Introduction
Colon cancer is one of the most lethal malignancies
worldwide, with its incidence rate rising each year due to the
lifestyle innovation and environment modification, especially in
the rapid urbanization regions such as China. Modern multimodality
treatments, including primary surgery resection and other adjuvant
therapies, have synergistically improved the overall 5-year
survival rate to ~60% (1,2). However, distant metastasis and drug
resistance restrain the efficacy of current medications. These
obstacles make it essential for the investigation of mechanisms
that contribute to the poor prognosis of advanced and resistant
cases.
MicroRNA (miRNA) is constituted by an endogenous
conserved class of small, non-coding RNAs exerting regulatory
function on multiple gene expressions via translational suppression
and mRNA degradation. Correlations between miRNAs and
carcinogenesis of colon cancer have been verified by emerging
evidence (3). By means of blocking
target oncogenes or tumor suppressor genes, miRNAs act
contradictory roles in modulating cancer proliferation, metastasis,
apoptosis and drug resistance (4).
miR-219-5p has been reported to have a role in many
biological procedures including myocardial regeneration (5) and lymphoblast immortalization
(6). Compelling evidence suggests
that miR-219-5p serves as a tumor manipulator in various types of
cancers, attenuating malignant features of liver cancer (7), papillary thyroid carcinoma (8) and glioblastoma (9) by targeting downstream oncogenic
molecules. Nevertheless, whether miR-219-5p is aberrantly expressed
and associated with oncogenesis in colon cancer remains
unclear.
Sall4 is a novel oncogene mediating origination and
development in different types of cancers. Its abnormal
overexpression in colon cancer leads to tumor progression and
metastasis (10), and Sall4 has
become a new biomarker for early diagnosis among colon malignancies
(11). Several upstream pathways
have been identified as modulators of Sall4, such as Wnt/β-catenin
signaling (12), and certain
miRNAs, including miR-107 (13),
directly regulate Sall4 expression, controlling cell proliferation
and invasiveness. In spite of all above evidence, the regulatory
mechanisms against Sall4 in colon cancer remain ill-defined.
In the present study, we investigated the aberrant
expression of miR-219-5p and Sall4 in colon cancer specimens, and
confirmed that Sall4 was the direct target of miR-219-5p.
Additionally, by aid of gain and loss of function assays,
miR-219-5p was observed to play an inhibitory effect on cell
proliferation, invasion and drug resistance. Our findings
demonstrate that miR-219-5p has potential clinical value against
colon cancer and may be an important therapy target in future
treatments.
Materials and methods
Tissue specimens
All human related studies were approved by the
Ethics Committee of Union Hospital, Tongji Medical College,
Huazhong University of Science and Technology and were performed in
accordance with the ethical standards of DECLaration of Helsinki.
Prior to the collection of specimens, a written informed consent
was given by each person involved.
The tissue specimens were collected from 20 patients
who had undergone surgical resection in the Gastroenterology
Department, Union Hospital, Tongji Medical College, Huazhong
University of Science and Technology in 2014. All patients had
surgical procedures prior to chemotherapy or radiation therapy.
Post-surgical pathology had verified the resection tissues as colon
cancer tissues and adjacent normal tissues from each patient were
applied as controls.
Cell culture
Normal human colon epithelial cell line NCM460 was
kindly donated by our laboratory instructor, and colon cancer cell
line HT-29, Caco-2, DLD-1, SW480, SW620 were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA).
Dulbecco's modified eagle's medium (DMEM; Life Technologies, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum (FBS;
SJQ, Zhejiang Tianhang Biotechnology Ltd., Hangzhou, China) was
applied for normal culture and the cells were incubated in a
humidified incubator at 37°C with 5% CO2.
Quantitative real-time PCR
Total RNA was isolated from tissue specimens and
cultured cells using TRIzol reagent (Takara, Dalian, China).
miR-219-5p was polyadenylated by One-Step PrimeScript miRNA cDNA
synthesis kit (D350A; Takara) under the manufacturer's protocol.
Then cDNA was quantified using SYBR Premix EX Taq ii (DRR081;
Takara) for real-time PCR quantification in Applied Biosystems 7500
Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA)
under standard procedures. U6 was used as an internal reference for
miR-219-5p. The forward primer sequence of miR-219-5p was:
5′-CGGTGATTGTCCAAACG CAATTC-3′; the reverse primer was Uni-miR qPCR
Primer provided by One-Step PrimeScript miRNA cDNA synthesis kit
(D350A; Takara). All reactions were perfomed in triplicate. The
2−ΔΔCT method was applied for relative expression
quantification.
