative activity and
the E2-induced cell growth is mediated through upregulation of H19
gene expression in MCF-7 breast cancer cells.
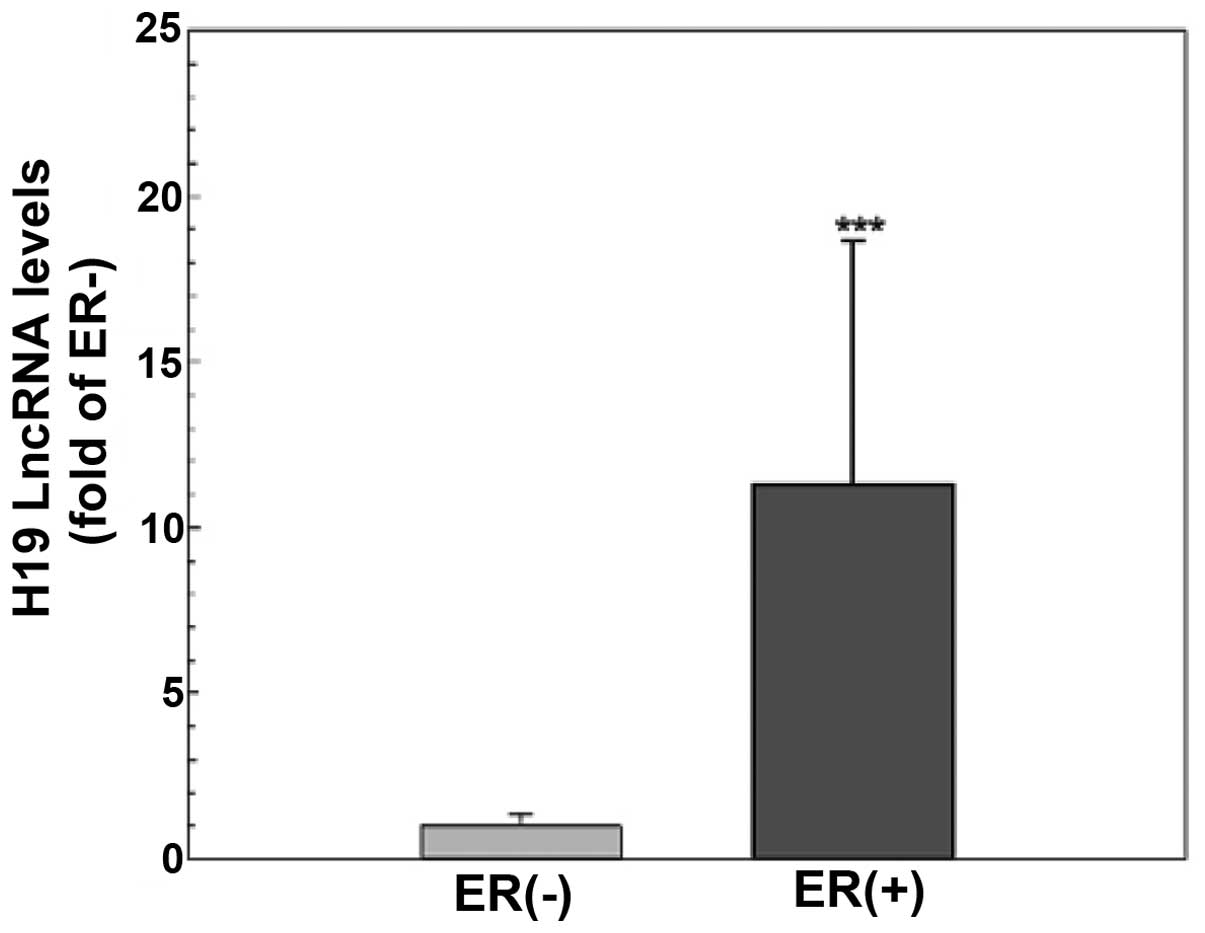
The level of H19 lncRNA is higher in the
ER-positive than that in the ER-negative breast cancer tissues
To explore the functional significance of H19 lncRNA
and the estrogen inducibility of H19 expression in vivo, the
levels of H19 lncRNA in primary tumors from breast cancer patients
(n=45) were evaluated using quantitative RT-PCR. As shown in
Fig. 5, tumor tissues from patients
with ER-positive breast cancer (n=30) had a significantly higher
H19 gene expression compared to those from ER-negative patients
(n=15, p<0.001), suggesting that H19 expression is dependent on
and/or induced by ER action in breast cancer cells.
