Introduction
Breast cancer is a common cancer and the second
leading cause of cancer-related mortality in women. The limits in
identifying patient subsets and the complexity of the disease
presentation lead to major difficulties in current breast cancer
diagnostic and therapeutic strategies (1). Investigations aiming to identify the
genes responsible for promoting the growth and malignancy of breast
cancer may provide insight into the nature of this disease.
The claudin family includes 24 related members which
are integral transmembrane proteins (2). These members are the major components
of the tight junction and are important in various cell activities
through interaction with a variety of proteins in signaling
pathways (3,4). The claudins are often
tissue-specifically expressed (5,6), and a
number of studies have described the abnormal expression of
claudins in various types of cancer, such as breast cancer,
hepatocellular carcinoma, colonic cancer, lung squamous cell and
bladder carcinoma (7–14). Moreover, claudins are considered to
participate in the pathology of these disorders (9–14). For
example, claudin-5 has been reported to be downregulated and
correlated with poor prognosis in patients with hepatocellular
carcinoma (9). Additionally,
claudin-1 is reduced in stage II colonic cancer and may be
associated with recurrence and poor patient survival (14). These results suggest that the
claudin family members are pivotal in tumorigenesis and cancer
progression.
The relationship of TJs dysfunction with disease
development in breast cancer has been shown. Claudin-1 has been
suggested to function as a tumor suppressor and a tumor
enhancer/facilitator in breast cancer (1,15,16).
Osanai et al (17) found
that claudin-6 knockout enhanced the migration and invasion of the
human MCF-7 breast cancer cells. As an important member of the
claudin family, claudin-4 has been investigated in breast cancer
(7,8). In a case-controlled study of breast
cancer, claudin-4 was found to be overexpressed and associated with
high tumor grade and poor prognosis of the disease (18). Moreover, claudin-4 positivity has
been associated with a shorter disease-free survival of patients
with luminal breast cancer (19,20).
Collectively, the abovementioned studies indicate that claudin-4 is
a potential molecular marker for breast cancer. However, the
functional role and regulation of claudin-4 in breast cancer
remains unknown.
In the present study, we aimed to investigate the
biological function and regulation of claudin-4 in breast cancer
cells in in vitro and in vivo experiments.
Materials and methods
Cells and animals
The human MCF-7 breast cancer cells were obtained
from the Shanghai Bioleaf Biotech Co., Ltd. The cells were grown in
RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with
10% fetal bovine serum (FBS; Gibco) and antibiotics at 37°C in the
presence of 95% air and 5% CO2. Female BALB/c nude mice
(4–6 weeks old) were purchased from the Experiment Animal Center of
Guangdong, and maintained in a pathogen-free facility.
Vector constructs
The 659-bp claudin-4 cDNA was amplified by PCR and
cloned into the EcoRI/BamHI sites of the green
fluorescence protein expression vector pEGFP-C1 [Beijing Genomics
Institute (BGI), Beijing, China], resulting in the
claudin-4-expressing construct pEGFP-C1-Cldn4. Short hairpin RNAs
(shRNAs) targeting the open reading frame of human claudin-4 was
cloned into the XhoI/HpaI sites of the green
fluorescence protein expressing vector PLL3.7 (BGI) to generate the
PLL3.7-siCldn4 vector. shRNA was purchased from BGI and the primer
sequences used were: Claudin-4 shRNA sense,
TGTGTACCAACTGCCTGGAGGATGA ATTCAAGAGATTCATCCTCCAGGCAGTTGGTACAC
TTTTTTC and antisense, TCGAGAAAAAAGTGTACCA
ACTGCCTGGAGGATGAATCTCTTGAATTCATCCTCC AGGCAGTTGGTACACA. Scrambled
shRNAs were used to generate the PLL3.7-scramble control vector.
The sequences used were: Scrambled shRNA sense, TGCCCTAGTGTAGAT
GGCTGCAAGAATTCAAGAGATTCTTGCAGCCATCTA CACTAGGGCTTTTTTC and
antisense, TCGAGAAAA AAGCCCTAGTGTAGATGGCTGCAAGAATCTCTTG
AATTCTTGCAGC CATCTACACTAGGGCA.
Generation of stable cell lines with
claudin-4 overexpression or knockdown
Cells were cultured to 80% confluency and
transfection was carried out using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) following the manufacturer’s instructions. To
generate the claudin-4-overexpressing MCF-7 cell clones, the
claudin-4-expressing vector pEGFP-C1-Cldn4 was transfected. The
cell clones with stable claudin-4 overex-pression were selected in
150 µg/ml G418 (MDBio, Qingdao, China) medium and were used
for proliferation, migration and apoptosis assays. MCF-7 cells
transfected with pEGFP-C1 empty vector were considered the
control.
For claudin-4 knockdown, the MCF-7 cells were
transfected with PLL3.7-siCldn4 or control vectors
(PLL3.7-scramble) using Lipofectamine 2000 following the
manufacturer’s instructions. The cell clones with stable knockdown
of claudin-4 were selected in G418 (150 µg/ml) medium and
were used for the in vitro proliferation, migration and
apoptosis assays, as well as in vivo nude mouse
experiment.
RT-PCR
Total RNA was isolated from the cell lines using a
TRIzol reagent kit (Invitrogen) according to the manufacturer’s
instructions. RNA samples were then reverse transcribed into cDNA
using an MML-V reverse transcription kit (Invitrogen) in a total
volume of 20 µl according to the manufacturer’s
instructions. Equal amounts of cDNA samples were used for RT-PCR to
detect the level of claudin-4 expression under the following
conditions: 3 min at 95°C, followed by a total of 40 cycles of two
temperature cycles (15 sec at 90°C and 1 min at 60°C). The primers
and annealing temperatures used for PCR are shown in Table I. GAPDH was used as an internal
control (housekeeping gene). The relative quantification of
claudin-4 was determined using the comparative CT method
(2−ΔΔCt).
 | Table IPrimer sequences and annealing
temperature for real-time PCR. |
Table I
Primer sequences and annealing
temperature for real-time PCR.
| Primer | Primer
sequence | Annealing
temperature (°C) |
|---|
| Claudin-4 | | 60 |
| F |
5′-TTCATCGGCAGCAACATTGTCACC-3′ | |
| R |
5′-AGTCGTACACCTTGCACTGCATCT-3′ | |
| GAPDH | | 60 |
| F |
5′-GAAGGTGAAGGTCGGAGTC-3′ | |
| R |
5′-CAGAGTCCGTGAAGGCAG-3′ | |
| MS-PCR | | |
| Methylated | | 57 |
| M–F |
5′-CTACCGATAAAAACCGTCACG-3′ | |
| M–R |
5′-GTGTATTTTGCGAACGTTAAGTTC-3′ | |
| Unmethylated | | 54 |
| U–F |
5′-AATATTACTACCAATAAAAACCATCACAC-3′ | |
| U–R |
5′-TGTATTTTGTGAATGTTAAGTTTGT-3′ | |
Western blotting
For the western blot analysis, 30 µg protein
lysates from the cells were separated by 10% SDS-PAGE on
Tris-glycine gels (Invitrogen) and transferred to polyvinylidene
difluoride membranes (Millipore Corporation, Bedford, MA, USA). The
membranes were blocked for 1 h at room temperature in TBS (50 mM
Tris, 150 mM NaCl, pH 7.6, 5% fat-free dry milk) and washed in 0.5%
Tween-20 in TBS (TBST), and 2 min at room temperature. Rabbit
anti-claudin-4 antibody (1:750; Invitrogen) was added for and the
cells were incubated overnight at 4°C and then washed in TBST.
β-actin (1:1,000; EarthOx, San Francisco, CA, USA) was used to
normalize protein loading. A secondary antibody (1:100,000;
EarthOx) was added for 1 h at room temperature. The membranes were
washed in TBST and signal detection was carried out using ECL
solution (Amersham, Sweden).
