Introduction
Skin squamous cell carcinoma (SSCC) is a malignancy
originating from epithelial keratinocyte, in which predisposing
factors are associated with ultraviolet radiation, exposure to
chemicals, the application of immunosuppressive drugs, trauma, scar
and certain precancerous diseases. At present, the pathogenesis of
the tumor is not entirely clear, but its ability for excessive
proliferation and inhibition of apoptosis has been identified. As
one of the most common skin malignant tumors, its incidence has
been on the increase worldwide (1),
and the age of patients is gradually becoming younger. Surgery is
the most common and effective method of treatment for SSCC.
However, radiotherapy, chemotherapy and scarring subsequent
following surgery are of great concern to most patients (2). Thus, the study of its mechanism,
prevention and treatment has become a hot topic in the field of
dermatology.
Dysregulation of the balance between cell
proliferation and apoptosis is a pro-tumorigenic principle in SSCC.
The NET-1 gene is a new member of the molecules of the tetraspan
superfamily (TM4SF), which is characterized by the existence of
four predicted transmembrane domains that delimit two extracellular
regions of unequal size (3). The
gene is involved in signal transduction, cell adhesion, migration,
proliferation and differentiation (4,5). It
was found to be overexpressed in certain tumors, such as SSCC
(6), gastric cancer (7), cervical carcinoma (8), colorectal adenocarcinoma (9) and ovarian carcinoma (10). Previous findings have shown that the
knockdown of NET-1 by RNA silencing and the antisense technique
inhibit proliferation and infiltration of human skin squamous
carcinoma cells. The NET-1 gene plays a role in the
proliferation of SSCC and is associated with cancer cell motility,
suggesting a function of the gene in the development of skin cancer
(11).
In recent years, it has been found that the
occurrence, development and recurrence of SSCC is closely
associated with cell apoptosis disorder (12). Survivin, a member of the inhibitor
of apoptosis protein (IAP) family, has been shown to inhibit
apoptosis, enhance proliferation and promote angiogenesis. Due to
its upregulation in malignancy and its key role in apoptosis,
proliferation and angiogenesis, survivin is currently attracting
considerable attention as a new target for anticancer therapies
(13). In several animal model
systems, the downregulation of survivin or inactivation of its
function has been shown to inhibit tumor growth. Strategies under
investigation to target survivin include antisense
oligonucleotides, siRNA, ribozymes, immunotherapy and small
molecular weight molecules. Survivin has been suggested as an
attractive target for new anticancer interventions.
RNA interference (RNAi) is the phenomenon in which
siRNA of 21–23 nt in length silences a target gene by binding to
its complementary mRNA and triggering its degradation. The potent
knockdown of specific gene sequences makes siRNA a promising
therapeutic strategy (14,15). Recent findings have shown that RNAi
targeting NET-1, survivin, vascular endothelial growth factor
(VEGF) and Bcl-2 transfected into A431 cells inhibited tumor cell
proliferation, promoted apoptosis, and achieved a good
tumor-suppressor effect in animal experiments.
Since the occurrence and development of cutaneous
squamous cell carcinoma is a multistep complex process involving a
variety of factors, there are some limitations for single oncogene
inhibition. In the present study, the bulk cutaneous squamous cell
carcinoma tissue chip was identified and the proliferation gene
NET-1 or anti-apoptotic gene survivin was found to be important in
the formation and development of cancer. Furthermore, a
one-chain-double-target siRNA was constructed aiming at the NET-1
and survivin genes on the basis of screening effective
single-target NET-1 siRNA and survivin siRNA sequences. The gene
silencing effect of the one-chain-double-target siRNA in cutaneous
squamous cell carcinoma and its effect on tumor proliferation and
apoptosis were measured in in vitro studies. Additionally,
the siRNA-NC group was established in the experiment to exclude the
effect of the transfection reagent on the results, and a control
group was established to demonstrate the effects of siRNA. The
study aimed to investigate the mechanism of action of
one-chain-double-target siRNA, expand RNAi technology, and provide
a foundation for the development of one-chain-double-target siRNA
technology. The results of the present study provided experimental
evidence and technical support for the continuous innovation of the
development of one-chain-multi-target siRNA nucleic acid drugs.
Additionally, the results identified a new approach for the
prevention and treatment of cutaneous squamous cell carcinoma.
Materials and methods
Patients and follow-up
This was a retrospective study based on archived
materials. The study group comprised 100 SSCC patients who were
diagnosed with SSCC and underwent curative resection in the
Affiliated Hospital of Nantong University and the Nantong Cancer
Hospital (Jiangsu, China) between January, 2007 and December, 2011.
The patients were selected according to the following criteria: ii)
primary SSCC; and ii) previously untreated with surgery as the
first treatment. Therefore, analysis of the data in this series
reflects the actual impact of the tumor biology on the clinical
outcome.
The study was approved by the Ethics Committee of
the Affiliated Hospital of Nantong University, and all the patients
provided written informed consent.
Tissue microarray (TMA) construction
A representative section of the SSCC specimens used
for creating tissue microarray were selected by two experienced
pathologists, using hematoxylin (Invitrogen-Life Technologies,
Carlsbad, CA, USA) and formalin-fixed and paraffin-embedded
eosin-stained sections. Detailed clinical and pathological
information of SSCC patients is provided in Table I. We used 1.0-mm core tissue
biopsies and took tissues from paraffin-embedded tissue blocks to
new recipient blocks (contained 100 samples). The recipient blocks
were cut and placed on slides.
 | Table IThe relationship between the
expression of NET-1 and survivin and clinicopathological parameters
in 100 cases of SSCC. |
Table I
The relationship between the
expression of NET-1 and survivin and clinicopathological parameters
in 100 cases of SSCC.