Discussion
In the present study, we demonstrated that
17β-estradiol mediated through ERα produced a dose- and
time-dependent induction of H19 lncRNA expression in the MCF-7
breast cancer cells. Most notably, knockdown of H19 lncRNA by RNAi
in the MCF-7 cells resulted in a decrease in viable cell number and
a blockade of estrogen-induced cell proliferation (Fig. 3). These results indicate that H19
lncRNA plays a significant role in cell survival and
estrogen-induced cell proliferation in MCF-7 cells.
H19 RNA is originally identified as a transcribed,
spliced and polyadenylated untranslatable RNA molecule (24), now termed as lncRNA. Functional
analysis of H19 lncRNA has linked it to both oncogenic and
tumor-suppressive activities (25).
Although H19 lncRNA has been shown to cause growth retardation and
morphological changes in tumor cells and abrogate the
tumorigenicity of G401 tumor cells in nude mice following ectopic
overexpression (26), growing
evidence suggests that H19 lncRNA possesses oncogenic,
proliferative and anti-apoptotic activity in various in
vitro and in vivo systems (26). Barsyte-Lovejoy et al
(27) reported that H19 lncRNA was
directly induced by the c-Myc oncogene, resulting in an
H19-associated promotion of clonogenicity and anchorage-independent
growth in lung and breast cancer cells. Lottin et al showed
that ectopic overexpression of H19 lncRNA in MDA-MB-231 breast
cancer cells promoted tumor progression in a xenograft animal model
(19). In the present study, we
demonstrated that ectopic overexpression of H19 lncRNA in MCF-7
cells significantly increased cell growth while H19 lncRNA
knockdown resulted in a decrease in cell survival (Figs. 3 and 4), further indicating the significance of
H19 lncRNA in tumor cell proliferation and survival. Moreover, we
observed that knockdown of estrogen-induced H19 lncRNA expression
completely blocked estrogen-induced cell growth in MCF-7 cells
(Figs. 3 and 4), suggesting that H19 lncRNA is a key
factor meditating estrogen-induced cell growth in ER-positive
breast cancer cells. Although the molecular mechanism of how H19
lncRNA stimulates cell proliferation remains to be investigated, a
previous study demonstrated that H19 promoted the cell entry into
S-phase in breast cancer cells through the E2F1 factor (20). Taken together, these data support
the concept that H19 lncRNA plays a critical role in breast cancer
development and progression, and it is therefore a valuable target
for breast cancer therapy.
Previous studies have shown that H19 lncRNA is
overexpressed in breast cancer cells (18,28,29).
