Introduction
Almost 85% of lung carcinoma patients suffer from
non-small cell lung cancer, while patients with lung squamous
carcinoma account for 25–30% of patients with non-small cell lung
cancer (1). At present, surgery
remains the main treatment method for lung squamous carcinoma.
However, should metastasis occur, treatment is rendered ineffective
and the prognosis is poor. Chemotherapy efficacy is relatively
limited in treating patients with metastasis lung squamous
carcinoma, and the patient 5-year survival rate remains at <15%
(2,3), while a targeted drug with good
efficacy for lung squamous carcinoma remains to be identified. As a
result, identification of a new and efficient antineoplastic agent
with small toxicity has become a research hotspot for lung squamous
carcinoma treatment.
Tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL) is a member of the tumor necrosis factor (TNF)
superfamily. Previous findings (4–6) have
shown that TRAIL induces apoptosis of a variety of tumor cells, but
exhibits no significant toxicity to normal tissues (7,8).
Therefore, studies have focused on the antitumor effect of TRAIL,
demonstrating a broad application prospect in the field of targeted
therapy for carcinoma. TRAIL-induced apoptosis of carcinoma cells
is mainly achieved by combining with death receptor 4 (DR4) and
death receptor 5 (DR5) to recruit the fas-associated death domain
(FADD) and form a death-inducing signaling complex (DISC),
initiating a cascade reaction and triggering cell apoptosis
(9–12). Therefore, a high expression of DR4
and DR5 can promote the TRAIL-induced apoptosis of carcinoma cells.
However, most carcinoma cells are resistant to TRAIL-induced
apoptosis, limiting its clinical application (13,14).
Previous in-depth investigations on the mechanism of tumor cells
for resisting TRAIL (15–19), have identified that drug resistance
may be associated with the hypermethylation of the DR4 gene
promoter. 5-Aza-2′-deoxycytidine (5-Aza-CdR), also known as
decitabine, is a specific methylation inhibitor, which is mainly
used for the clinical treatment of myelodysplastic syndrome and
chronic myelogenous leukemia, and has achieved significant results
(20–22). It has been reported that 5-Aza-CdR
is capable of reversing the hypermethylation status of carcinoma
cells, recover the expression of related genes and reverse
carcinoma drug resistance (23–25).
Thus, the aim of the present study was to examine lung squamous
carcinoma and investigate whether the DR4 gene promoter methylation
status affected the TRAIL-induced apoptosis of lung squamous
carcinoma cells.
Materials and methods
Materials used in the in vitro
experiments
H226, SK-MES-1 and H520 lung squamous carcinoma cell
lines were procured from Beinuo Biotechnology Co., Ltd. (Shanghai,
China), the main reagent TRAIL was donated by Qiaer Biotechnology
Co., Ltd.(Shanghai, China), 5-Aza-CdR, MTT and DMSO were purchased
from Sigma (St. Louis, MO, USA), RPMI-1640 and DMEM high-glucose
cell culture medium as well as fetal bovine serum were purchased
from Gibco-Life Technologies (Carlsbad, CA, USA). The EZ DNA
Methylation-Gold™ kit was procured from Zymo (Orange, CA, USA), the
DNA extraction kit, TRIzol reagent, reverse transcription kit,
TaqPCR SuperMix and DNA markers were purchased from Beijing
TransGen Biotech Co., Ltd. and the Annexin-V FITC/PI apoptosis
detection kits were purchased from BD Biosciences (Bedford, MA,
USA). Rabbit anti-human DR4 polyclonal antibodies and rabbit
anti-human β-actin polyclonal antibodies are purchased from Abcam
(Cambridge, MA, USA). Horseradish peroxidase-labeled goat
anti-rabbit IgG (H + L) antibodies were purchased from Beyotime
Biotechnology Co., Ltd. (Shanghai, China) and rat/rabbit universal
secondary antibodies were purchased from MXB Biotechnology Co.,
Ltd. (Fuzhou, China). PCR primers were designed and produced by
Shanghai Invitrogen Biotechnology Co., Ltd. (Shanghai, China).
Patients participating in the in vivo
experiments
Thirty-six surgical specimens were obtained from
patients diagnosed with lung squamous carcinoma at the Fuzhou
General Hospital of Nanjing Military Command (Fujian, China)
between March 2010 and March 2013. There were 35 male patients and
1 female patient (age range, 42–82 years; median, 61 and average
age, 61.6). Of the 36 patients, 32 had a history of smoking.