Cell proliferation assay
The proliferative capabilities of cells with
claudin-4 knockdown/overexpression and corresponding controls were
measured using a
3-[4,5-dimethylthiazyol-2yl]-2,5-diphenyltetrazolium bromide (MTT)
assay (5 mg/ml; Sigma, St. Louis, MO, USA). Cells were seeded in
96-well plates at a concentration of 2×103 cells/well
containing 200 µl of RPMI-1640 cell culture medium
supplemented with 10% FBS. Every 24 h of culture in 5 days, 20
µl of MTT solution was added and cells were incubated for 4
h at 37°C. Then 150 µl DMSO was added to each well, and the
cells were incubated for 20 min at 37°C. The optical density (OD)
value of cells was read at 490 nm in a microplate reader (BioTek,
Winooski, VT, USA). Cells in triplicate wells were counted at each
time point, and the experiment was repeated in three independent
experiments.
Cell cycle profile
Cell cycle profiles were analyzed using flow
cytometry after the propidium iodide labeling of DNA, as previously
described (21,22).
Cell migration assay
The migration rate of breast cancer cells was
measured by a wound-healing assay. The cells were grown to
confluence on 60-mm cell culture dishes. A scratch was made through
the cell monolayer using a pipette tip. After washing with
phosphate-buffered saline (PBS), 0.5% FBS maintenance medium was
added. Images of the wounded area were captured immediately after
making the scratch (0 h time-point) and the invasion of cells into
the wounded area was monitored once every 8 h for 48 h using an
inverted microscope (LEICA DFC500, Germany).
Apoptosis assay
MCF-7 cells with claudin-4 overexpression or
knockdown and the corresponding controls were seeded into 60-mm
cell culture dishes at a concentration of 4×105
cells/well. To induce apoptosis, the cells were treated with 0.1
µg/ml chemotherapeutic drug 5-fluoro-2,4 (1 and 3 H)
pyrimidinedione (5-FU) (Sigma) for 24 h. Apoptotic cells were
detected by the PE Annexin V apoptosis detection kit (BD
Biosciences, San Diego, CA, USA). The cells were labeled with
Annexin V-PE and 7-AAD following the manufacturer’s instructions.
The percentage of Annexin V-labeled cells was measured by flow
cytometric analysis using FACSCalibur flow cytometry (BD
Biosciences).
Methylation-specific PCR
The methylation status was detected using
methylation-specific PCR (MSPCR) as previously described (23). The primers and annealing
temperatures used for MSPCR are shown in Table I. Genomic DNA was extracted and
purified from the cell lines using a QIAamp DNA Mini kit (Qiagen,
Germany) according to the manufacturer’s instructions. Bisulfite
modification was performed according to the manufacturer’s
instructions of the EpiTect® Plus DNA Bisulfite kit
(Qiagen). The PCR mixture contained 10X Maxima Hot Start Taq
buffer, 5 mmol/l MgCl2, 2 µl dNTP 10 mmol/l,
0.125 µl Maxima Hot Start Taq DNA Polymerase (all
reagents from Thermo Scientific, UK), 1 µl each of forward
and reverse primers, and 2 µl of DNA brought to a total
volume of 25 µl by the addition of autoclaved deionized
water. PCR reactions were hot started at 95°C for 5 min, followed
by 35 cycles (30 sec at 95°C, 30 sec at the annealing temperature,
30 sec at 72°C) and a final 7-min elongation at 72°C. Each PCR
product was loaded into a 1% agarose gel, stained with ethidium
bromide and visualized under UV illumination.
To assess the effect of DNA methylation, the cell
lines were treated with 10 µmol/l 5-AZA-2V-deoxycytidine
(5-AZA; Sigma), an inhibitor of DNA methylation. Treatment of cell
cultures started at 30–40% confluence for a total of 3 days. The
medium and drug were changed every 24 h. RT-qPCR and western
blotting were then performed to detect the level of claudin-4.
In vivo tumor growth
Animal experiments were carried out according to the
protocol approved by the Animal Studies Committee at Guangdong
Medical College. Six-week-old female BALB/c nude mice were housed
in sterile microisolators, and were randomly divided into two
groups (n=6) each for subcutaneous injection of
2×106/100 µl MCF-7 cells with claudin-4 knockdown
or control MCF-7 cells into the breast fat. After 14 days, the mice
were sacrificed for tumor dissection. The length and width were
measured with metric calipers for tumor volume calculation using
the equation: Volume = le ngth × width × width/2.
Statistical analysis
Data are presented as the mean ± SD. Comparisons for
two groups were performed using a Student’s t-test (GraphPad Prism
5 software). Multiple comparisons were performed by one- or two-way
ANOVA. P<0.05 was considered to indicate a statistically
significant result.
Results
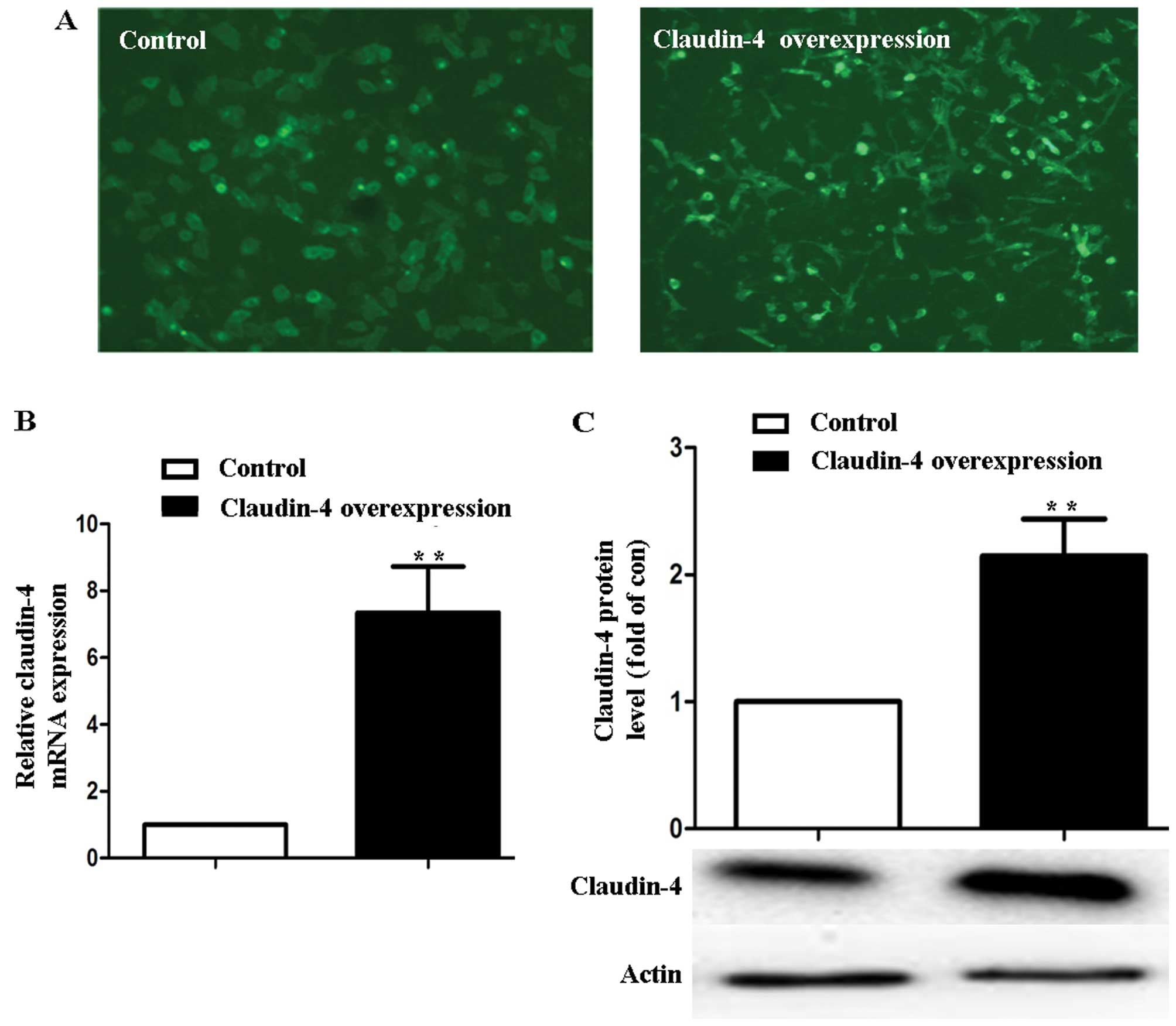
Overexpression of claudin-4 in MCF-7
cells
The MCF-7 cells with pEGFP-C1 and pEGFP-C1-Cldn4
stable transfection were observed by fluorescence microscope
(Fig. 1A). In MCF-7 cells treated
with vector pEGFP-C1-Cldn4 (claudin-4 over-expression), claudin-4
expression increased (7.34±1.38)-fold at the mRNA level and
(2.41±0.69)-fold at the protein level in comparison to the
pEGFP-C1-transfected cells (control) (P<0.01, Fig. 1B and C).