To determine whether H19 expression is associated with estrogen
action, we compared the expression levels of 83 lncRNAs including
H19 between an ER-positive and an ER-negative breast cancer cell
line, and evaluated the inducibility of the 83 lncRNAs upon
17β-estradiol treatment in ER-positive MCF-7 cells. We revealed
that the H19 lncRNA level was much higher in the ER-positive MCF-7
cells compared to the ER-negative MDA-MB-231 cells, and
17β-estradiol produced a dose- and time-dependent induction of H19
gene expression in the ER-positive MCF-7 yet not in the ER-negative
MDA-MB-231 cells (Figs. 1 and
2, Tables I and II). The demonstration of estrogen
induction of H19 expression is consistent with a previous report by
Adriaenssens et al, in which an estrogen-associated
induction of H19 expression was observed in mouse mammary gland and
uterus as well as in MCF-7 cells (21). Using both chemical and genetic
approaches, we further elucidated that the estrogen induction of
H19 gene expression was mediated through ERα (Fig. 2). ICI 182780, a specific ER
antagonist, inhibited 17β-estradiol-induced H19 expression in a
dose-dependent manner; and knockdown of ERα by either a specific
siRNA or an shRNA blocked this estrogen action. Furthermore,
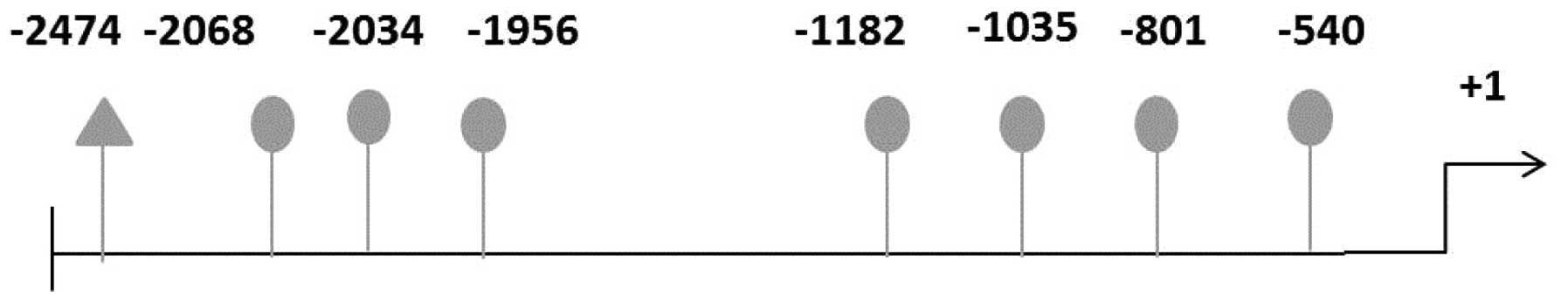
bioinformatic analysis of the H19 promoter region showed that
multiple potential estrogen-response elements (EREs) are present in
the promoter region of the H19 gene (Fig. 6) although the functional
significance of these elements remains to be elucidated
experimentally. Based on current and previous studies, we propose
that similar to the estrogen regulation of protein-encoding genes,
estrogen upregulation of lncRNA H19 expression is mediated through
the traditional genomic pathway via the estrogen-ERα complex
interacting with EREs in the H19 promoter region.
The biological significance of H19 lncRNA is further
illustrated by quantification of H19 lncRNA levels in breast cancer
tissues. Consistent with a previous observation of a steroid
receptor dependency in H19 expression (18) and the screening result of a higher
level of H19 lncRNA in ER-positive MCF-7 cells (Table I), we demonstrated that H19 lncRNA
expression was >10-fold higher in the ER-positive tumor tissues
compared to the ER-negative tissues (Fig. 5). These results not only provide
evidence to support that H19 expression is dependent on estrogen-ER
action, yet also indicate that H19 lncRNA is a valuable target for
the diagnosis and treatment of ER-positive breast cancers.
In summary, lncRNAs have emerged as a critical field
in cancer development and progression, and H19 lncRNA has been
shown to be an oncogenic factor in breast cancer. The present study
provides additional valuable evidence that H19 lncRNA plays a
crucial role in cell survival and proliferation in ER-positive
breast cancer cells. Our results indicate that H19 expression is
ER-dependent and induced by estrogen through the traditional
genomic pathway of estrogen-ER interaction with the gene. Since H19
is an ER-dependent gene, and mediates estrogen-induced cell
proliferation, it may serve as a potential biomarker for breast
cancer diagnosis and progression and as a valuable target for
breast cancer therapy.
Acknowledgments
We are grateful to Dr A. Wakeling (Zeneca
Pharmaceuticals, UK) who provided the ICI 182780. The present study
was supported in part by grants from the National Natural Science
Foundation of China (no. 81403021), the Natural Science Foundation
of Fujian Province (no. 2013J01364), and the Central South
University Special Talents Fund.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999.PubMed/NCBI
|
|
3
|
Stender JD, Frasor J, Komm B, Chang KC,
Kraus WL and Katzenellenbogen BS: Estrogen-regulated gene networks
in human breast cancer cells: Involvement of E2F1 in the regulation
of cell proliferation. Mol Endocrinol. 21:2112–2123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Osborne CK: Steroid hormone receptors in
breast cancer management. Breast Cancer Res Treat. 51:227–238.