According to AJCC staging in 2010, 6 patients were classified as
stage-IA lung squamous carcinoma, 4 as stage-IB carcinoma, 14 as
stage-IIA carcinoma, 2 as stage-IIB carcinoma, 8 as stage-IIIA
carcinoma, 1 as stage-IIIB carcinoma and 1 as stage-IV carcinoma.
According to the pathological differentiation degree, 3 patients
exhibited high degree, 6 patients high and middle, 14 patients
middle, 9 patients low and middle, and 4 patients exhibited low
degree. The patients did not undergo radio- or chemotherapy prior
to surgery. However, all 36 patients accepted radio- and
chemotherapy as adjuvant therapy following surgery, with 29 of the
patients eventually exhibiting disease progression or succumbed to
the disease.
Immunohistochemistry
The Elivision two-step method was employed to detect
the expression of clinical tissue DR4. The detection procedure was
as follows: After deparaffinization and hydration,
paraffin-embedded tissue sections were placed into citrate buffer
(10 mmol/l, pH 6.0) for heating in a water bath for 15 min for
antigen retrieval. After cooling, the section was soaked and
incubated with 3% H2O2 for 10 min, to block
the activity of endogenous peroxidase. After rinsing three times
with PBS (3 min each time), the section was immersed in rabbit
anti-human DR4 polyclonal antibodies (purchased from Abcam),
diluted at a ratio of 1:25 and then incubated for 3 h at room
temperature. After rinsing three times with PBS, with a drop of
polymer enhancer (reagent A) was added to each section, incubated
for 20 min at room temperature and rinsed with PBS three times. A
drop of HRP anti-rat/rabbit polymer (reagent B) was added to each
slice, incubated for 30 min at room temperature, rinsed with PBS,
developed with DAB, re-stained with hematoxylin, differentiated
with 0.1% HCl, rinsed with tap water, stained with Acian blue
solution, dehydrated and dried with graded alcohol, sealed with
neutral gum and dried for observation. A DR4-positive signal was
considered brown granular, located in the cytoplasm with staining
intensity and divided into four grades: +, ++, +++ and ++++.
Methylation-specific PCR (MSP)
The cell/tissue DNA was extracted according to the
DNA extraction kit. After quantification of DNA, 500 ng DNA was
extracted for methylation modification using the EZ DNA
Methylation-Gold™ kit. Taking it as a template, methylated and
unmethylated primers were applied for PCR amplification.
Methylation-specific primers (23,24)
used were: upstream, 5′-TTCGAATTTCGGGAGCGTAGC-3′ and downstream,
5′-GTAATTCAATCCTCCCCG CGA-3′ (fragments, 91 bp). Unmethylated
specific primers used were: upstream,
5′-GTAGTGATTTTGAATTTTGGGAGTGTAGT-3′ and downstream,
5′-CTCATAATTCAATCCCCACAA-3′ (fragments, 102 bp). The PCR reaction
conditions were: 95°C for 5 min, 95°C for 30 sec, 57°C for 30 sec
and 72°C for 40 sec, with a total of 30 cycles, and 72°C for an
extension of 5 min. Amplified products were analyzed with 2%
agarose gel electrophoresis and gel imaging and observed and
photographed under ultraviolet light. It was found that methylated
primers were positive after amplification and unmethylated primers
were negative after amplification, and were deemed exhaustive
methylation. Additionally, methylated and unmethylated primers were
found to have positive bands with partial methylation after
amplification, but remained positive in methylation. By contrast,
when methylated primers were negative after amplification,
methylation was deemed to be negative only when unmethylated
primers were positive after amplification. This experiment was
repeated three times.
Cell and adherent culture
H226 and H520 cells were cultured in RPMI-1640
culture medium containing 10% FBS and 10 U/ml penicillin and
streptomycin, while SK-MES-1 cells were cultured in DMEM (high
glucose) culture medium containing 10% FBS and 100 U/ml penicillin
and streptomycin. The cells were incubated in 5% CO2 at
37°C, with the culture medium replaced once every two days. After
passage for 1 day, 5 µmol/l 5-azacytidine was, respectively,
added to the culture flasks to continue culturing for 2 days. After
culture termination, the cells without intervention by adding
5-azacytidine were considered the control group.