| Characteristics | NET-1
| P-valuea | Survivin
| P-valuea |
|---|
| No. | (−) | (+) | (++) | (+++) | (−) | (+) | (++) | (+++) |
|---|
| Age (years) |
| ≤63 | 45 | 2 | 7 | 16 | 20 | 0.762 | 3 | 6 | 16 | 20 | 0.811 |
| >63 | 55 | 2 | 9 | 19 | 25 | | 2 | 8 | 21 | 24 | |
| Diseased
region |
| Head and face | 74 | 3 | 10 | 26 | 35 | 0.619 | 2 | 8 | 31 | 33 | 0.735 |
| Body | 5 | 0 | 1 | 2 | 2 | | 1 | 1 | 1 | 2 | |
| Arms and legs | 7 | 0 | 2 | 3 | 2 | | 1 | 1 | 2 | 3 | |
| Externalia | 14 | 1 | 3 | 4 | 6 | | 1 | 4 | 3 | 6 | |
| Tumor thickness
(mm) |
| ≤5 | 87 | 4 | 16 | 33 | 34 | 0.004 | 5 | 14 | 36 | 32 | 0.002 |
| >5 | 13 | 0 | 0 | 2 | 11 | | 0 | 0 | 1 | 12 | |
| TNM staging |
| I | 42 | 3 | 12 | 12 | 15 | 0.021 | 4 | 11 | 16 | 11 | 0.006 |
| II | 51 | 1 | 4 | 22 | 24 | | 1 | 3 | 21 | 26 | |
| III | 5 | 0 | 0 | 1 | 4 | | 0 | 0 | 0 | 5 | |
| IV | 2 | 0 | 0 | 0 | 2 | | 0 | 0 | 0 | 2 | |
| Pattern of
organization |
| Classic SSCC | 55 | 3 | 6 | 18 | 28 | 0.032 | 4 | 5 | 18 | 28 | 0.039 |
| Acantholysis | 20 | 0 | 3 | 11 | 6 | | 0 | 1 | 13 | 6 | |
| Spindle cell | 14 | 0 | 0 | 3 | 11 | | 0 | 0 | 4 | 10 | |
| Verrucous
SSCC | 11 | 1 | 7 | 3 | 0 | | 1 | 8 | 2 | 0 | |
| Lymph node
metastasis |
| Present | 4 | 0 | 0 | 1 | 3 | 0.047 | 0 | 0 | 0 | 4 | 0.001 |
| Absent | 96 | 4 | 16 | 34 | 42 | | 5 | 13 | 36 | 40 | |
Immunohistochemical (IHC) staining and
scoring system
After deparaffinizing and rehydrating, the slides
were treated with 3% H2O2 solution for 15 min
at room temperature to block endogenous peroxidase. The slides were
then soaked in sodium citrate buffer (10 mM sodium citrate, 0.05%
Tween-20, pH 6.0) at 96°C for 5 min for antigen retrieval. After
blocking using BSA (Sigma, St. Louis, MO, USA), the following
antibodies were used: mouse monoclonal antibody for NET-1 (1:100),
mouse polyclonal antibody for survivin (1:100) (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Antibodies were added to
the slides, kept overnight at 4°C and then incubated at room
temperature with biotinylated secondary antibody (Santa Cruz
Biotechnology, Inc.) for 30 min, and then HRP-streptavidin for 15
min. After 3,3′-diaminobenzidine (DAB) (Sigma) staining, the
results were graded for intensity (1, pale yellow; 2, yellow and 3,
brown) and the percentage of positive cells [0 (≤5%), 1 (6–25%), 2
(26–50%), 3 (51–75%) and 4 (>75%)] with discrepancies was
resolved by consensus. The grades were multiplied to determine a
score. The scores of tumors were defined as: 0 (−), 1–3 (+), 4–5
(++), >5 (+++).
Cell culture
A431 cells were grown in Dulbecco’s modified Eagle’s
medium (DMEM; Invitrogen-Life Technologies) supplemented with 10%
fetal bovine serum (FBS; Gibco-Life Technologies, Carlsbad, CA,
USA), 2 mM L-glutamine, 100 U/ml of penicillin and 100 µg/ml
of streptomycin. The cell lines were purchased from the Institute
of Cell Biology, Chinese Academy of Sciences. The cell cultures
were maintained at 37°C in a humidified incubator (Fuyilian, China)
with 5% CO2.
siRNA, shRNA expression vector design,
construct and shRNA expression vector stable transfection
On the basis of an optimization principle of siRNA,
a sequence-specific siRNA targeting NET-1 (NET-1 siRNA) or survivin
(survivin siRNA) was designed, and a dual gene targeting siRNA
(one-chain-double-target siRNA) for NET-1 and survivin was assessed
in vitro. Chemically produced oligonucleotides were obtained
from Biomics Biotechnology Corp. (Nantong, China), and identified
using PCR and DNA sequencing. Sequences of all the siRNAs and shRNA
are shown in Table II.
 | Table IIsiRNA sequences. |
Table II
siRNA sequences.
| Gene name | Strand | Sequence
(5′-3′) |
|---|
| siRNA-NET-1 | Positive-sense |
5′-UGUGGUCUUUGCUCUUGGUUUCCdTdT-3′ |
| Antisense |
5′-ACACCAGAAACGAGAACCAAAGGdTdT-3′ |
| siRNA-survivin | Positive-sense |
5′-UCUUUGUGACCCGGUUCAGdTdT-3′ |
| Antisense |
5′-AGAAACACUGGGCCAAGUCdTdT-3′ |
| siRNA-NET-1 and
survivin | Positive-sense |
5′-UGUGGUCUUUGCUCUUGGUUUCCUCUUUGUGACCCGGUUCAGdTdT-3′ |
| Antisense |
5′-ACACCAGAAACGAGAACCAAAGGdTdT-3′ |
|
5′-AGAAACACUGGGCCAAGUCdTdT-3′ |
| siRNA-NC | Positive-sense |
5′-GAGTGATTGGAGGTTGGGGAC-3′ |
| Antisense |
5′-CTCACTAACCTCCAACCCCCTG-3′ |
Briefly, A431 1.5×105 cells/ml (Bogoo,
China) were seeded in multi-well plates (Corning Costar Corp.,
Cambridge, MA, USA) and were grown overnight until they reached
70–80% confluence. The abovementioned siRNA was transfected into
A431 cells with Lipofectamine™ 2000 reagent (Invitrogen-Life
Technologies) according to the manufacturer’s instructions. A431
cells were transfected with siRNA-survivin, siRNA-NET-1,
siRNA-NET-1 and survivin or siRNA-NC, while the negative control
was cultured using DMEM with 10% FBS.
Immunoprecipitation assay
A431 cells in the logarithmic growth phase were
collected with trypsin and lysed in a pre-chilled RIPA buffer (1
ml/107 cells; Beyotime, Jiangsu, China) for 1 h
agitation at 4°C. Protein G agarose beads (Sigma) were prepared by
washing twice with phosphate-buffered saline (PBS) (KeyGen Biotech,
Jiangsu, China) and restoring to a 50% slurry bead suspension with
a RIPA buffer. Prior to immunoprecipitation, the cell lysate was
pre-cleared by adding 50 µl of bead slurry/ml, incubated at
4°C, agitated for 10 min and centrifuged for 10 min at 10,000 × g
at 4°C. The supernatant was transfered to a new tube. Mouse
anti-human NET-1 antibody (4 µg) was then added to 0.5 ml
pre-cleared cell lysate followed by incubation with agitation for 3
h on ice. Normal rabbit IgG and no IgG were used as controls. Fifty
microliters of 50% slurry beads were added and rocked for 1 h at
4°C. After the samples were centrifuged at 10,000 × g for 15 sec,
the beads were washed twice with 1 ml RIPA buffer and then three
times with 1 ml PBS to remove detergents. The beads were then
resuspended in a 60 µl sample buffer, and boiled at 95°C for
5 min. Western blotting was performed with mouse anti-human
survivin and mouse anti-human NET-1 antibodies to examine whether
survivin was combined with beads and NET-1 was precipitated with
survivin, respectively.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from A431 cells using TRIzol
reagent (Invitrogen-Life Technologies), and then submitted to a 25
µl PCR reaction in the presence of 12.5 µl of 2X
Master Mix, 1 µl of each Primer Mix (10 µM/ml), 0.5
µl of 50X SYBR-Green I and 4 µl RNA according to the
One-Step kit (Quantace, Australia). The PCR mixtures were first
subjected to 30 min at 42°C for reverse transcription and initially
denatured for 10 min at 95°C and then to 40 cycles of amplification
with the following cycling parameters: 20 sec at 95°C, 30 sec at
55°C, and 30 sec at 72°C. The primer pairs for each gene were
designed with Primer Premier 5.0 software.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an
internal control for PCR. Primer sequences are provided in Table III.