1998. View Article : Google Scholar
|
|
5
|
Katzenellenbogen BS, Montano MM, Ediger
TR, et al: Estrogen receptors: Selective ligands, partners, and
distinctive pharmacology. Recent Prog Horm Res. 55:163–195.
2000.PubMed/NCBI
|
|
6
|
Frasor J, Danes JM, Komm B, Chang KC,
Lyttle CR and Katzenellenbogen BS: Profiling of estrogen up- and
down-regulated gene expression in human breast cancer cells:
Insights into gene networks and pathways underlying estrogenic
control of proliferation and cell phenotype. Endocrinology.
144:4562–4574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stossi F, Barnett DH, Frasor J, Komm B,
Lyttle CR and Katzenellenbogen BS: Transcriptional profiling of
estrogen-regulated gene expression via estrogen receptor (ER) alpha
or ERbeta in human osteosarcoma cells: Distinct and common target
genes for these receptors. Endocrinology. 145:3473–3486. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pauli A, Rinn JL and Schier AF: Non-coding
RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136–149.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ariel I, de Groot N and Hochberg A:
Imprinted H19 gene expression in embryogenesis and human cancer:
The oncofetal connection. Am J Med Genet. 91:46–50. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glaser T, Housman D, Lewis WH, Gerhard D
and Jones C: A fine-structure deletion map of human chromosome 11p:
Analysis of J1 series hybrids. Somat Cell Mol Genet. 15:477–501.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Adriaenssens E, Dumont L, Lottin S, Bolle
D, Leprêtre A, Delobelle A, Bouali F, Dugimont T, Coll J and Curgy
JJ: H19 overexpression in breast adenocarcinoma stromal cells is
associated with tumor values and steroid receptor status but
independent of p53 and Ki-67 expression. Am J Pathol.
153:1597–1607. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lottin S, Adriaenssens E, Dupressoir T,
Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ:
Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adriaenssens E, Lottin S, Dugimont T,
Fauquette W, Coll J, Dupouy JP, Boilly B and Curgy JJ: Steroid
hormones modulate H19 gene expression in both mammary gland and
uterus. Oncogene. 18:4460–4473. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsang WP and Kwok TT: Riboregulator H19
induction of MDR1-associated drug resistance in human
hepatocellular carcinoma cells. Oncogene. 26:4877–4881. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan C, Cai LQ, Wu W, Qiao Y,
Imperato-McGinley J, Chen GQ and Zhu YS: NSC606985, a novel
camptothecin analog, induces apoptosis and growth arrest in
prostate tumor cells. Cancer Chemother Pharmacol. 63:303–312. 2009.
View Article : Google Scholar
|
|
24
|
Brannan CI, Dees EC, Ingram RS and
Tilghman SM: The product of the H19 gene may function as an RNA.
Mol Cell Biol. 10:28–36. 1990.PubMed/NCBI
|
|
25
|
Gabory A, Jammes H and Dandolo L: The H19
locus: Role of an imprinted non-coding RNA in growth and
development. BioEssays. 32:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hao Y, Crenshaw T, Moulton T, Newcomb E
and Tycko B: Tumour-suppressor activity of H19 RNA. Nature.
365:764–767. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barsyte-Lovejoy D, Lau SK, Boutros PC,
Khosravi F, Jurisica I, Andrulis IL, Tsao MS and Penn LZ: The c-Myc
oncogene directly induces the H19 noncoding RNA by allele-specific
binding to potentiate tumorigenesis. Cancer Res. 66:5330–5337.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Doucrasy S, Coll J, Barrois M, Joubel A,
Prost S, Dozier C, Stehelin D and Riou G: Expression of the human
fetal bac h19 gene in invasive cancers. Int J Oncol. 2:753–758.
1993.PubMed/NCBI
|
|
29
|
Dugimont T, Curgy JJ, Wernert N, Delobelle
A, Raes MB, Joubel A, Stehelin D and Coll J: The H19 gene is
expressed within both epithelial and stromal components of human
invasive adenocarcinomas. Biol Cell. 85:117–124. 1995. View Article : Google Scholar : PubMed/NCBI
|