Reverse transcription-PCR (RT-PCR)
Three lung squamous carcinoma cells at the
exponential phase prior to and following treatment with 5-Aza-CdR
were used. The total RNA of cells was extracted using TRIzol
reagent and reverse transcribed to become cDNA using a reverse
transcription kit. This was utilized as a template for the PCR
amplification. DR4 primers used were: upstream,
5′-AAGTCCCTGCACCACGAC-3′ and downstream, 5′-CCACAACCTGAGCCGATG-3′
(fragments, 256 bp), and GAPDH primers: upstream,
5′-TGCACCACCAACTGCTTAGC-3′ and downstream,
5′-GGCATGGACTGTGGTCATGAG-3′ (fragments, 100 bp). The PCR reaction
conditions were: 96°C for 5 min, 96°C for 30 sec, 57°C for 30 sec
and 72°C for 40 sec, with a total of 35 cycles, and 72°C for an
extension of 5 min. Amplified products were analyzed using 1.5%
agarose gel electrophoresis and gel imaging, and observed and
photographed under ultraviolet light. Quantity One software was
employed to analyze the gray value of the electrophoretic band and
GAPDH was used as a standardized internal control.
Western blot analysis
Three lung squamous carcinoma cells at the
exponential phase prior to and following treatment with 5-Aza-CdR
were used. Proteins were extracted by adding protein lysate to lyse
cells and quantified using the BCA method. The cell protein was
separated by electrophoresis using 10% SDS-PAGE and transmembrane
after electrophoresis. After successful transfer, the membranes
were blocked for 1 h with 5% skim milk and incubated overnight at
4°C with DR4 and β-actin primary antibodies, respectively. Primary
antibodies were washed, while secondary antibodies were incubated
for 1.5 h. ECL was used for color development and β-actin was
considered as a standardized internal control.
MTT detection
H226 and SK-MES-1 cells at the exponential phase
prior to and following treatment with 5-Aza-CdR were removed and
inoculated into the 96-well plate at 1×105/ml and 100
µl/well. Six parallel wells were used for each group and
cultivated for 24 h, after which the supernatant was discarded.
Culture medium (100 µl) containing different concentrations
of the drug (TRAIL concentrations of 0.01, 0.05, 0.10, 0.50, 1.00
and 5.00 µg/ml) was added. Each experiment established the
blank and control groups. After the drug treatment for 24 and 48 h,
100 µl 0.5 mg/ml MTT solution was added to each well. After
termination of culture after 4 h of continuous incubation, the
liquid in each well was gently aspirated and 150 µl DMSO was
added to each well. The liquid was then agitated for 10 min to
dissolve the crystal completely. Subsequently, the absorbance value
(A) of each well was measured at 490 nm wavelength of the
enzyme-linked immunosorbent assay. The experiment was repeated
three times. The cell proliferation inhibition rate was calculated
according to the drug concentration, using the formula: (1-average
A value of the experimental group/average A value of the control
group) ×100%.
Flow cytometry
H226 and SK-MES-1 cells were treated with 0.5
µg/ml TRAIL (TRAIL group) and 5 µmol/l 5-Aza-CdR
(5-Aza-CdR group) for 24 h, while the joint group was treated with
1 µg/ml TRAIL for 24 h after previously being treated with 5
µmol/l 5-Aza-CdR for 48 h. The cells were collected and
rinsed with PBS twice. The cell apoptotic rate was detected
according to the instructions of the Annexin-V FITC/PI apoptosis
detection kit. The experiment was repeated three times.
Statistical analysis
Using SPSS18 statistical software, the measurement
data were presented as mean ± standard deviation (means ± SD). The
Spearman rank correlation analysis was used to determine the
dose-effect relationship. The t-test was used to compare the
differences among the various groups. The χ2 or Fisher's
exact test was employed to detect the relationship between DR4 gene
promoter methylation statuses and clinico pathological
characteristics of patients with lung squamous carcinoma. The κ
value was used to assess the relationship between the DR4 gene
promoter methylation and DR4 protein expression of patients with
lung squamous carcinoma. To determine the relationship between the
expression of DR4 protein and prognosis of the patients with lung
squamous carcinoma, the Kaplan-Meier was employed to assess the
survival rate, while the log-rank and Breslow tests were utilized
to assess the survival rate differences of patients from different
groups. The bilateral probability test denoted that P<0.05
indicates statistical significance.