Western blotting
Total proteins were extracted from tissue specimens
and cultured cells using RIPA buffer. BCA assay was applied for
protein concentration evaluation using Pierce™ BCA protein assay
kit (23225; Life Technologies) following the manufacturer's
protocol. Total protein (40 μg/lane) was separated on 10%
SDS polyacrylamide gels and transferred onto PVDF membranes. After
blocking in 5% fat-free milk for 2 h, the membranes were then
incubated with primary antibody overnight at 4°C. The primary
antibodies used were: anti-Sall4 (ab29112; Abcam, Cambridge, MA,
USA); anti-GAPDH (ab37168; Abcam); anti-p21 (sc-397; Santa Cruz
Biotechnology, Santa Cruz, CA, USA); anti-cyclin D1 (sc-753; Santa
Cruz Biotechnology); anti-MMP-9 (sc-10737; Santa Cruz
Biotechnology); anti-e-cadherin (sc-7870; Santa Cruz
Biotechnology); anti-N-cadherin (sc-7939; Santa Cruz
Biotechnology); anti-BAX (sc-493; Santa Cruz Biotechnology);
anti-cleaved caspase-3 (sc-22171-R; Santa Cruz Biotechnology);
anti-cleaved caspase-9 (#7237; Cell Signaling Technology, Danvers,
MA, USA); anti-P-gp (ab129450; Abcam); anti-MRP1 (sc-13960; Santa
Cruz Biotechnology). After washing with TBST, the membrane was
incubated with anti-rabbit IgG-HRP secondary antibody (074–1506;
KPL, Gaithersburg, MD, USA) for 2 h. Protein bands were visualized
by X-ray film using Pierce™ ECL Western blotting substrate (32209;
Life Technologies).
Dual-luciferase reporter assay
miR-219-5p mimic, inhibitor, negative control and
wild-type or mutant pGL3-Sall4-3′UTR were co-transfected into
cultured cells seeded in a 96-well. Dual-luciferase reporter assay
system (Promega, Madison, WI, USA) was applied for analyzing
luciferase activity 48 h post-transfection. The firefly and
Renilla luciferase activities were measured and firefly
luciferase activity was normalized to the Renilla luciferase
activity. All experiments were conducted in triplicate.
Cell transfection
miR-219-5p mimic, miR-219-5p inhibitor, scrambled
negative control and siRNA of Sall4 were synthesized by Guangzhou
RiboBio Co., Ltd. (Guangzhou, China); pcDNA3.1-Sall4 expression
vector was constructed by CIYA Technologies (Wuhan, China). Cells
were incubated in 6-well plates at a density of 105.
When the cell confluence reached 70%, miR-219-5p mimic, miR-219-5p
inhibitor, scrambled negative control, siRNA of Sall4 and
pcDNA3.1-Sall4 expression vector were transfected respectively or
co-transfected to cells in different groups with
Lipofectamine® 2000 transfection reagent (11668-019;
Life Technologies) following standard procedures. The testing of
transfection efficiency on qRT-PCR and collection of cells for
assays were conducted 48 h post-transfection.
Cell proliferation assay
Cells were seeded in 96-well plates in a
concentration of 104/well, each group had 6 parallel
wells. CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) was
applied to detect the proliferation of cells after 24-, 48- and
72-h incubation. CCK-8 reagent (10 μl) was added into each
well at different time-points before 2-h incubation. Then the
absorbance at 450 nm was measured. Three independent experiments
were conducted.
Cell migration and invasion assays
Wound healing assay
An artificial wound was made by a 200-μl
pipette tip scratching on the well surface. To eliminate
proliferation effect, basic DMEM medium without fetal bovine serum
was used to culture cells for 24 h until evaluating the final
efficacy.
Transwell assay
Cell invasion was examined using 24-well Matrigel
invasion chamber with pore size 8 μm (Corning, Incorporated,
Corning, NY, USA). Cells (105) were added to the upper
compartment of the chamber with serum-free medium, while the lower
compartment was filled with complete medium with 10% fetal bovine
serum as a chemo-attractant. After 48-h incubation, non-invading
cells on the upper surface were wiped with a cotton swab. Ethanol
(95%) and 0.1% crystal violet solution were used to fix and stain
the invasive cells. The numbers of invasive cells were counted from
four randomly selected fields (magnification, ×200). All
experiments were conducted in triplicate.