 | Table IIIOligonucleotide primers used for
RT-qPCR analyses. |
Table III
Oligonucleotide primers used for
RT-qPCR analyses.
| Gene | Primer sequence
(strand) | Product size
(bp) |
|---|
| NET-1 |
5′-GTGGCTTCACCAACTATACG-3′ (+) | 191 |
|
5′-GACTGCATTAGTTCGGATGT-3′ (−) |
| Survivin |
5′-ACCGCATCTCTACATTCAAG-3′ (+) | 113 |
|
5′-CAAGTCTGGCTCGTTCTC-3′ (−) |
| VEGF |
5′-GACATCTTCCAGGAGTACC-3′ (+) | 197 |
|
5′-TGCTGTAGGAAGCTCATATCTC-3′ (−) |
| Cortactin |
5′-AGCCGTCGCCCTGTACGACT-3′ (+) | 127 |
|
5′-GTACCGGCCCTTGCACACCC-3′ (−) |
| Bcl-2 |
5′-GGCTGGGATGCCTTTGTG-3′ (+) | 64 |
|
5′-GCCAGGAGAAATCAAACAGAGG-3′ (−) |
| Caspase-3 |
5′-AGAACTGGACTGTGGCATTGAG-3′ (+) | 191 |
|
5′-GCTTGTCGGCATACTGTTTCAG-3′ (−) |
| Caspase-8 |
5′-CGCAAAGGAAGCAAGAAC-3′ (+) | 361 |
|
5′-TTGAGCCCTGCCTGGTGT-3′ (−) |
| GAPDH |
5′-TGCACCACCAACTGCTTAGC-3′ (+) | 87 |
|
5′-GGCATGGACTGTGGTCATGAG-3′ (−) |
Western blot analysis
After siRNA transfection for 48 h, A431 cells in
6-well plates (Corning Costar Corp.) were lysed in RIPA buffer
(Beyotime Institute of Biotechnology, China). The amount of total
cell proteins was determined by BCA kit (Beyotime). Protein (25
µg) was separated by sodium dodecyl sulfate-polyacrylamide
(SDS-page) separation gel and elec-troblotted onto PVDF membranes
(Pharrnacai, Covington, KY, USA). For A431 proteins, the membrane
was blocked with 5% (wt/vol) milk and probed with a
mouse-anti-NET-1 monoclonal antibody (1:500 dilution) and a
mouse-anti-survivin polyclonal antibody (1:1,000 dilution), (Santa
Cruz Biotechnology, Inc.) at 4°C overnight. The membrane was washed
with Tris-buffered saline with Tween 20 (TBST; KeyGen Biotech),
incubated with DyLight 800-labeled antibody to mouse IgG (1:5,000)
for 2 h, and the membrane was scanned by the Bio-Rad imaging system
for semi-quantitative analysis. An enhanced chemiluminescence (ECL)
reagent was used to incubate the immune complex and signals were
collected by ChemiDoc XRS (Bio-Rad, Hercules, CA, USA). Developed
membranes were semi-quantitatively analyzed by scanning volume
density using an ImageJ densitometer (National Institutes of
Health). Results were expressed as optical volume density corrected
by β-actin for loading. The size of target protein was measured by
a comparison with protein molecular weight markers (Bio-Rad
Laboratories, Ltd.).
Cell proliferation assay
Proliferation of the A431 cell line was measured
using a CCK-8 detection kit (Dojindo, Japan). CCK-8 was applied at
10 µl/well on the 1–3 days after transfection followed by 2
h incubation at 37°C. The absorbance at 450 nm was determined by a
microplate reader, model 680 (Bio-Rad). The samples were assessed
in triplicate and differences among the controls and test groups
were analyzed.
Annexin V-FITC apoptosis assay and flow
cytometry
Briefly, after incubation for 48 h following
transfection, the cells were collected and washed with PBS. The
extent of apoptosis was measured by an Annexin V-FITC apoptosis
detection kit (Beyotime Institute of Biotechnology) according to
the manufacturer’s instructions. The cells were washed with PBS
twice, gently resuspended in an Annexin V binding buffer and
incubated with Annexin V-FITC/PI in the dark for 15 min and
analyzed by flow cytometry using CellQuest software (BD
Biosciences, San Jose, CA, USA). The fraction of the cell
population in different quadrants was analyzed using quadrant
statistics.
Immunohistochemical staining
After incubation for 48 h following transfection,
the A431 cells were incubated with primary antibodies [mouse
monoclonal antibody for NET-1 (1:100) and mouse polyclonal antibody
for survivin (1:100) (Santa Cruz Biotechnology, Inc.) for 30 min
RT] at 4°C overnight followed by the standard
avidin-biotin-peroxidase complex technique. Staining was visualized
using a DAB+ substrate chromogen solution and
hematoxylin QS counter-stain. Images were captured as five fields
of view.
Immunofluorescence assay
The expression of NET-1 and survivin was determined
by immunofluorescence using specific antibodies (Cell Signaling
Technology, Beverly, MA, USA). Briefly, A431 cells were grown into
60–80% confluence on coverslips in 6-well plates. The cells were
then fixed with 4% paraformaldehyde (KeyGen Biotech) for 30 min.
The fixed cells were permeabilized in 1% BSA-supplemented PBS
containing 0.5% Triton X-100 (Sigma) for 30 min, washed and
incubated overnight at 4°C with mouse-anti-NET-1 monoclonal
antibody (1:200 dilution) and mouse-anti-survivin polyclonal
antibody (1:200 dilution) (Santa Cruz Biotechnology, Inc.) as the
primary antibodies. The cells were then washed extensively and
incubated for 1 h at room temperature with secondary antibody. A
goat anti-rabbit IgG-conjugated with FITC was used as the secondary
antibody at a dilution of 1:100. The samples were counterstained
with Hoechst 33258 and photographed using a confocal microscope
(BX51; Olympus, Tokyo, Japan).
Statistical analysis
Experiments were performed independently at least
three times. One-way ANOVA and t-test analyses were utilized to
identify differences between groups. Statistical analysis was
performed with SPSS software 16.0. P<0.05 was considered to
indicate a statistically significant result.