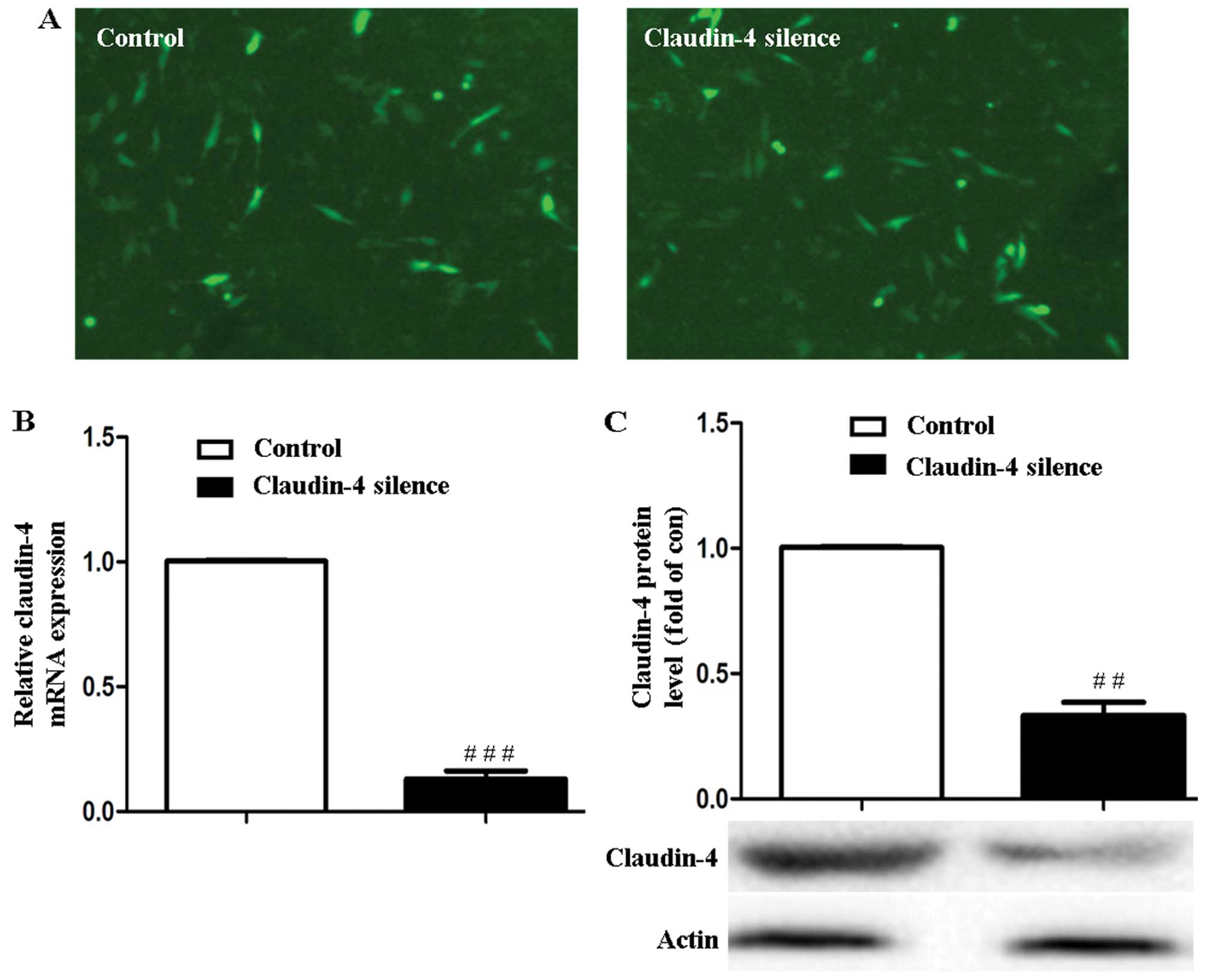
Knockdown of claudin-4 in MCF-7
cells
The green fluorescence of the PLL3.7-scramble and
PLL3.7-siCldn4 stably transfected MCF-7 cells were observed using
fluorescence microscope (Leica, Germany) (Fig. 2A). Claudin-4 mRNA and protein levels
were downregulated to (15.95±1.34) and (32.49±3.19)% in PLL
3.7-siCldn4 stably transfected MCF-7 cells (claudin-4 silencing)
compared to the PLL3.7-scramble-transfected cells (control)
(P<0.001 or P<0.01, Fig. 2B and
C).