Cell cycle and apoptosis analysis
Cell cycle
The cells were collected and fixed in 70% ethanol
overnight at −20°C, then treated with DNA staining solution
containing 3.4 mM Tris-Cl (pH 7.4), propodium iodide, 0.1% Triton
X-100 buffer and 100 mg/ml RNase A. Cell cycle analysis was
conducted in FACS flow cytometry (BD Biocsiences, San Jose,
USA).
Apoptosis
Annexin V-FITC/PI dual staining kit (KGA108; Nanjing
Keygen Biotech Co., Ltd., Nanjing, China) was used for evaluation
of apoptosis. Cells (3×105) were collected before
analyzing by FACS flow cytometry (BD Biocsiences) following
recommended steps. All experiments were performed in
triplicate.
Drug resistance analysis
CCK-8 assay was used for analyzing the cell
viability after exposure to each drug for 72 h. Fluorouracil (Yabao
Pharmaceutical Co., Ltd., Beijing, China) and oxaliplatin (Hangzhou
Sanofi-Aventis Minsheng Co., Ltd., China) were used to confirm the
drug sensitivity of the different cells.
Statistical analysis
The linear analysis between miR-219-5p and Sall4 in
specimens was explored by Spearman's correlation. Other data were
analyzed by the Student's t-test. P<0.05 was regarded as
statistically significant. Excel software and GraphPad Prism 5 were
used for statistical analysis.
Results
Sall4 expression is elevated and
miR-219-5p is down-regulated in colon cancer tissue specimens and
cell lines compared to controls
By analyzing specimens from 20 colon cancer
patients, Sall4 expression in the tumor tissues was found elevated
compared to adjacent normal tissues (P<0.001; Fig. 1A), while miR-219-5p level was
reduced in the malignant tissues compared to controls (P<0.001;
Fig. 1B). Then, the linear
regression analysis displayed a possible relevance between Sall4
and miR-219-5p in tissue specimens with R2=0.7119 (Fig. 1C). Cell line assays coordinated with
tissue analysis indicating that Sall4 level was elevated in colon
cancer cell line HT-29 (P<0.01), Caco-2 (P<0.01), DLD-1
(P<0.01), SW480 (P<0.01) and SW620 (P<0.01) compared to
normal colon epithelial cell line NCM460 (Fig. 1D) while miR-219-5p level was
decreased in HT-29 (P<0.001), Caco-2 (P<0.001), DLD-1
(P<0.001), SW480 (P<0.001) and SW620 (P<0.001) (Fig. 1E). The results revealed that Sall4