Results
NET-1 and survivin expression in
SSCC
Preparation of tissue microarray
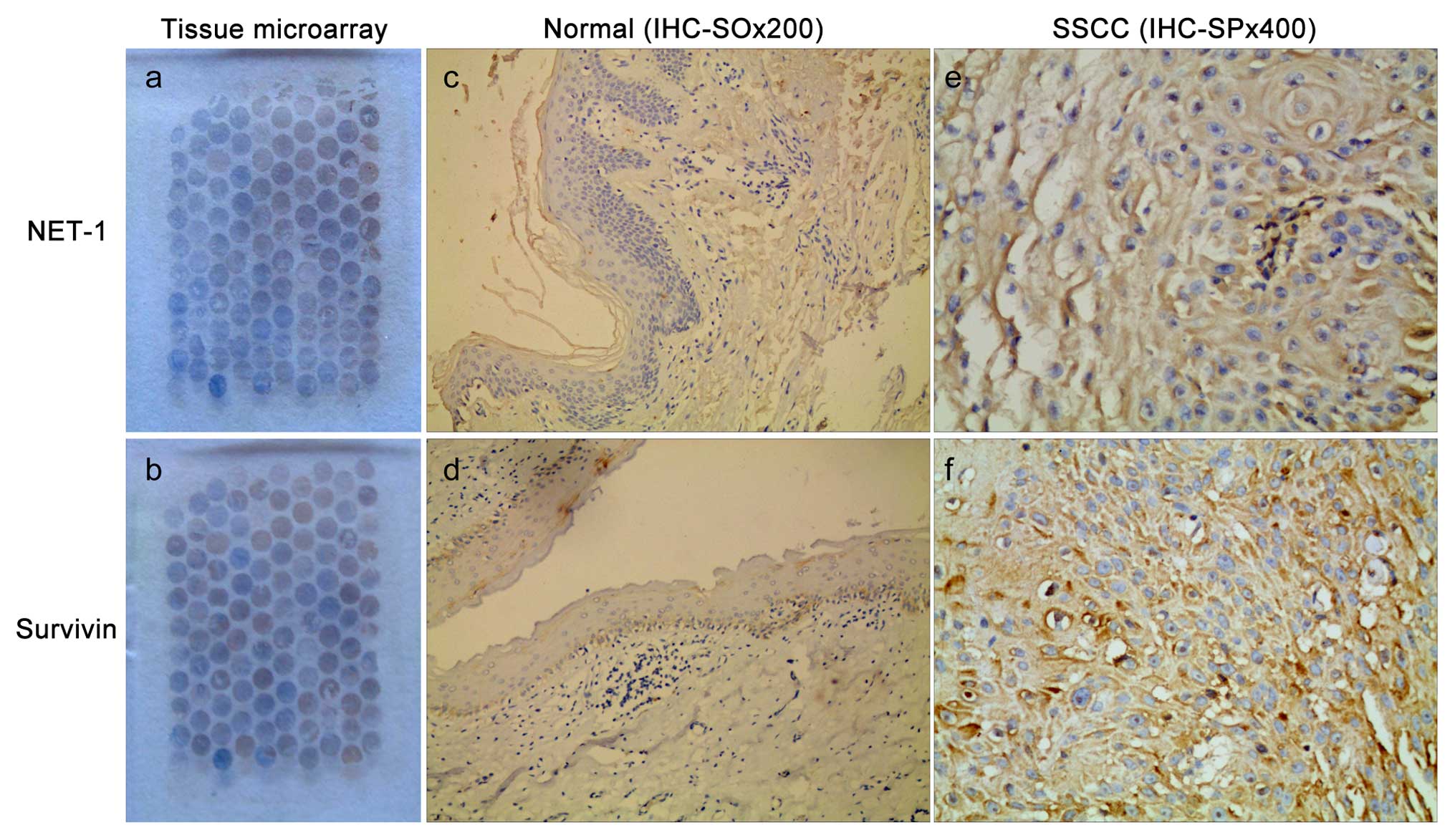
We successfully prepared the SSCC tissue microarray
paraffin blocks, and each case was selected as a representative
area of H&E sections. Each tissue microarray contained 100
sites. The tissue microarray results showed that the organization
structure of the sites was well preserved, without any obvious
necrosis organization. Additionally, the chips were arranged in an
orderly manner (Fig. 1a and b).
NET-1 and survivin expression in SSCC
and surgically removed breast cancer tissue
NET-1 and survivin were positively expressed in the
cytoplasm and/or the cell membrane, stained as tan or brown
(Fig. 1e and f). NET-1 and survivin
were not expressed in surgically removed breast cancer tissue
(Fig. 1c and d). The NET-1 positive
rate was 96% in the SSCC group, but negative in the control group
(p<0.001). The survivin positive rate was 95% in the SSCC group,
but negative in the control group (p<0.001).
Relationship between the expression of
NET-1 and survivin and clinicopathological characteristics in
SSCC
The results showed that there were statistical
differences between the expression intensity of NET-1 and survivin
and tumor thickness, tumor-node-metastasis (TNM) stage, tissue
types, and lymph node metastasis in SSCC (p<0.001). However,
there were no significant associations between NET-1, survivin
expression and other clinicopathological characteristics
(p>0.05) (Table I).
Correlation between NET-1 and
surviving
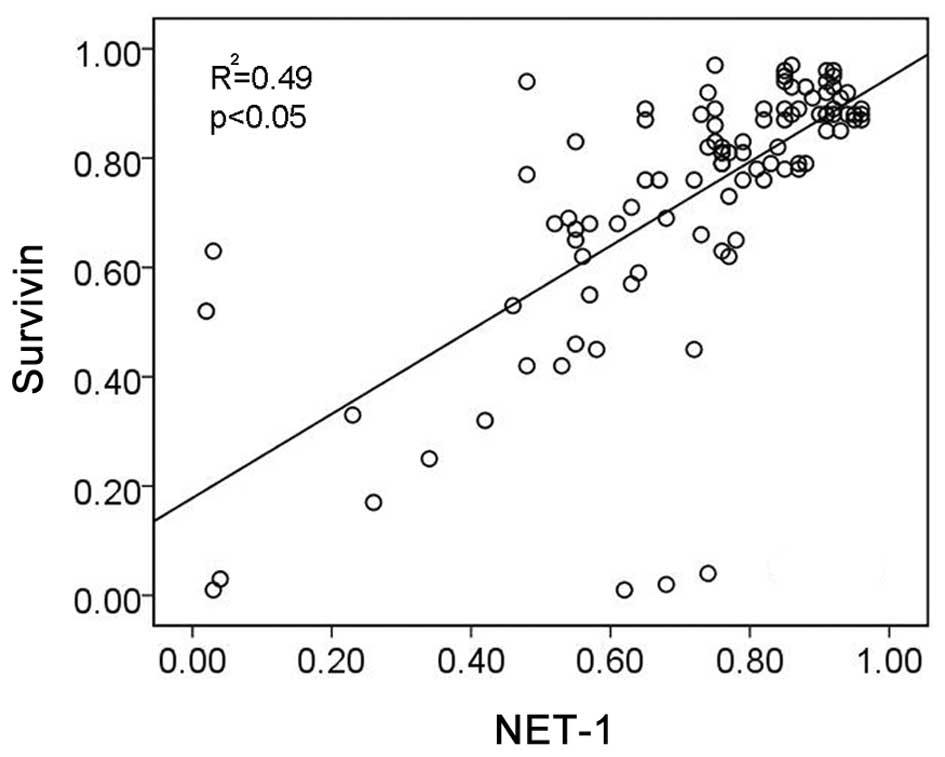
Due to the high expression of NET-1 and survivin in
SSCC, we used Pearson’s correlation analysis to assess the
correlation between NET-1 and survivin protein expression. The
percentage of positivity of NET-1 and survivin was detected and the
result showed that there was positive correlation between the
expression of NET-1 and survivin (r2=0.49, p<0.05,
Fig. 2).