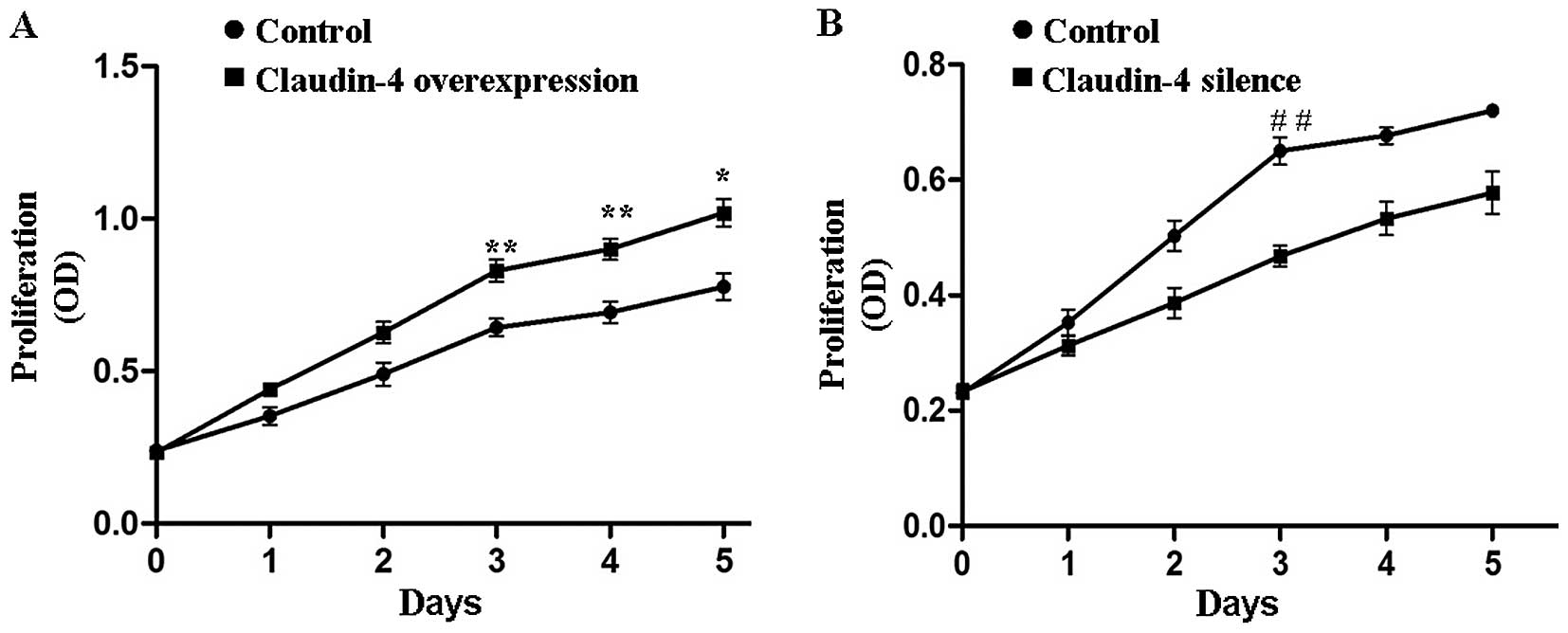
Claudin-4 is involved in MCF-7 cell
proliferation
To determine whether claudin-4 protein influenced
the proliferation of breast cancer cell lines, an MTT assay was
carried out on the claudin-4-overexpressing MCF-7 cells and the
corresponding negative control cells. The proliferation capability
of claudin-4 overexpressed MCF-7 cells was significantly stronger
(claudin-4 overexpression vs. control, P<0.05 or P<0.01,
Fig. 3A). By contrast, when we
knocked down the expression of claudin-4 in MCF-7 cells, the cell
proliferation rate was significantly retarded as compared to the
control cells (claudin-4-silence vs. control, P<0.05 or
P<0.01, Fig. 3B). The results
showed that claudin-4 significantly enhanced proliferation of the
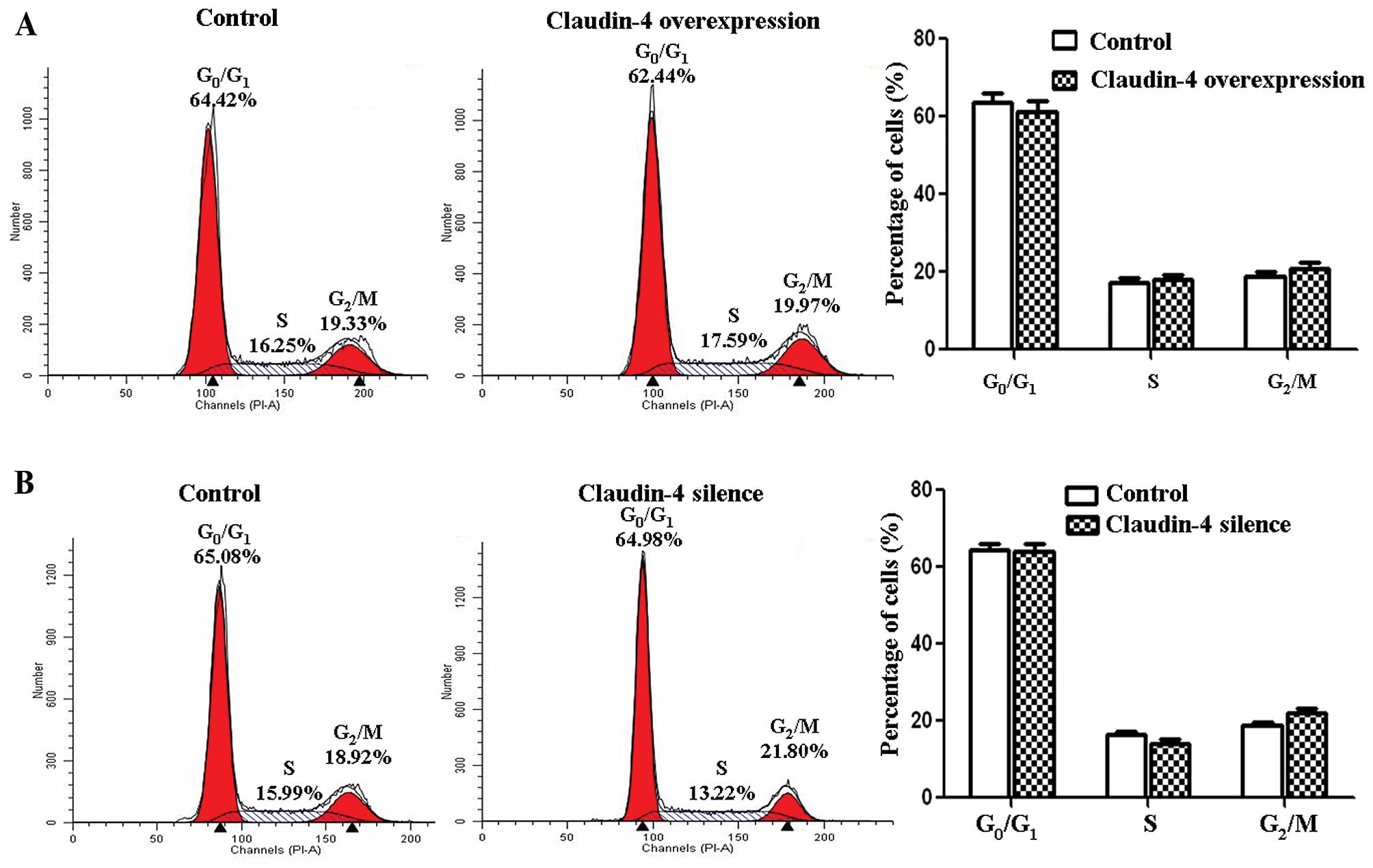
breast cancer cells. However, claudin-4 did not influence the cell
cycle of the breast cancer cells according to our flow cytometric
results (Fig. 4).