expression was elevated in cancer tissues while miR-219-5p was
reversely downregulated. There was potential interaction between
Sall4 and miR-219-5p based on expression analysis.
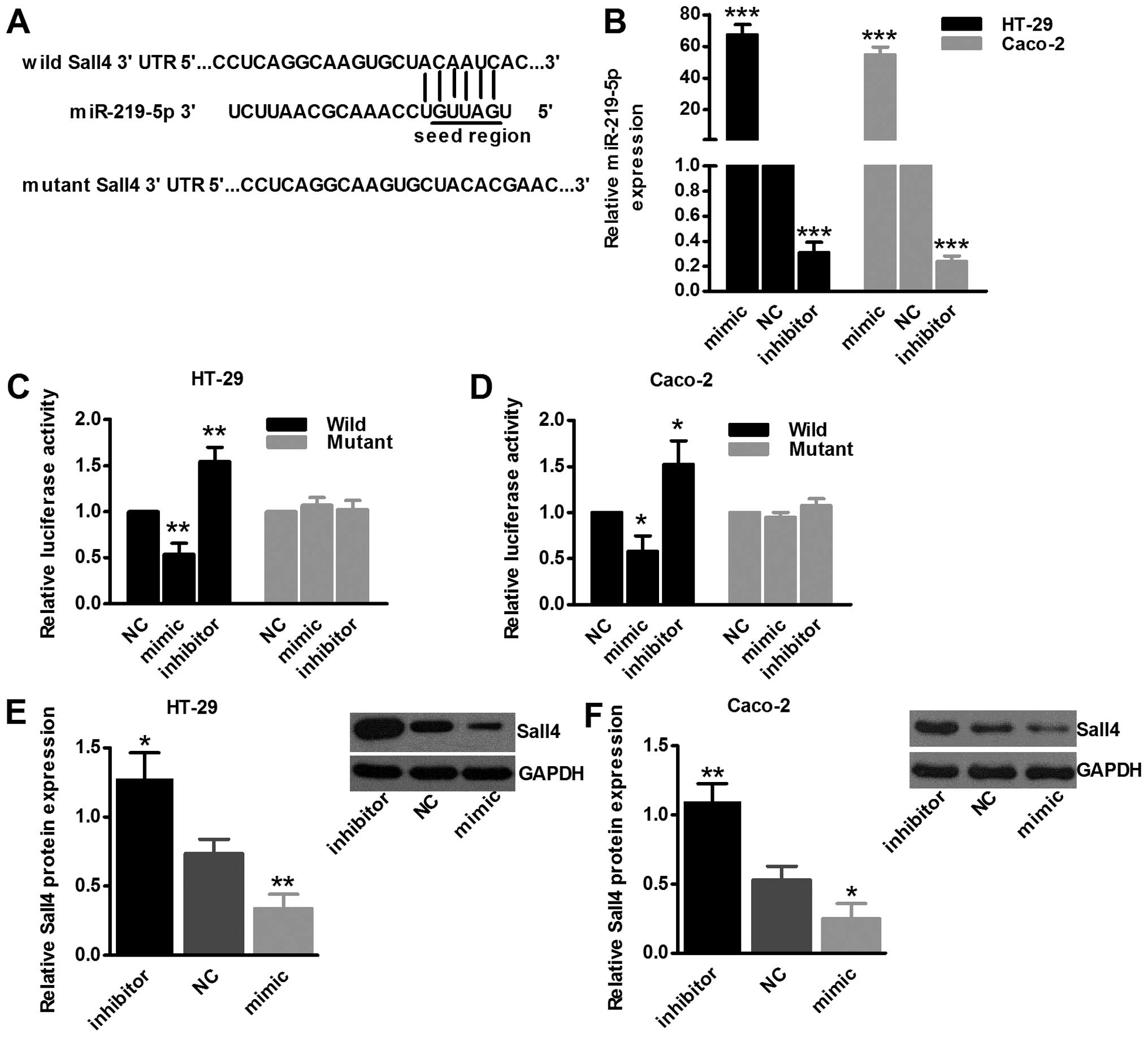
Sall4 is a direct target of
miR-219-5p
miR-219-5p was predicted by TargetScan as a
potential regulator of oncogene Sall4 (Fig. 2A). To clarify the interplay between
Sall4 and miR-219-5p, we used the dual luciferase assay in HT-29
and Caco-2 cell lines. Transfection efficiency detection is shown
in Fig. 2B, miR-219-5p mimic
upregulated its expression dramatically (P<0.001) and vice versa
(P<0.001). Dual luciferase assay revealed that upregulation of
miR-219-5p inhibited luciferase activity compared to negative
control group in wild-type 3′UTR, but not in mutant type, which
strongly proved that miR-219-5p could indeed interact with Sall4
3′UTR sequence (Fig. 2C and D;
P<0.05). Moreover, western blot assay additionally supported the
results of dual luciferase assay that upregulation of miR-219-5p
inhibited Sall4 expression and vice versa (Fig. 2E and F; P<0.05), confirming Sall4
was a direct target of miR-219-5p.
miR-219-5p inhibited colon cancer
proliferation and G0/G1 cell cycle arrest by targeting Sall4
To accurately find out the interaction of miR-219-5p
and Sall4 on colon cancer proliferation, we divided hT-29 and
Caco-2 into five groups respectively: 1, miR-219-5p mimic and Sall4
activation vector co-transfection; 2, miR-219-5p; 3, negative
control; 4, miR-219-5p inhibitor; 5, miR-219-5p inhibitor and siRNA
of Sall4 co-transfection. By CCK-8 test, we discovered that in both
cell lines, miR-219-5p inhibited cell proliferation dramatically
compared to negative control, which then could be antagonized by
Sall4 activation (P<0.05; Fig. 3A
and B). Additionally, reduction on miR-219-5p expression
increased cell proliferation and those effects were inhibited by
Sall4 siRNA transfection (P<0.05; Fig. 3A and B). In cell cycle analysis,
both cell lines displayed G0/G1 arrest and S phase percentage
reduction by miR-219-5p activation and vice versa (P<0.05;
Fig. 3C and D). p21 was a G1 phase
checkpoint protein acting as a tumor suppressor while cyclin D1
promoted cells into S phase functioning as a proliferative role.