Association between NET-1 and survivin
in A431 cells is identified using an immunoprecipitation assay
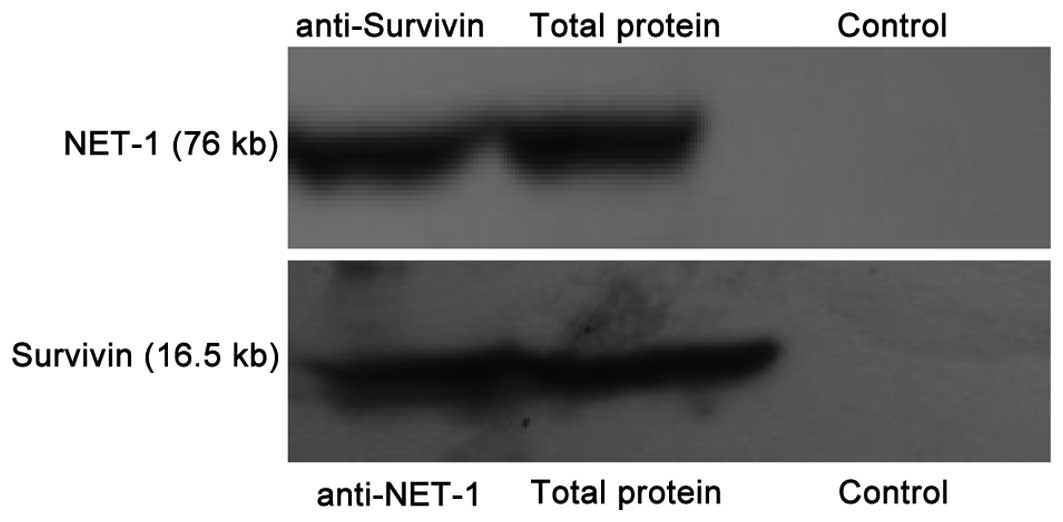
The relationship between NET-1 and survivin was
investigated in A431 cells using an immunoprecipitation assay
(Fig. 3). NET-1 protein was
recognized by the NET-1 antibody in the anti-survivin group. The
specificity was further supported by the fact that no corresponding
protein bands were observed in any IgG group. The total protein was
used as a positive control for NET-1 and survivin proteins.
Survivin was recognized by the survivin antibody in the anti-NET-1
group, confirming a direct interaction between NET-1 and survivin
in A431 cells. This also confirmed the correlation between the
expression of NET-1 and survivin at the SSCC organization level in
the present study.
Effects of NET-1 siRNA, survivin siRNA
and one-chain-double-target siRNA on NET-1 and survivin expression
in A431 cells
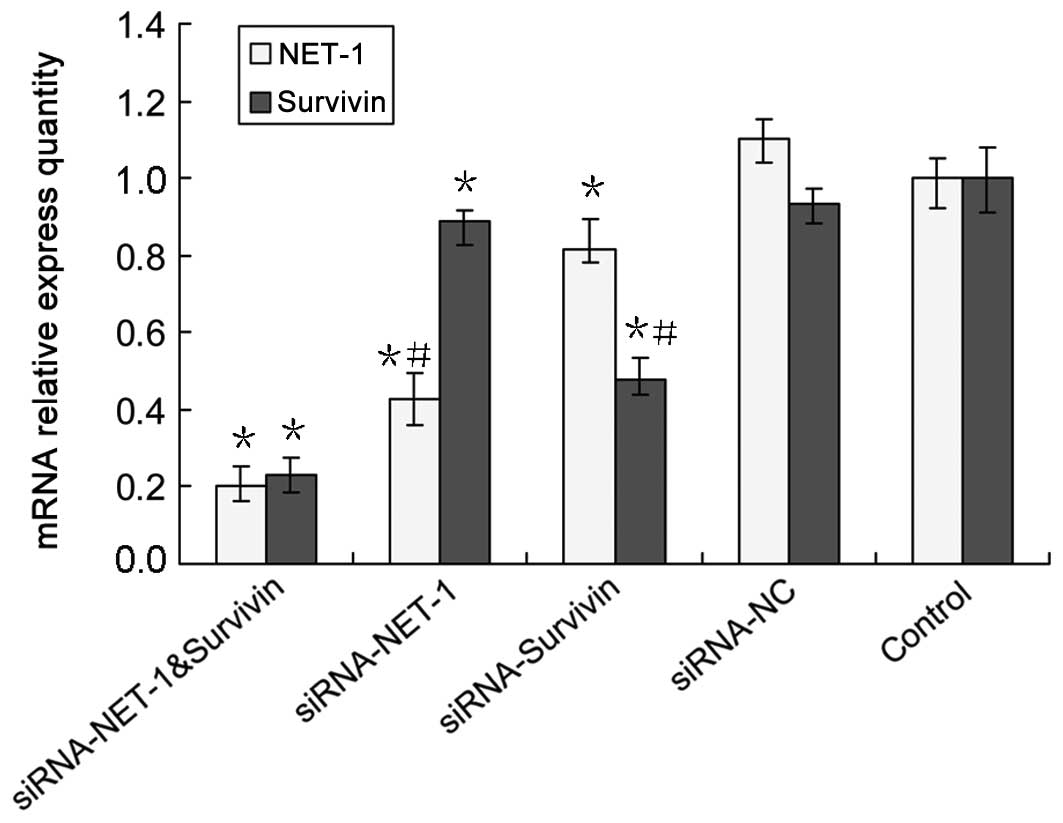
After the A431 cells were transfected with siRNA for
48 h, the levels of mRNA were determined by RT-qPCR while the
protein expression of NET-1 and survivin was assessed using western
blot analysis. When compared to the untreated group, NET-1
expression was decreased by 57% at the mRNA level for the cells
transfected with NET-1 siRNA, and by 54% at the protein level
(Figs. 4 and 5), indicating the effective silencing at
the mRNA and protein levels. Similarly, survivin expression was
effectively inhibited by survivin siRNA at the mRNA and protein
levels by 52 and 58% (Figs. 4 and
5), respectively. Notably, we found
that the NET-1 siRNA significantly decreased survivin expression at
the mRNA level by 11% (Fig. 4) and
protein level by 27% (Fig. 5).
Similarly, survivin siRNA significantly decreased NET-1 expression
at the mRNA level by 19% (Fig. 4)
and the protein level by 15% (Fig.
5). This result showed for the first time that, inhibiting
NET-1 signaling reduced survivin expression and inhibiting survivin
signaling reduced NET-1 expression.
Subsequently, we examined the effect of
one-chain-double-target siRNA on the expression of NET-1 and
survivin, respectively. The results showed that NET-1 mRNA and
protein levels were downregulated by 80% (Fig. 4) and 85% (Fig. 5), respectively, which was in
concordance with that of the protein level of NET-1 siRNA. When
compared to NET-1 siRNA or survivin siRNA alone, the
one-chain-double-target siRNA showed a higher inhibition of
survivin mRNA expression of up to 77% (Fig. 4) and of the protein level of up to
82% (Fig. 5), suggesting a marked
effect of dual gene-targeted siRNA on survivin protein expression.
No statistically significant difference was observed in the
expression of the mRNA and protein siRNA-NC and blank control
groups.