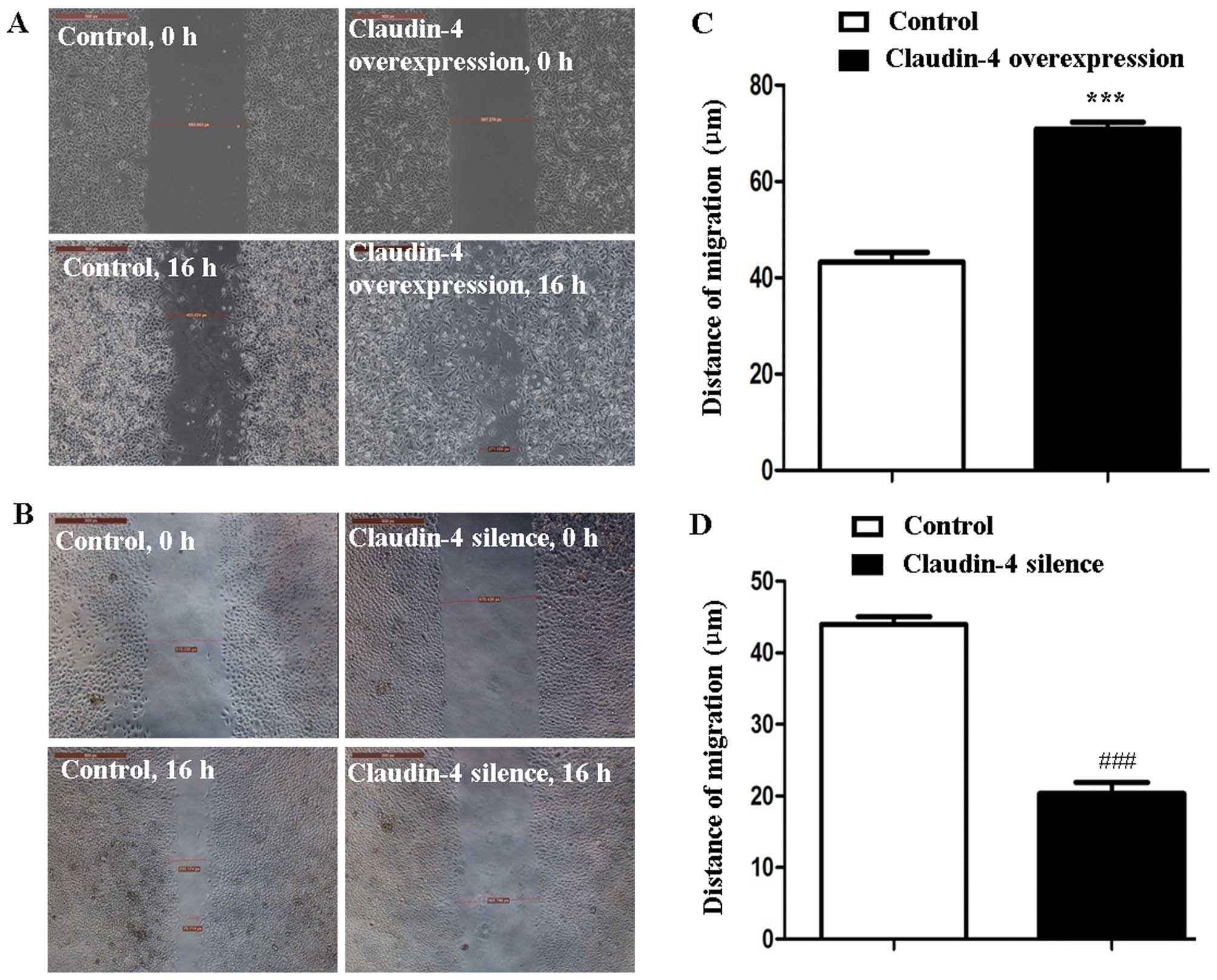
Claudin-4 regulates MCF-7 cell
migration
A scrape injury assay and time-lapse analysis were
carried out to assess the effect of claudin-4 on breast cancer cell
migration. The result revealed a significant increase in the
average migration distance of claudin-4 overexpression MCF-7 cells
compared to the control cells (70.89±2.41 and 43.25±3.39 µm,
claudin-4 over-expression vs. control, P<0.001, Fig. 5A and C). Conversely, the average
migration distance of claudin-4 knockdown MCF-7 cells was
significantly decreased compared to the control cells (20.35±2.70
and 43.98±1.89 µm, claudin-4 silenced vs. control,
P<0.001, Fig. 5B and D).
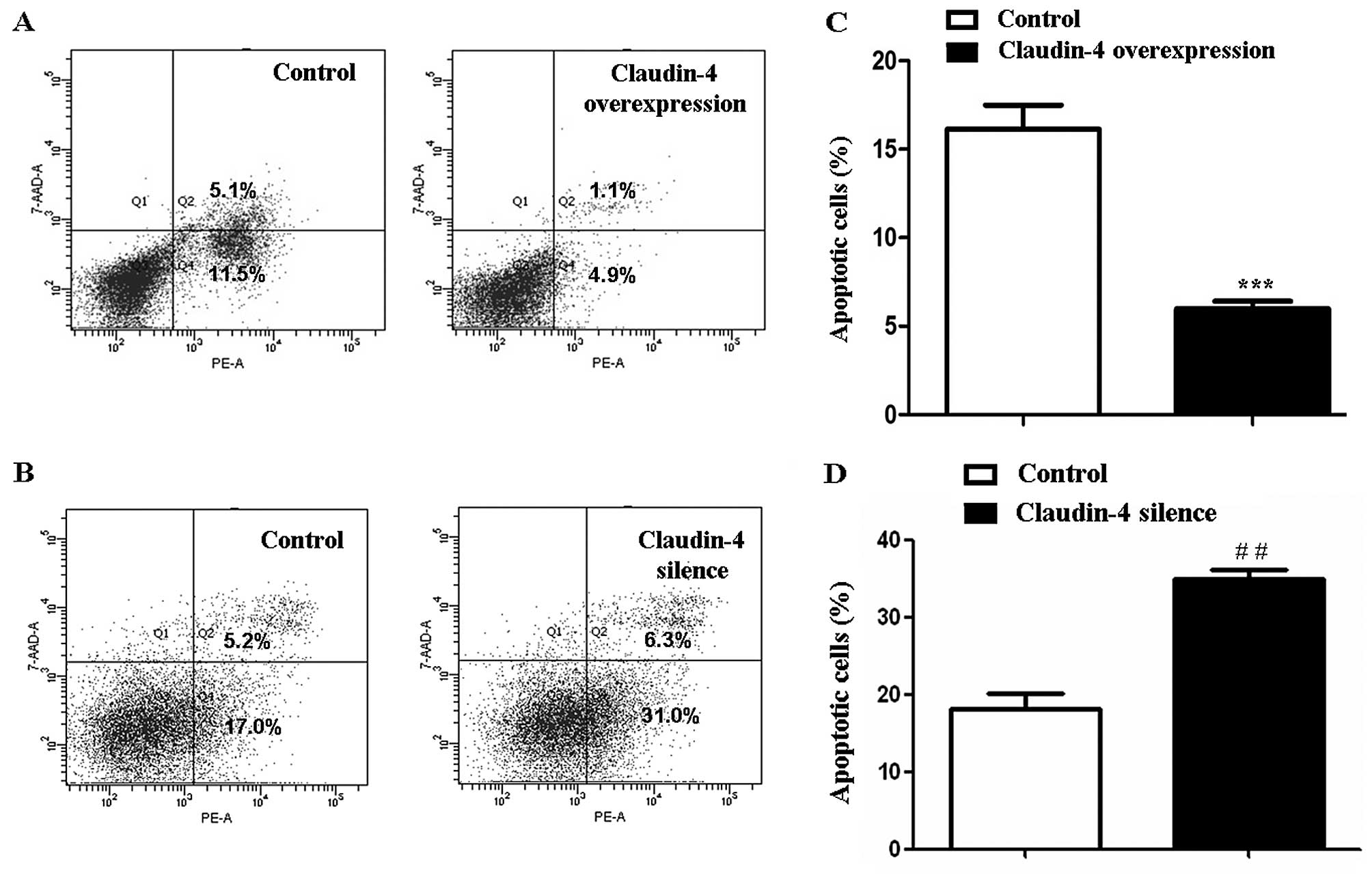
Claudin-4 affects MCF-7 cell
apoptosis
MCF-7 cell apoptosis was assessed using flow
cytometry. 5-FU treatment for 24 h significantly increased the rate
of cell apoptosis in claudin-4 overexpressed/silenced cells and
corresponding control cells. However, 5-FU-induced apoptosis in
claudin-4 overexpressed MCF-7 cells was significantly lower
(6.00±0.72 and 16.60±2.79%, claudin-4 overexpression vs. control,
P<0.001, Fig. 6A and C). By
contrast, 5-FU-induced apoptosis of MCF-7 cells was increased after
claudin-4 knockdown (34.90±2.14 and 18.07±3.65%, claudin-4 silence
vs. control, P<0.01, Fig. 6B and
D). These results showed that claudin-4 inhibited apoptosis of
breast cancer cells.