Moreover, by activation of miR-219-5p, p21 increased its expression
while cyclin D1 decreased, these results could be antagonized by
targeting Sall4 (P<0.05; Fig. 3E and
F). All the outcomes confirmed that miR-219-5p inhibited colon
cancer proliferation and G0/G1 cell cycle arrest by targeting
Sall4.
miR-219-5p inhibits colon cancer
migration and invasion by targeting Sall4
Wound healing assay was routinely used to analyze
cell migration and Transwell assay was applied as a classical
measure for cancer invasiveness. Our experiments confirmed that
cell migration and invasion were reduced dramatically by miR-219-5p
activation, which were promoted by miR-219-5p inhibition. The Sall4
expression level was able to influence the efficacy, suggesting
miR-219-5p inhibited colon cancer migration and invasion by
targeting Sall4 (P<0.05; Fig. 4C and
F). MMP-9, E-cadherin and N-cadherin are representative
invasion-related protein in cancer research increasing their
expression in more invasive cases (E-cadherin decreasing). By
western blot assay, the expression level of targeted protein
revealed similar results to the above experiments (P<0.05;
Fig. 4G and H).
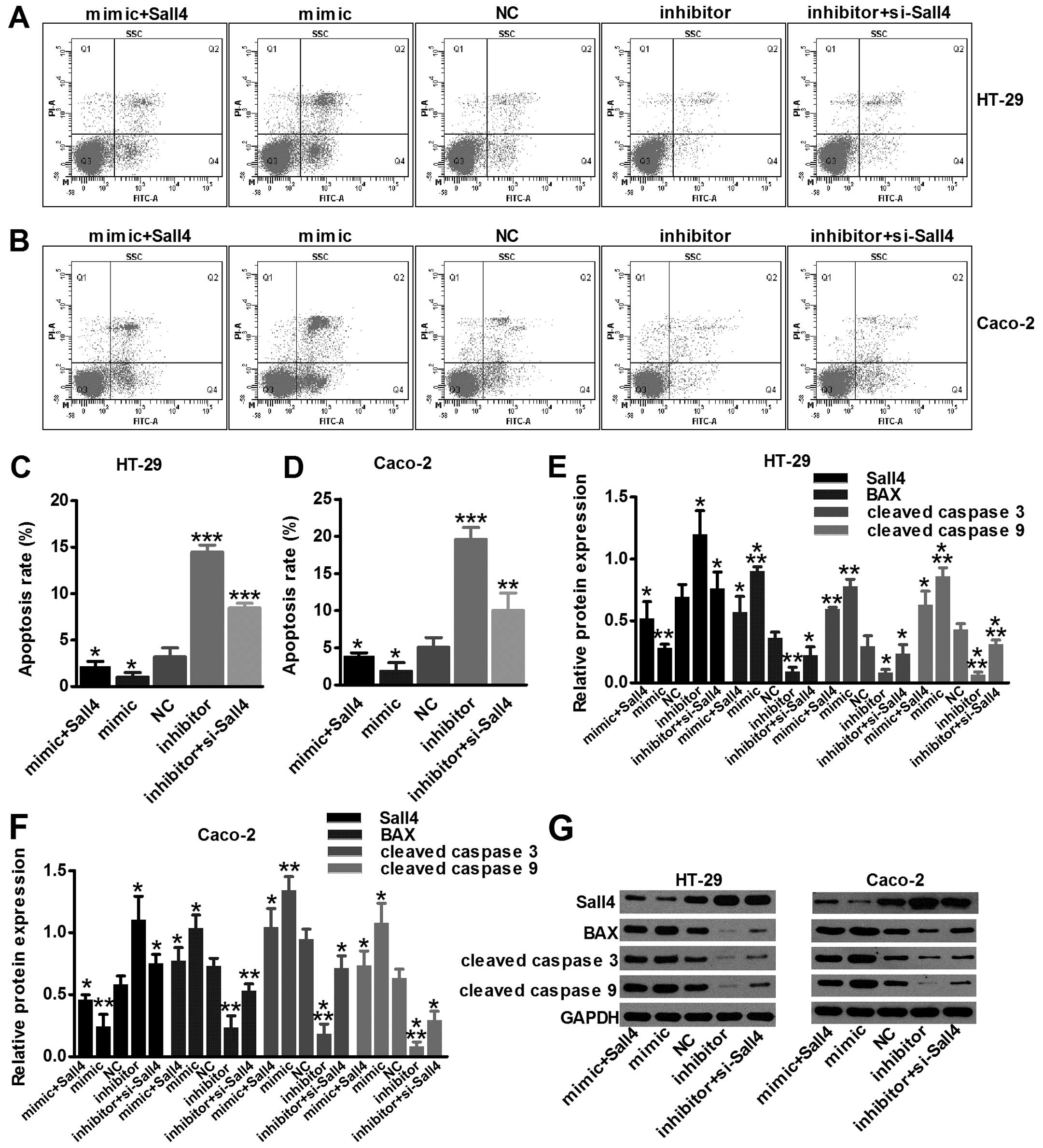
miR-219-5p induces colon cancer apoptosis
by targeting Sall4
Our experiments confirmed by flow cytometry
analysis, that miR-219-5p strongly increased cell apoptosis rate,
while the rate was inhibited by miR-219-5p inhibition (P<0.05;
Fig. 5C and D). Sall4 was
identified as a target of miR-219-5p-induced apoptosis in colon
cancer cell lines. BAX, cleaved caspase-3 and cleaved caspase-9 are
three common apoptosis-inducing proteins. By western blot assay,
all three proteins were upregulated in miR-219-5p activation,
according to the results of the apoptosis analysis (P<0.05;
Fig. 5E and F). Thus, we proved
that miR-219-5p induced colon cancer apoptosis by targeting
Sall4.