Effects of NET-1 siRNA, survivin siRNA
and one-chain-double-target siRNA on VEGF, cortactin, Bcl-2,
caspase-3 and -8 mRNA expression in A431 cells
As shown in Fig. 6,
NET-1 siRNA, survivin siRNA and one-chain-double-target siRNA
inhibited the VEGF, cortactin and Bcl-2 expression at the mRNA
level in comparison to the untreated group (p<0.05). By
contrast, NET-1 siRNA, survivin siRNA and one-chain-double-target
siRNA increased caspase-3 and -8 expression at mRNA level in
comparison to the untreated group (p<0.05). In addition,
compared to NET-1 siRNA or survivin siRNA alone, the
one-chain-double-target siRNA was more effective in inhibiting and
increasing the mRNA gene expression (p<0.05). There was no
statistically significant difference in the expression of mRNA and
protein between siRNA-NC and blank control groups (p>0.05).
Effect of siRNAs on A431 cell
proliferation and apoptosis Effect of NET-1 siRNA, survivin siRNA
and one-chain-double-target siRNA on A431 cell proliferation
We examined the effect of silencing of NET-1 and
survivin on cell proliferation of A431 cells. The absorbance values
of the A431 cells at 24, 48 and 72 h after the transfection with
NET-1 siRNA or survivin siRNA were significantly lower than those
of the untreated cells (Fig. 7).
There was no significant difference between the absorbance values
of cells treated with NET-1 siRNA and that of survivin siRNA. The
absorbance value of A431 cells treated with one-chain-double-target
siRNA was significantly lower than the cells treated with NET-1
siRNA or survivin siRNA at 24, 48 and 72 h, respectively. There was
no statistically significant difference in A431 cell proliferation
between the siRNA-NC and blank control groups.
Effect of NET-1 siRNA, survivin siRNA
and one-chain-double-target siRNA on A431 cell apoptosis
Annexin V-FITC staining and flow cytometric analysis
were performed to evaluate the effect of NET-1 siRNA, survivin
siRNA and one-chain-double-target siRNA on inducing A431 cell
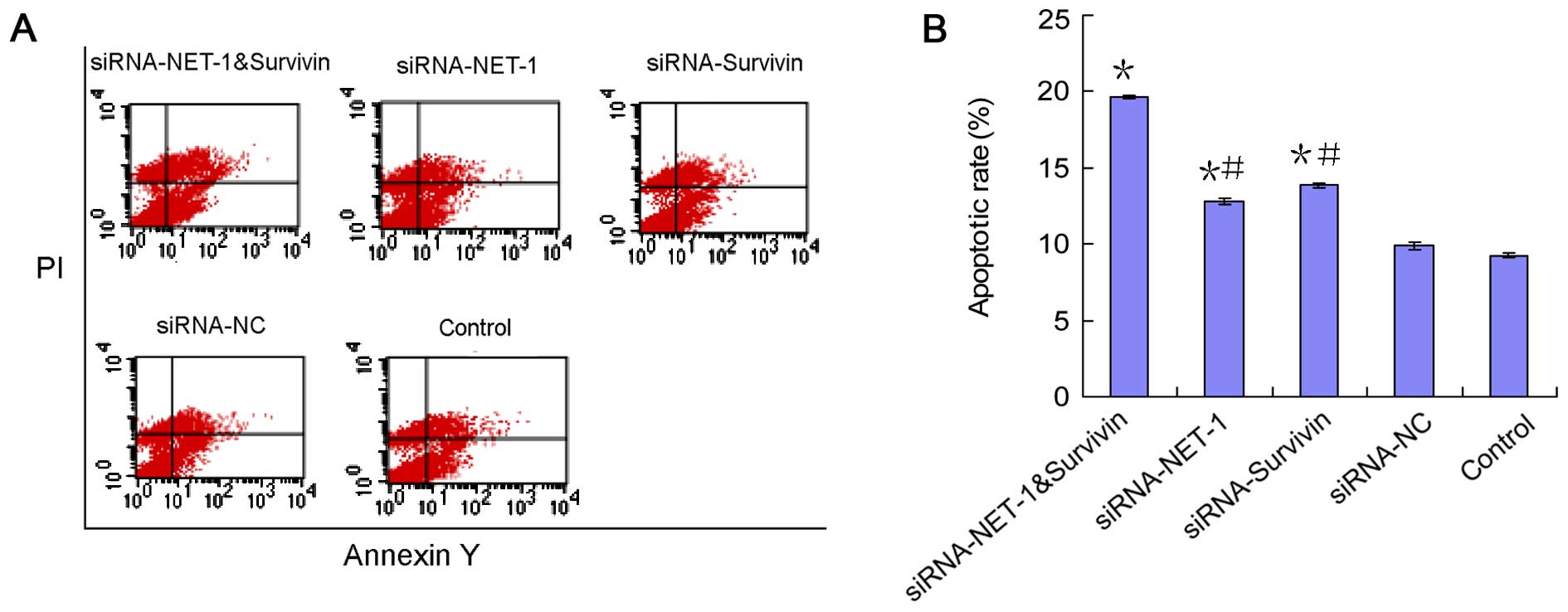
apoptosis. As shown in Fig. 8,
treatment with NET-1 siRNA resulted in a significant increase by
1.39-fold of apoptosis compared with that of the untreated cells
(p<0.05). Similarly, an increase in apoptotic cells was
identified after the survivin siRNA transfection compared to the
untreated group. Furthermore, one-chain-double-target siRNA-treated
cells showed a higher increase of apoptosis, by 2.12-fold, compared
to the untreated cells (p<0.05). There were significant
differences in the apoptotic rates between one-chain-double-target
siRNA-treated and NET-1 siRNA, or survivin siRNA-treated cells (all
p<0.05). No statistically significant difference was observed in
A431 cell apoptosis between the siRNA-NC and blank control
groups.
Detection of the protein expression by
immunofluorescence staining and immunohistochemical staining in
each group
As shown in Figs. 9
and 10, the expression of NET-1
and survivin was increased in SSCC cells in the untreated group,
whereas the location of NET-1 and survivin varied from the membrane
to cytoplasm, or mixed. These results were consistent with those of
the gene microarray. A lower expression was observed in the NET-1
shRNA, survivin shRNA and dual-shRNAs groups as compared to that in
the siRNA-NC and untreated groups. The one-chain-double-target
siRNA therefore showed a higher inhibition of NET-1 and survivin
expression as compared to that of NET-1 and survivin siRNA (all
p<0.05). The difference between the expression of siRNA-NC and
blank control groups was not statistically significant (p>0.05).
The results showed that the immunofluorescence and
immunohistochemical stainings were identical in the experiment.
Discussion
As a new member of the tetraspanins group, NET-1 is
a recently identified tumor-associated gene. NET-1 is closely
associated with the development and prognosis of several malignant
tumors (7–10,16).
The overexpression of NET-1 in SSCC is involved in cancer cell
proliferation (11).
Survivin is one of the most cancer-specific proteins
identified thus far, being upregulated in almost all human tumors.
Biologically, survivin has been shown to inhibit apoptosis, enhance
proliferation and promote angiogenesis (13). Due to its upregulation in malignancy
and its key role in apoptosis, proliferation and angiogenesis,
survivin has been attracting considerable attention as a new target
for anticancer therapies (17).