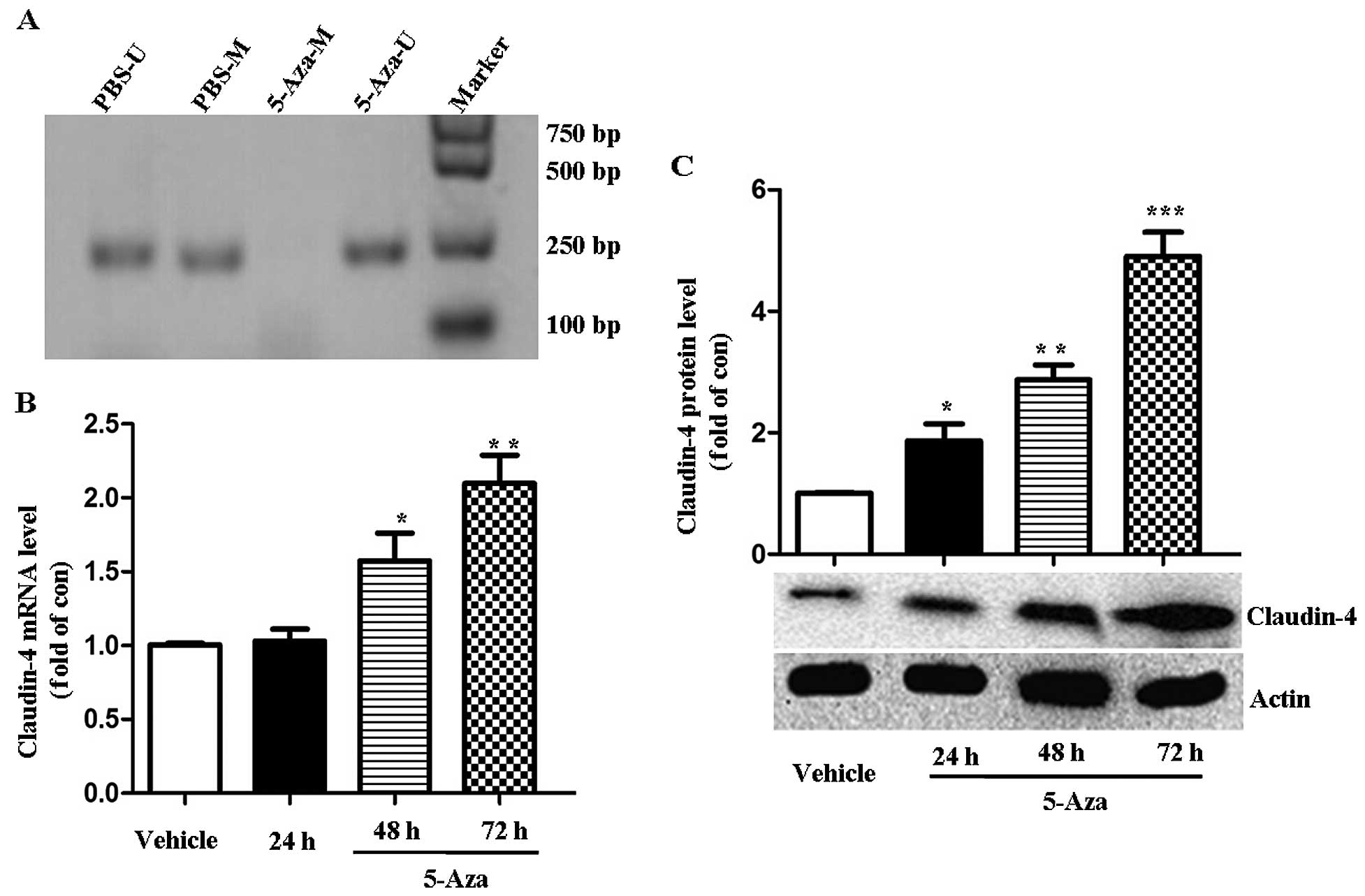
Expression of claudin-4 is regulated by
methylation in MCF-7 cells
The methylation status of claudin-4 was determined
in MCF-7 cells by MSPCR. As shown in Fig. 7A, normal MCF-7 cells exhibited
methylated and unmethylated PCR products. Following treatment with
5-AZA, an inhibitor of DNA methylation for 24 h, an unmethylated
allele only was identified. 5-AZA treatment induced an increased
claudin-4 expression in MCF-7 cells, as demonstrated by RT-PCR and
western blotting. After 48 and 72 h of 5-AZA incubation, the
expression of claudin-4 mRNA increased (1.53±0.36)- and
(2.1±0.33)-fold in comparison to the untreated vehicle cells,
respectively (P<0.05 or 0.01, Fig.
7B). Claudin-4 protein expression significantly increased
(1.85±0.48)-, (2.86±0.18)- and (4.89±0.44)-fold after 24, 48 and 72
h following 5-AZA treatment (P<0.05, 0.01 or 0.001, Fig. 7C). This result indicated that the
expression of claudin-4 expression was regulated by the methylation
status.
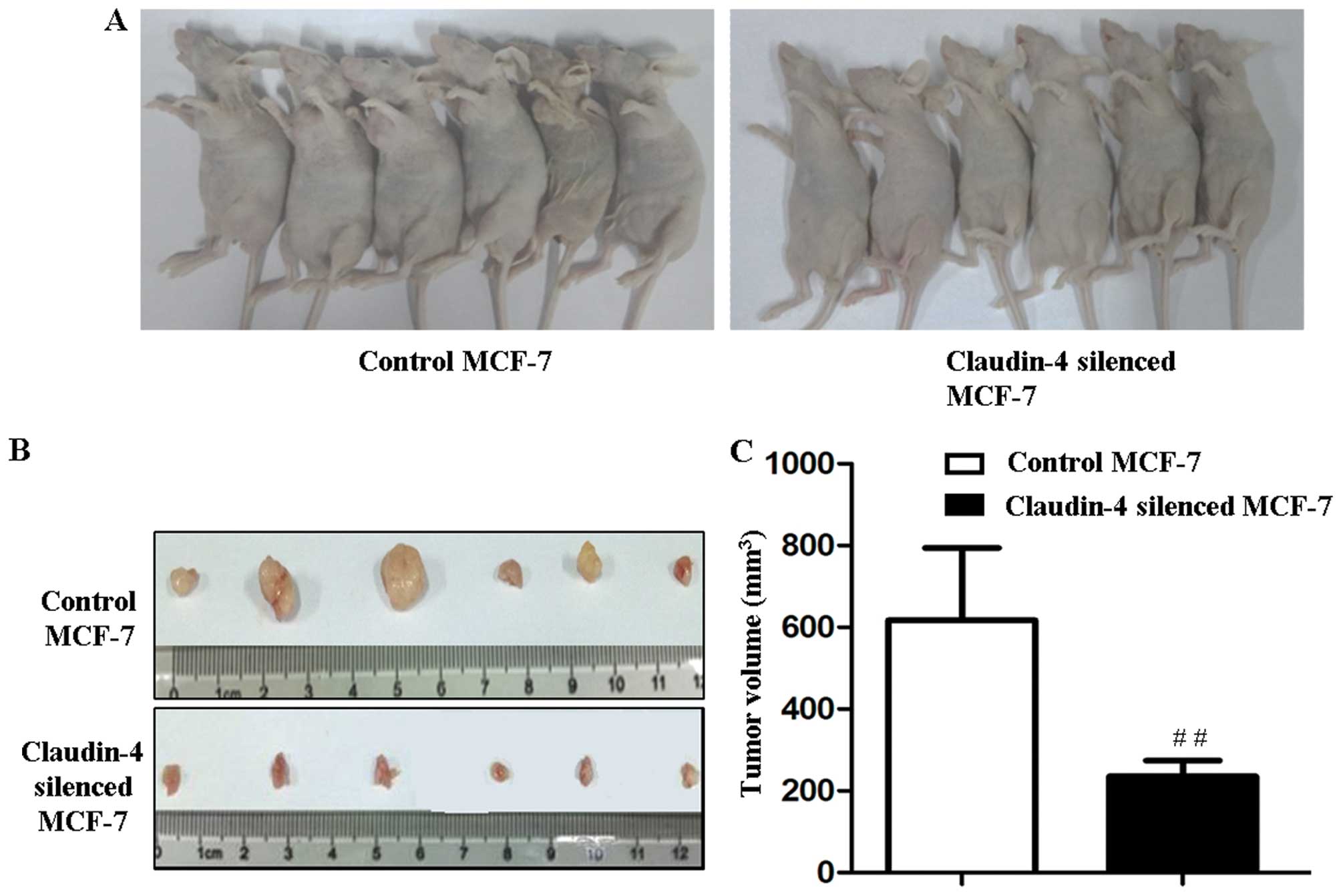
Silencing of claudin-4 inhibits the tumor
formation of MCF-7 cells in vivo
To explore the role of claudin-4 in in vivo
tumor formation, claudin-4-silenced MCF-7 and corresponding control
MCF-7 cells (2×106 cells) were injected subcutaneously
into BALB/c nude mice. Palpable tumors were formed 7 days after
injection in control and claudin-4-silenced MCF-7 groups. On day
14, the tumors derived from claudin-4-silenced MCF-7 were
significantly smaller than those formed in the control cells
(202.2±82.3 and 616.7±177.2 mm3, claudin-4-silenced
MCF-7 vs. control MCF-7, P<0.01, Fig. 8A-C). The results suggested that
knockdown of claudin-4 inhibited the oncogenicity of MCF-7
cells.