miR-219-5p reduces colon cancer drug
resistance by targeting Sall4
Drug resistance of colon cancer greatly restricted
the clinical efficacy. We analyzed resistance of fluorouracil and
oxaliplatin among five different groups in both cell lines, with
these commonly used chemotherapy drugs in clinical practice. The
results suggested that in both cell lines, miR-219-5p reduced colon
cancer drug resistance by targeting Sall4 with greatly less viable
cells after drug intervention (P<0.05; Fig. 6A and B). P-gp and MRP1 are two
representative proteins participating in chemotherapy resistance.
Our western blot results confirmed the results of cell survival
after drug intervention showing more expressed resistant proteins
(P<0.05; Fig. 6C and D).
Discussion
Although surgical operation functions as a preferred
method in most colon cancer cases, those who suffer of locally
advanced tumors or distant metastasis may be treated with internal
medications, such as chemotherapy and specific targeted drugs.
Targeted therapy is a future trend for advanced colon cancer
medication as it is more specific and has less adverse effect
compared to traditional chemotherapy drugs (14,15).
However, unlike the revolutionary role of imatinib on
gastrointestinal stromal tumor (16) and trastuzumab on breast carcinoma
treatment (17), curative targeted
drugs for advanced colon cancer are still lacking. Classical
chemotherapy drugs fluorouracil and oxaliplatin still dominate the
adjunctive treatment (18), while
recent clinical evidence have disclosed conclusions that certain
indicative targeted drugs such as cetuximab (19,20)
and bevacizumab (21) demonstrate
disappointing nonprofit survival rate in advanced patients despite
the much expected clinical success on other malignancies such as
lung cancer (22).
Search for available targets is a hotspot on colon
cancer research, and miRNAs are a group of non-coding RNAs, acting
as a broad regulation medium to connect cell signaling transduction
and control cancer gene expression. Moreover, they can be readily
transfected into target tissues by current techniques, facilitating
its potential clinical usage. Those features have turn the miRNAs
into a new favorite on targeted therapy research, which is proven
by accumulating evidence (3,4). The
expression and function of miR-219-5p on colon cancer has not been
published yet, and our experiments first revealed the antitumor
properties of miR-219-5p on colon cancer via inhibiting the
ßoncogene Sall4 expression. This result is in coordination with the
tumor suppressor role of miR-219-5p in liver cancer (7), papillary thyroid carcinoma (8) and glioblastoma (9) published already, suggesting its
extensive anticancer efficacy. Many miRNAs have bipolar regulatory
mechanisms in different types of cancers via targeting opposite
functional genes, such as the oncogenic effect of miR-182 on
colorectal cancer (23) while
suppressing the proliferation of gastric cancer as well (24). Current evidence merely unveils the
tumor inhibitory role of miR-219-5p, whether it still has the
oncogenic role in diverse tumors or certain sub-types of colon
cancer remains unclear. Additionally, miRNAs can be partially
regulated by upstream effectors to precisely control its biological
behavior, especially some classic developmental signaling pathways
such as Sonic hedgehog (25) and
Wnt/β-catenin signaling (26). The
network in which miR-219-5p is located and fits into all require
further exploration before its clinical applications.