RNA interference (RNAi) is a sequence-specific
posttranscriptional gene silencing mechanism that is triggered by
double-stranded RNA (dsRNA) and causes degradation of homologous
mRNA in sequence to the dsRNA. RNAi has emerged as one of the most
important findings in the field of molecular biology (18). Due to its high efficacy and
specificity in downregulating gene expression, RNAi was considered
to be a potential therapeutic strategy against human cancer.
Various individual oncogenes have been targeted by RNAi technology
in different tumor cell models, leading to successful silencing of
the protein and subsequently, cancer impairment. However, a number
of these recent studies have selected a single gene rather than
multiple genes as their targets. It is now generally accepted that
there are many genes that are abnormally expressed in malignant
tumors. In most cancers, silencing a single gene may be
insufficient to therapeutically treat cancer cells. Therefore,
simultaneously blocking multiple genes that are abnormally
expressed may be more effective in the treatment of cancer cells
than silencing a single gene. Previous findings have shown that
simultaneously blocking multiple genes has a high protein
inhibition as compared to silencing a single gene in cancer
(19). Therefore, the application
of vector-based RNAi technology involving multiple targets may be a
promising therapeutic modality in the gene therapy of various types
of cancer.
Due to the importance of the proliferation of NET-1
gene and anti-apoptotic survivin gene in tumor formation, the
expression of cutaneous squamous cell carcinoma tissue chip was
detected in 100 cases and a high expression of NET-1 and survivin
was identified. Furthermore, this expression was positively
correlated. This result indicated that the expression of NET-1 and
survivin was closely associated with the degree of malignancy and
biological behavior of cutaneous squamous cell carcinoma,
indicating that the imbalance of the co-expression of NET-1 and
survivin may be one of the mechanisms involved in the occurrence
and development of SSCC. Previous results have shown that: the main
role of NET-1 is to promote the proliferation of tumor cells,
downregulate NET-1 gene expression and inhibit cell proliferation.
By contrast, survivin has a strong anti-apoptotic role,
downregulates survivin gene expression, and functions to promote
cell apoptosis (pro-apoptotic and antiproliferative). Thus, a set
of one-chain-double-target siRNAs was constructed aiming at the
NET-1 and survivin genes in order to screen for effective
single-target NET-1 siRNA and survivin siRNA sequences. The gene
silencing effect of one-chain-double-target siRNA in cutaneous
squamous cell carcinoma and its effect on tumor proliferation and
apoptosis were assessed in in vitro studies.
In the present study, the RT-qPCR, western blotting,
immuno histochemical and immunofluorescent assays showed that
one-chain-double-target siRNA significantly inhibited the
expression of NET-1, survivin mRNA and protein and the inhibitory
effect was better than that of the single-target siRNA group,
suggesting that one-chain-double-target siRNA was capable of
closing two target genes simultaneously, and thus downregulating
the expression of mRNA and protein more effectively. CCK-8 and flow
cytometry showed that after transfection with
one-chain-double-target siRNA, the apoptotic rate of A431 cells was
markedly increased and cell proliferation was significantly
inhibited. The effects were better than those of the single target
group, and the difference was of statistical significance. In
addition, the RT-qPCR results showed that when NET-1 and survivin
gene expression was inhibited, the expression of the angiogenic
gene VEGF, proliferation gene cortactin and anti-apoptotic gene
Bcl-2 were all decreased, while the expression of apoptotic genes
caspase-3 and -8 were increased.
VEGF is the most potent pro-angiogenic signal and
was identified as a key angiogenic stimulator in cancer. Therefore,
as VEGF is an effective target for SSCC therapy, a new approach to
inhibit VEGF expression has to be developed (20). SSCC is a vascular tumor, with high
invasion and metastasis, and the significant increase of
angiogenesis in cancers may be closely associated with the
malignant biological behavior of the tumor. Therefore, in the
present study, the RT-qPCR result showed that VEGF levels of the
double-target siRNA group were obviously decreased.
Cortactin has been described as an actin-associated
scaffolding protein. It binds and activates the Arp2/3 complex and
regulates the branched actin networks in the formation of dynamic
cortical actin-associated structures (21). Cortactin participates in the
regulation of actin cytoskeleton formation, and thereby the
mechanism for controlling tumor cell migration, invasion and
metastasis. The cortactin gene is the critical gene in the
amplified chromosome 11q13 region responsible for increasing
carcinoma motility and invasion (22). Cortactin excess expression may be
associated with the deterioration of the tumor. Therefore, in the
present study, the RT-qPCR result showed that cortactin levels of
the double-target siRNA group were obviously decreased.
Bcl-2 is a prominent member of a protein family that
is responsible for the dysregulation of apoptosis and prevention of
death in cancer cells (23). It
controls of caspase-3 and -8 mRNA was the pathways leading to the
release of cytochrome c from the mitochondrial membrane, the
activation of caspase cascade and eventually the execution of
apoptosis. The flow cytometry results showed the percentage of
apoptotic SSCC was significantly increased in all the treated
groups compared with the untreated group. RT-qPCR showed the
expression of caspase-3 and -8 mRNA was significantly increased in
the double-target group compared with the untreated group, which
was consistent with the flow cytometric results, suggesting that
NET-1 inhibits cell apoptosis through inactivation of the cell
membrane receptor. NET-1 interference may lead to multimerization
after Fas binding with FasL on SSCC surface and then bind with the
cytoplasmic death domain binding protein, activate caspase-8 and
-3, and induce SSCC apoptosis (24). Notably, the RT-qPCR results showed
that the expression of Bcl-2 mRNA was markedly decreased in the
double-target group, suggesting that NET-1 also inhibits cell
apoptosis through the deactivation of mitochondria. NET-1
deficiency may cause the release of cytochrome c from SSCC,
which stimulates caspase-3 and -9, leading to protein hydrolysis,
DNA shear and activation of the intrinsic apoptosis pathway
(25). Therefore, NET-1 inhibited
the intrinsic and extrinsic apoptotic pathways of SSCC.
The above results suggest that there was a common
signaling pathway between the NET-1 and survivin gene and the
signals played a key role in processes such as tumor angiogenesis,
cell proliferation, and apoptosis. Therefore, the downregulation or
inhibition of the expression of NET-1 and survivin became an
important strategy for the treatment of malignant tumors. On the
other hand, NET-1 and survivin belong to the genes responsible for
regulating cell proliferation and apoptosis. The
immunohistochemistry results confirmed the existence of a
correlation between NET-1 and survivin, and the
co-immunoprecipitation results confirmed the existence of an
interaction between NET-1 and survivin, suggesting that there was a
common signaling pathway between the two genes and they were
effective when combined. The combination can be synergized to
improve the antitumor effect. Therefore, NET-1 and survivin may be
combined as diagnostic and treatment indices for patients suffering
from cutaneous squamous cell carcinoma.
In the present study, A431 cells were transfected
with one-chain-double-target siRNA, NET-1 single-target siRNA and
survivin single-target siRNA. The NC siRNA constructed by Biomics
Biotechnology Co., Ltd was used as the empty vector control group
to exclude the effect of vector transfection on cell viability. The
untransfected cells were used as a vehicle control group to exclude
the effect of reagents on cell viability. The hypothesis of the
present study was based on the understanding that excessive cell
proliferation and apoptosis inhibition were key to tumor formation
of tumors, and therefore the one-chain-double-target siRNA designed
for the proliferation of NET-1 and anti-apoptotic survivin genes
may be important. The one-chain-double-target siRNA sequence had
the properties of specificity, uniqueness and innovation and is
conducive to the formation of therapeutic siRNA targeting cutaneous
squamous cell carcinoma. One-chain-double-target siRNA targeting
NET-1 and survivin genes expanded the RNAi technology, providing a
foundation and experimental basis for the development of
one-chain-multi-target siRNA technology.