Discussion
Tight junctions exist in the junctional complexes of
epithelial and endothelial cells, where they play important roles
in various cell activities such as cell adhesion, permeability
proliferation and differentiation (3,4,24).
Claudins are the major components of tight junctions for backbone
and barrier formation (3). The
abnormality of claudins has been shown to contribute to tumor
development (25). Previous studies
have identified the overexpression of claudin-4 in breast cancer
(18–20), suggesting that claudin-4 be a
biomarker for the detection and diagnosis of breast cancer. In the
present study, to the best of our knowledge we found for the first
time that claudin-4 promoted the proliferation and migration of
breast cancer cells, while inhibiting the apoptosis of these cells
in vitro, resulting in a more aggressive phenotype.
Claudin-4 inhibition repressed the tumorigenesis of MCF-7 breast
cancer cells in nude mice. Moreover, the expression of claudin-4
may be regulated by the methylation status according to our
preliminary study.
A high proliferative potential is one of the most
important characteristics of malignant tumors (26). Members of the claudin family have
been proven to regulate the proliferation of human cancer cells
(27). Based on the findings that
the expression of claudin-4 was increased in breast cancer
(18–20), we hypothesized that claudin-4
overexpression contributes to the proliferative potential of cancer
cells in breast cancer. As expected, we found that cell number
expansion was promoted by claudin-4 in the breast cancer cell
lines. The cell-cycle progression of breast cancer cells was not
affected by the upregulation or downregulation of claudin-4
expression in the present study. Thus, the precise mechanisms by
which claudin-4 regulates cell expansion should be studied. It has
been shown that claudin proteins interact with various proteins
through the PDZ domain-binding motif existing in the COOH-terminal
of claudins (28). These
interactions can serve as adapters for regulatory proteins such as
Rab3b and Rab13, and transcription factors such as ZONAB (29,30),
regulating cell proliferation.
Apoptosis is another essential factor for
maintaining cell/tissue homeostasis. Dysregulation of cell
apoptosis is one of the leading mechanisms in tumor formation
(31). Recent studies have
described the role of claudin proteins in cell apoptosis (32,33).
In breast cancer, the anti-apoptotic effect of claudin-1 has been
identified (34). In the present
study, our results show that claudin-4 may also inhibit the
apoptosis of breast cancer cells.
Most malignant tumors exhibit a highly invasive and
migratory ability, which is closely associated with tumor
metastasis. Previous studies have demonstrated the association
between claudin dysregulation and cancer cell metastasis alteration
(7). Claudin-4 overexpression has
been identified to reduce the invasiveness of pancreatic cancer
cells (35). In breast cancer
cells, the upregulation of claudin-6 expression decreases cell
invasiveness and migration (22).
Claudin-7 downregulation contributes to the increased cellular
discohesion and the ability of cancer cells to disseminate
(21). Those results indicate that
tight junction overexpression decreases cell invasion and motility.
However, our results have shown that claudin-4 increased the
migration of breast cancer cells. This discrepancy may be that
different types of claudins function differently in different
cells/tissues due to tissue-specific molecular mechanisms (7). Our finding is consistent with those of
a previous study showing that knockdown of claudin-4 in ovarian
cancer cell lines results in a decrease in the invasion of these
cells, which is associated with increased matrix
metal-loproteinase-2 activity (36).
Based on our in vitro studies, claudin-4
overexpression promotes the aggressive behavior of human breast
cancer cells, possibly by inhibiting apoptosis and promoting cell
proliferation and migration. To confirm our results, we assessed
the role of claudin-4 in tumorigenesis in nude mice. We found that
claudin-4 silencing inhibited the tumorigenesis of MCF-7 cells,
suggesting that claudin-4 overexpression promotes tumorigenesis in
breast cancer. These results are consistent with our in
vitro findings, and provides the preclinical evidence for
claudin-4 to be a potential candidate for therapeutic target for
breast cancer. It has been demonstrated that immunotoxin-mediated
targeting of claudin-4 inhibited the proliferation of
claudin-4-positive cancers (37).
All of the results suggest that claudin-4 is
important in the development of breast cancer. However, the
mechanisms of claudin-4 regulation in breast cancer remain to be
elucidated. Overexpression of claudin-4 may be mediated through
multiple mechanisms, one of which is gene epigenetic modification
(23). Methylation of 5′-cytosines
in CpG islands is an important epigenetic modulator of gene
expression (38). Alterations in
the methylation status have been proven to be associated with
aberrant gene expression of claudin-4 in pancreatic carcinomas and
ovarian cancer (23,39). In the present study, we found that
claudin-4 expression was increased in MCF-7 breast cancer cells
after unmethylated treatment. The results suggest that the
increased expression of claudin-4 was associated with the
hypomethylated status in breast cancer. As cultured cells may have
an altered methylation pattern compared to their tissue
counterparts (21), breast cancer
tissue samples have to be examined to confirm our results.
In summary, the present study demonstrates a
potential role of claudin-4 in the pathogenesis of breast cancer
via the control of cancer cell proliferation, apoptosis and
migration. In addition, our data reveal that methylation controls
claudin-4 expression in breast cancer. However, further in-depth
investigations on the role and the precise underlying mechanisms of
claudin-4 regulation in breast cancer are necessary.
Acknowledgments
This study was supported by funding from the
Scientific Research Foundation of Guangdong Medical College (no.
XB1225), and the Affiliated Hospital of Guangdong Medical College
(no. BK201210).
References
|
1
|
Myal Y, Leygue E and Blanchard AA: Claudin
1 in breast tumorigenesis: Revelation of a possible novel ‘claudin
high’ subset of breast cancers. J Biomed Biotechnol.
2010:9568972010. View Article : Google Scholar
|
|
2
|
Singh AB, Sharma A and Dhawan P: Claudin
family of proteins and cancer: An overview. J Oncol.
2010:5419572010. View Article : Google Scholar
|
|
3
|
Tsukita S, Furuse M and Itoh M:
Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol.
2:285–293. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matter K and Balda MS: Signalling to and
from tight junctions. Nat Rev Mol Cell Biol. 4:225–236. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Furuse M and Tsukita S: Claudins in
occluding junctions of humans and flies. Trends Cell Biol.
16:181–188. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Van Itallie CM and Anderson JM: Claudins
and epithelial para-cellular transport. Annu Rev Physiol.
68:403–429. 2006. View Article : Google Scholar
|
|
7
|
Morin PJ: Claudin proteins in human
cancer: Promising new targets for diagnosis and therapy. Cancer
Res. 65:9603–9606. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soini Y: Expression of claudins 1, 2, 3,
4, 5 and 7 in various types of tumours. Histopathology. 46:551–560.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sakaguchi T, Suzuki S, Higashi H, Inaba K,
Nakamura S, Baba S, Kato T and Konno H: Expression of tight
junction protein claudin-5 in tumor vessels and sinusoidal
endothelium in patients with hepatocellular carcinoma. J Surg Res.