Sall4, a novel zinc finger transcriptional factor,
behaves as an oncogene mediating tumorigenesis in different types
of cancers, especially in liver cancer (27)and leukemia (28). The transcriptional role of Sall4
expands its downstream cancer gene expression to fulfill oncogenic
efficacy, such as directly stimulating the expression of ubiquitous
oncogene Bmi-1 in various cancers (29). Apart from the reliable evidence
revealing its role in colon carcinogenesis and diagnosis (10,11),
Our experiments further verified that Sall4 overexpression
correlated positively with chemo-resistance by upregulation of
certain drug resistant genes including P-gp and MRP1, exhibiting a
broader pro-oncogenic function of Sall4. Sall4 treatment has
displayed inspiring tumor attenuating effect on leukemia cells,
suggesting its potentials on clinical practice (30). Clarification of the mechanism
network mediating its oncogenic role and regulating its expression
is essential before launching the Sall4-targeted therapy. Several
upstream signaling and miRNAs have been confirmed to participate in
regulating expression of Sall4 including Wnt/β-catenin signaling
(12). miR-107 is reported to
target Sall4 and to inhibit cell proliferation in glioma (13) and our experiment result verifies
that miR-219-5p targets Sall4 in colon cancer to inhibit tumor
progression and to reduce malignant properties. Furthermore, an
oncogene can always be regulated by several different miRNAs such
as YAP1 targeted by miR-15a (31),
miR-375 (32) and miR-200a
(33). Thus, our laboratory will
testi more potential miRNAs of Sall4 according to TargetScan
prediction, offering more options of future miRNA mimic treatment
against Sall4 on colon cancer.
The first miRNA targeted drug SPC3649 (anti-miR-122)
was launched for clinical testing in 2008 and exhibits good
clinical outcome against hepatitis C patients (34). Along with the breakthrough on
pharmaceutical technology, certain miRNAs may appear on the market
for clinical practice facilitating patients and clinicians. Thus,
we need to make great efforts to elaborate the working systems of
miR-219-5p and Sall4. The actual clinical efficacy of
anti-miR-219-5p or anti-Sall4 therapy compared to current
first-line medications needs further investigation under strict and
comparable trials. We hope that it will bring benefits to patients
in the future.
Acknowledgments
We sincerely appreciate what our team members have
done for these experimente and the accomplishment is the fruit of
great effort. We are also thankful for financial support by the
National Natural Science Foundation of China grant number
81172294.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li L and Ma BB: Colorectal cancer in
Chinese patients: Current and emerging treatment options. Onco
Targets Ther. 7:1817–1828. 2014.PubMed/NCBI
|
|
3
|
Tokarz P and Blasiak J: The role of
microRNA in metastatic colorectal cancer and its significance in
cancer prognosis and treatment. Acta Biochim Pol. 59:467–474.
2012.PubMed/NCBI
|
|
4
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar
|
|
5
|
Cui Y, Bai Y, Wang XD, Liu B, Zhao Z and
Wang LS: Differential expression of miRNA in rat myocardial tissues
under psychological and physical stress. Exp Ther Med. 7:901–906.
2014.PubMed/NCBI
|
|
6
|
Shim SM, Jung SY, Nam HY, Kim HR, Lee MH,
Kim JW, Han BG and Jeon JP: Network signatures of cellular
immortalization in human lymphoblastoid cell lines. Biochem Biophys
Res Commun. 441:438–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang C, Cai Z, Huang M, Mao C, Zhang Q,
Lin Y, Zhang X, Tang B, Chen Y, Wang X, et al: miR-219-5p modulates
cell growth of papillary thyroid carcinoma by targeting estrogen
receptor α. J Clin Endocrinol Metab. 100:e204–e213. 2015.
View Article : Google Scholar
|
|
9
|
Rao SA, Arimappamagan A, Pandey P, Santosh
V, Hegde AS, Chandramouli BA and Somasundaram K: miR-219-5p
inhibits receptor tyrosine kinase pathway by targeting EGFR in
glioblastoma. PLoS One. 8:e631642013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Forghanifard MM, Moghbeli M, Raeisossadati
R, Tavassoli A, Mallak AJ, Boroumand-Noughabi S and Abbaszadegan
MR: Role of SALL4 in the progression and metastasis of colorectal
cancer. J Biomed Sci. 20:62013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ardalan Khales S, Abbaszadegan MR,
Abdollahi A, Raeisossadati R, Tousi MF and Forghanifard MM: SALL4
as a new biomarker for early colorectal cancers. J Cancer Res Clin
Oncol. 141:229–235. 2015. View Article : Google Scholar
|
|
12
|
Böhm J, Sustmann C, Wilhelm C and Kohlhase
J: SALL4 is directly activated by TCF/LEF in the canonical Wnt
signaling pathway. Biochem Biophys Res Commun. 348:898–907. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prenen H, Vecchione L and Van Cutsem E:
Role of targeted agents in metastatic colorectal cancer. Target
Oncol. 8:83–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oestreicher P: Sequencing therapies: The
role of targeted agents in metastatic colorectal cancer. ONS
Connect. 22(Suppl 8): 37–38. 2007.PubMed/NCBI
|
|
16
|
Iqbal N and Iqbal N: Imatinib: A
breakthrough of targeted therapy in cancer. Chemother Res Pract.