In conclusion, the one-chain-double-target siRNA
targeting NET-1 and survivin genes constructed in the present study
was highly specific. The effect of double silencing of NET-1 and
survivin genes was significantly enhanced compared with the
single-target siRNA. The results may provide insight into the
mechanisms and clinical treatment of cutaneous squamous cell
carcinoma and other tumors that may be applied in further
investigations. Future studies are required to create a model of
nude mice bearing human squamous cell carcinoma, and examine the
silencing effect of target genes caused by one-chain-double-targets
siRNA in vivo by transdermal peptide transmission pathway.
The ability of the tumor cells treated with
one-chain-double-targets siRNA to resist chemotherapeutic drugs by
ATP-TCA tumor susceptibility testing should also be investigated.
We examined the manner in which the instability and in vivo
complexity of one-chain-double-targets siRNA may be overcome,
thereby delivering siRNA accurately and safely to the target cells
in order for RNAi to take effect in the local skin, which may
contribute to prevention of problems in the circulatory system, be
beneficial in the reduction of the administration dose and promote
the development of one-chain-double-target siRNA technology. These
methods may result in new treatment for patients suffering from
skin cancer and other tumors.
Acknowledgments
The present study was supported by the foundation of
the production-study-research prospective joint research programs
of Jiangsu Province, China (BY 2013042-06).
References
|
1
|
Smoller BR: Squamous cell carcinoma: From
precursor lesions to high-risk variants. Mod Pathol. 19(Suppl 2):
S88–S92. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bowen GM, White GL Jr and Gerwels JW: Mohs
micrographic surgery. Am Fam Physician. 72:845–848. 2005.PubMed/NCBI
|
|
3
|
Serru V, Dessen P, Boucheix C and
Rubinstein E: Sequence and expression of seven new tetraspans.
Biochim Biophys Acta. 1478:159–163. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
5
|
Yauch RL and Hemler ME: Specific
interactions among transmembrane 4 superfamily (TM4SF) proteins and
phosphoinositide 4-kinase. Biochem J. 351:629–637. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen L, Wang JL, Li H, Qin J and Wu YY:
Functions of NET-1 gene in skin squamous cell carcinoma cell line
(A431): A siRNA study. Zhonghua Bing Li Xue Za Zhi. 38:691–696.
2009.In Chinese.
|
|
7
|
Chen L, Li X, Wang GL, Wang Y, Zhu YY and
Zhu J: Clinicopathological significance of overexpression of
TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 94:531–538.
2008.PubMed/NCBI
|
|
8
|
Wollscheid V, Kühne-Heid R, Stein I,
Jansen L, Köllner S, Schneider A and Dürst M: Identification of a
new proliferation-associated protein NET-1/C4.8 characteristic for
a subset of high-grade cervical intraepithelial neoplasia and
cervical carcinomas. Int J Cancer. 99:771–775. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Zhu YY, Zhang XJ, Wang GL, Li XY,
He S, Zhang JB and Zhu JW: TSPAN1 protein expression: A significant
prognostic indicator for patients with colorectal adenocarcinoma.
World J Gastroenterol. 15:2270–2276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scholz CJ, Kurzeder C, Koretz K, Windisch
J, Kreienberg R, Sauer G and Deissler H: Tspan-1 is a tetraspanin
preferentially expressed by mucinous and endometrioid subtypes of
human ovarian carcinomas. Cancer Lett. 275:198–203. 2009.
View Article : Google Scholar
|
|
11
|
Chen L, Zhu Y, Li H, Wang GL, Wu YY, Lu
YX, Qin J, Tuo J, Wang JL and Zhu J: Knockdown of TSPAN1 by RNA
silencing and antisense technique inhibits proliferation and
infiltration of human skin squamous carcinoma cells. Tumori.
96:289–295. 2010.PubMed/NCBI
|
|
12
|
Kim GY, Lim SJ and Kim YW: Expression of
HuR, COX-2, and survivin in lung cancers; cytoplasmic HuR
stabilizes cyclooxy-genase-2 in squamous cell carcinomas. Mod
Pathol. 24:1336–1347. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ryan BM, O’Donovan N and Duffy MJ:
Survivin: A new target for anti-cancer therapy. Cancer Treat Rev.
35:553–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng K and Mahato RI: Gene modulation for
treating liver fibrosis. Crit Rev Ther Drug Carrier Syst.
24:93–146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mahato RI, Cheng K and Guntaka RV:
Modulation of gene expression by antisense and antigene
oligodeoxynucleotides and small interfering RNA. Expert Opin Drug
Deliv. 2:3–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L, Wang Z, Zhan X, Li DC, Zhu YY and
Zhu J: Association of NET-1 gene expression with human
hepatocellular carcinoma. Int J Surg Pathol. 15:346–353. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zaffaroni N, Pennati M and Daidone MG:
Survivin as a target for new anticancer interventions. J Cell Mol
Med. 9:360–372. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu JB, Fu HQ, Huang LZ, Liu AW and Zhang
JX: Effects of siRNA-targeting BMP-2 on the abilities of migration
and invasion of human liver cancer SMMC7721 cells and its
mechanism. Cancer Gene Ther. 18:20–25. 2011. View Article : Google Scholar
|
|
19
|
Chen SM, Wang Y, Xiao BK and Tao ZZ:
Effect of blocking VEGF, hTERT and Bcl-xl by multiple shRNA
expression vectors on the human laryngeal squamous carcinoma
xenograft in nude mice. Cancer Biol Ther. 7:734–739. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takahashi H and Shibuya M: The vascular
endothelial growth factor (VEGF)/VEGF receptor system and its role
under physiological and pathological conditions. Clin Sci.
109:227–241. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada S, Yanamoto S, Kawasaki G, Mizuno A
and Nemoto TK: Overexpression of cortactin increases invasion
potential in oral squamous cell carcinoma. Pathol Oncol Res.
16:523–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu KF, Lin CK, Yu CP, Tzao C, Lee SC, Lee
YY, Tsai WC and Jin JS: Cortactin, fascin, and survivin expression
associated with clinicopathological parameters in esophageal
squamous cell carcinoma. Dis Esophagus. 22:402–408. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thomas S, Quinn BA, Das SK, Dash R, Emdad
L, Dasgupta S, Wang XY, Dent P, Reed JC, Pellecchia M, et al:
Targeting the Bcl-2 family for cancer therapy. Expert Opin Ther
Targets. 17:61–75. 2013. View Article : Google Scholar
|
|
24
|
Ghavami S, Hashemi M, Ande SR, Yeganeh B,
Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, et
al: Apoptosis and cancer: Mutations within caspase genes. J Med
Genet. 46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeong SY and Seol DW: The role of
mitochondria in apoptosis. BMB Rep. 41:11–22. 2008. View Article : Google Scholar : PubMed/NCBI
|