147:123–131. 2008. View Article : Google Scholar
|
|
10
|
Escudero-Esparza A, Jiang WG and Martin
TA: The Claudin family and its role in cancer and metastasis. Front
Biosci. 16:1069–1083. 2011. View
Article : Google Scholar
|
|
11
|
Boireau S, Buchert M, Samuel MS, Pannequin
J, Ryan JL, Choquet A, Chapuis H, Rebillard X, Avancès C, Ernst M,
et al: DNA-methylation-dependent alterations of claudin-4
expression in human bladder carcinoma. Carcinogenesis. 28:246–258.
2007. View Article : Google Scholar
|
|
12
|
Liu Y, Sun W, Zhang K, Zheng H, Ma Y, Lin
D, Zhang X, Feng L, Lei W, Zhang Z, et al: Identification of genes
differentially expressed in human primary lung squamous cell
carcinoma. Lung Cancer. 56:307–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paschoud S, Bongiovanni M, Pache JC and
Citi S: Claudin-1 and claudin-5 expression patterns differentiate
lung squamous cell carcinomas from adenocarcinomas. Mod Pathol.
20:947–954. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Resnick MB, Konkin T, Routhier J, Sabo E
and Pricolo VE: Claudin-1 is a strong prognostic indicator in stage
II colonic cancer: A tissue microarray study. Mod Pathol.
18:511–518. 2005. View Article : Google Scholar
|
|
15
|
Morohashi S, Kusumi T, Sato F, Odagiri H,
Chiba H, Yoshihara S, Hakamada K, Sasaki M and Kijima H: Decreased
expression of claudin-1 correlates with recurrence status in breast
cancer. Int J Mol Med. 20:139–143. 2007.PubMed/NCBI
|
|
16
|
Blanchard AA, Skliris GP, Watson PH,
Murphy LC, Penner C, Tomes L, Young TL, Leygue E and Myal Y:
Claudins 1, 3, and 4 protein expression in ER negative breast
cancer correlates with markers of the basal phenotype. Virchows
Arch. 454:647–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Osanai M, Murata M, Chiba H, Kojima T and
Sawada N: Epigenetic silencing of claudin-6 promotes
anchorage-independent growth of breast carcinoma cells. Cancer Sci.
98:1557–1562. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lanigan F, McKiernan E, Brennan DJ,
Hegarty S, Millikan RC, McBryan J, Jirstrom K, Landberg G, Martin
F, Duffy MJ, et al: Increased claudin-4 expression is associated
with poor prognosis and high tumour grade in breast cancer. Int J
Cancer. 124:2088–2097. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kolokytha P, Yiannou P, Keramopoulos D,
Kolokythas A, Nonni A, Patsouris E and Pavlakis K: Claudin-3 and
claudin-4: Distinct prognostic significance in triple-negative and
luminal breast cancer. Appl Immunohistochem Mol Morphol.
22:125–131. 2014. View Article : Google Scholar
|
|
20
|
Szasz AM, Nemeth Z, Gyorffy B, Micsinai M,
Krenacs T, Baranyai Z, Harsanyi L, Kiss A, Schaff Z, Tokes AM, et
al: Identification of a claudin-4 and E-cadherin score to predict
prognosis in breast cancer. Cancer Sci. 102:2248–2254. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kominsky SL, Argani P, Korz D, Evron E,
Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP and Sukumar S:
Loss of the tight junction protein claudin-7 correlates with
histological grade in both ductal carcinoma in situ and invasive
ductal carcinoma of the breast. Oncogene. 22:2021–2033. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Q, Liu Y, Ren Y, Xu X, Yu L, Li Y and
Quan C: Tight junction protein, claudin-6, downregulates the
malignant phenotype of breast carcinoma. Eur J Cancer Prev.
19:186–194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Litkouhi B, Kwong J, Lo CM, Smedley JG
III, McClane BA, Aponte M, Gao Z, Sarno JL, Hinners J, Welch WR, et
al: Claudin-4 overexpression in epithelial ovarian cancer is
associated with hypomethylation and is a potential target for
modulation of tight junction barrier function using a C-terminal
fragment of Clostridium perfringens enterotoxin. Neoplasia.
9:304–314. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abuazza G, Becker A, Williams SS,
Chakravarty S, Truong HT, Lin F and Baum M: Claudins 6, 9, and 13
are developmentally expressed renal tight junction proteins. Am J
Physiol Renal Physiol. 291:F1132–F1141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oliveira SS and Morgado-Díaz JA: Claudins:
Multifunctional players in epithelial tight junctions and their
role in cancer. Cell Mol Life Sci. 64:17–28. 2007. View Article : Google Scholar
|
|
26
|
Lazarevich NL and Fleishman DI:
Tissue-specific transcription factors in progression of epithelial
tumors. Biochemistry. 73:573–591. 2008.PubMed/NCBI
|
|
27
|
Zavala-Zendejas VE, Torres-Martinez AC,
Salas-Morales B, Fortoul TI, Montaño LF and Rendon-Huerta EP:
Claudin-6, 7, or 9 overexpression in the human gastric
adenocarcinoma cell line AGS increases its invasiveness, migration,
and proliferation rate. Cancer Invest. 29:1–11. 2011. View Article : Google Scholar
|
|
28
|
Itoh M, Furuse M, Morita K, Kubota K,
Saitou M and Tsukita S: Direct binding of three tight
junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH
termini of claudins. J Cell Biol. 147:1351–1363. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Balda MS, Garrett MD and Matter K: The
ZO-1-associated Y-box factor ZONAB regulates epithelial cell
proliferation and cell density. J Cell Biol. 160:423–432. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamamoto Y, Nishimura N, Morimoto S,
Kitamura H, Manabe S, Kanayama HO, Kagawa S and Sasaki T: Distinct
roles of Rab3B and Rab13 in the polarized transport of apical,
basolateral, and tight junctional membrane proteins to the plasma
membrane. Biochem Biophys Res Commun. 308:270–275. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo Y, Xu X, Liu Z, Zhang T, Zhang X, Wang
L, Wang M, Liu Y, Lu Y, Liu Y, et al: Apoptosis signal-regulating
kinase 1 is associated with the effect of claudin-6 in breast
cancer. Diagn Pathol. 7:1112012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Armstrong SM, Wang C, Tigdi J, Si X,
Dumpit C, Charles S, Gamage A, Moraes TJ and Lee WL: Influenza
infects lung microvascular endothelium leading to microvascular
leak: Role of apoptosis and claudin-5. PLoS One. 7:e473232012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akasaka H, Sato F, Morohashi S, Wu Y, Liu
Y, Kondo J, Odagiri H, Hakamada K and Kijima H: Anti-apoptotic
effect of claudin-1 in tamoxifen-treated human breast cancer MCF-7
cells. BMC Cancer. 10:5482010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Michl P, Barth C, Buchholz M, Lerch MM,
Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Löhr M, et al:
Claudin-4 expression decreases invasiveness and metastatic
potential of pancreatic cancer. Cancer Res. 63:6265–6271.
2003.PubMed/NCBI
|
|
36
|
Agarwal R, D’Souza T and Morin PJ:
Claudin-3 and claudin-4 expression in ovarian epithelial cells
enhances invasion and is associated with increased matrix
metalloproteinase-2 activity. Cancer Res. 65:7378–7385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hashimi SM, Yu S, Alqurashi N, Ipe DS and
Wei MQ: Immunotoxin-mediated targeting of claudin-4 inhibits the
proliferation of cancer cells. Int J Oncol. 42:1911–1918.
2013.PubMed/NCBI
|
|
38
|
Esteller M: Relevance of DNA methylation
in the management of cancer. Lancet Oncol. 4:351–358. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sato N, Maitra A, Fukushima N, van Heek
NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C and Goggins M:
Frequent hypomethylation of multiple genes overexpressed in
pancreatic ductal adenocarcinoma. Cancer Res. 63:4158–4166.
2003.PubMed/NCBI
|