2014:3570272014.PubMed/NCBI
|
|
17
|
Balduzzi S, Mantarro S, Guarneri V,
Tagliabue L, Pistotti V, Moja L and D'Amico R:
Trastuzumab-containing regimens for metastatic breast cancer.
Cochrane Database Syst Rev. 6:CD0062422014.PubMed/NCBI
|
|
18
|
Gustavsson B, Carlsson G, Machover D,
Petrelli N, Roth A, Schmoll HJ, Tveit KM and Gibson F: A review of
the evolution of systemic chemotherapy in the management of
colorectal cancer. Clin Colorectal Cancer. 14:1–10. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alberts SR, Sargent DJ, Nair S, Mahoney
MR, Mooney M, Thibodeau SN, Smyrk TC, Sinicrope FA, Chan E, Gill S,
et al: Effect of oxaliplatin, fluorouracil, and leucovorin with or
without cetuximab on survival among patients with resected stage
III colon cancer: A randomized trial. JAMA. 307:1383–1393. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tveit KM, Guren T, Glimelius B, Pfeiffer
P, Sorbye H, Pyrhonen S, Sigurdsson F, Kure E, Ikdahl T, Skovlund
E, et al: Phase III trial of cetuximab with continuous or
intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic
FLOX) versus FLOX alone in first-line treatment of metastatic
colorectal cancer: The NORDIC-VII study. J Clin Oncol.
30:1755–1762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seymour MT: Adjuvant bevacizumab in colon
cancer: Where did we go wrong? Lancet Oncol. 13:1176–1177. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sculier JP, Berghmans T and Meert AP:
Advances in target therapy in lung cancer. Eur Respir Rev.
24:23–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Wang X, Wang Z, Tang H, Fan H and
Guo Q: miR-182 promotes cell growth and invasion by targeting
forkhead box F2 transcription factor in colorectal cancer. Oncol
Rep. 33:2592–2598. 2015.PubMed/NCBI
|
|
24
|
Tang L, Chen F, Pang EJ, Zhang ZQ, Jin BW
and Dong WF: MicroRNA-182 inhibits proliferation through targeting
oncogenic ANUBL1 in gastric cancer. Oncol Rep. 33:1707–1716.
2015.PubMed/NCBI
|
|
25
|
Jiang Z, Cushing L, Ai X and Lü J: miR-326
is downstream of Sonic hedgehog signaling and regulates the
expression of Gli2 and smoothened. Am J Respir Cell Mol Biol.
51:273–283. 2014.PubMed/NCBI
|
|
26
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-Catenin activates MiR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yong KJ, Gao C, Lim JS, Yan B, Yang H,
Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, et al: Oncofetal
gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med.
368:2266–2276. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao C, Kong NR and Chai L: The role of
stem cell factor SALL4 in leukemogenesis. Crit Rev Oncog.
16:117–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang J, Chai L, Liu F, Fink LM, Lin P,
Silberstein LE, Amin HM, Ward DC and Ma Y: Bmi-1 is a target gene
for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci
USA. 104:10494–10499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao C, Dimitrov T, Yong KJ, Tatetsu H,
Jeong HW, Luo HR, Bradner JE, Tenen DG and Chai L: Targeting
transcription factor SALL4 in acute myeloid leukemia by
interrupting its interaction with an epigenetic complex. Blood.
121:1413–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang W, Tong JH, Lung RW, Dong Y, Zhao J,
Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al: Targeting of YAP1
by microRNA-15a and microRNA-16-1 exerts tumor suppressor function
in gastric adenocarcinoma. Mol Cancer. 14:522015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang ZW, Men T, Feng RC, Li YC, Zhou D
and Teng CB: miR-375 inhibits proliferation of mouse pancreatic
progenitor cells by targeting YAP1. Cell Physiol Biochem.
32:1808–1817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di
GH, Wu J, Shen ZZ, Song HY and Shao ZM: MicroRNA-200a promotes
anoikis resistance and metastasis by targeting YAP1 in human breast
cancer. Clin Cancer Res. 19:1389–1399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gebert LF, Rebhan MA, Crivelli SE, Denzler
R, Stoffel M and Hall J: Miravirsen (SPC3649) can inhibit the
biogenesis of miR-122. Nucleic Acids Res. 42:609–621. 2014.
View Article : Google Scholar
|