Results
The relationship between DR4 gene
promoter methylation statuses and clinicopathological
characteristics of patients with lung squamous carcinoma
The MSP results showed that 80.6% of 36 lung
squamous carcinoma patients exhibited positive methylation status,
1 patient exhibited hypermethylation status, 28 patients exhibited
partial methylation status, while the remaining 7 patients did not
exhibit positive methylation status, indicating lung squamous
carcinoma exhibited a high methylation modification (Fig. 1). At the same time, the results
confirmed that the DR4 gene promoter methylation status did not
correlate with the clinicopathological characteristics, such as
age, gender, smoking, pathological grade, TNM stage and lymph node
metastasis, of patients suffering from lung squamous carcinoma
(P>0.05) (Table I).
 | Table IRelationship between DR4 gene promoter
methylation status and clinicopathological characteristics of
patients with lung squamous carcinoma. |
Table I
Relationship between DR4 gene promoter
methylation status and clinicopathological characteristics of
patients with lung squamous carcinoma.
| Clinicopathological
parameters | n | m | u | P-value |
|---|
| Age (years) | | | | 1.000 |
| ≤60 | 17 | 14 | 3 | |
| >60 | 19 | 15 | 4 | |
| Gender | | | | 1.000 |
| Male | 35 | 28 | 7 | |
| Female | 1 | 10 | | |
| Smoking
history | | | | 0.408 |
| ≤20 | 19 | 14 | 5 | |
| >20 | 17 | 15 | 2 | |
| Pathological degree
of differentiation | | | | 0.333 |
| High-middle
differentiation | 9 | 6 | 3 | |
| Low-middle
differentiation | 27 | 23 | 4 | |
| TNM stage | | | | 0.076 |
| Early | 26 | 23 | 3 | |
| Stage-I | 10 | 8 | 2 | |
| Stage -II | 16 | 16 | 1 | |
| Middle and
advanced | 10 | 6 | 4 | |
| Stage -III | 9 | 5 | 4 | |
| Stage -IV | 1 | 1 | 0 | |
| Lymph node
metastasis | | | | 0.674 |
| Positive | 21 | 16 | 5 | |
| Negative | 15 | 13 | 2 | |
The relationship between DR4 gene
promoter methylations and DR4 protein expression of the patients
with lung squamous carcinoma
DR4 protein expression was assessed using
immunohistochemistry based on the staining intensity. Four cases of
36 clinical samples exhibited 0 DR4 protein expression, 17
exhibited +, 10 exhibited ++ to +++ and 5 exhibited ++++ (Fig. 2). The probability of DR4 low
expression of patients with lung squamous carcinoma was 58.3% (21
of 36 samples). The results confirmed that the DR4 protein
expression levels of the patients with lung squamous carcinoma were
associated with their gene promoter methylation degrees (P<0.05)
(Table II). This finding suggested
that the probability of low DR4 expression of patients with lung
squamous carcinoma was relatively high, which may be associated
with gene promoter methylation.
 | Table IIRelationship between DR4 gene
promoter methylation and DR4 protein expressions of patients with
lung squamous carcinoma. |
Table II
Relationship between DR4 gene
promoter methylation and DR4 protein expressions of patients with
lung squamous carcinoma.
| IHC | Methylation status
| κ-value | P-value |
|---|
| n | m | u |
|---|
| Low expression (−
to +) | 21 | 20 | 1 | 0.381 | 0.008 |
| High expression (++
to ++++) | 15 | 9 | 6 | | |
The relationship between DR4 protein
expression and prognosis of patients with lung squamous
carcinoma
In the group of low DR4 expression, 21 patients with
lung squamous carcinoma had a median PFS of 11 months (95% CI:
9.505–12.495); while in the high DR4 expression group, 15 patients
with lung squamous carcinoma had a median PFS of 25.653 months (95%
CI: 19.631–31.676), based on results of the log rank test,
χ2=9.494, P=0.002 and Breslow test,
χ2=10.515, P=0.001; or combined, P<0.05 (Fig. 3). Thus, the two groups of PFS time
difference were statistically significant. The result showed that
the prognosis of patients with a high DR4 expression of lung
squamous carcinoma was lower than that of the patients with a low
DR4 expression.
DR4 gene promoter region CpG island
methylation status
The MSP result showed that the DR4 promoter region
in H226 and SK-MES-1 cells was partly methylated while H520
exhibited a non-methylated state. Following treatment with
5-Aza-CdR, the DR4 promoter region in H226 and SK-MES-1 cells was
altered from a partly methylated to a non-methylated status, while
H520 remained in the non-methylated state (Fig. 4).
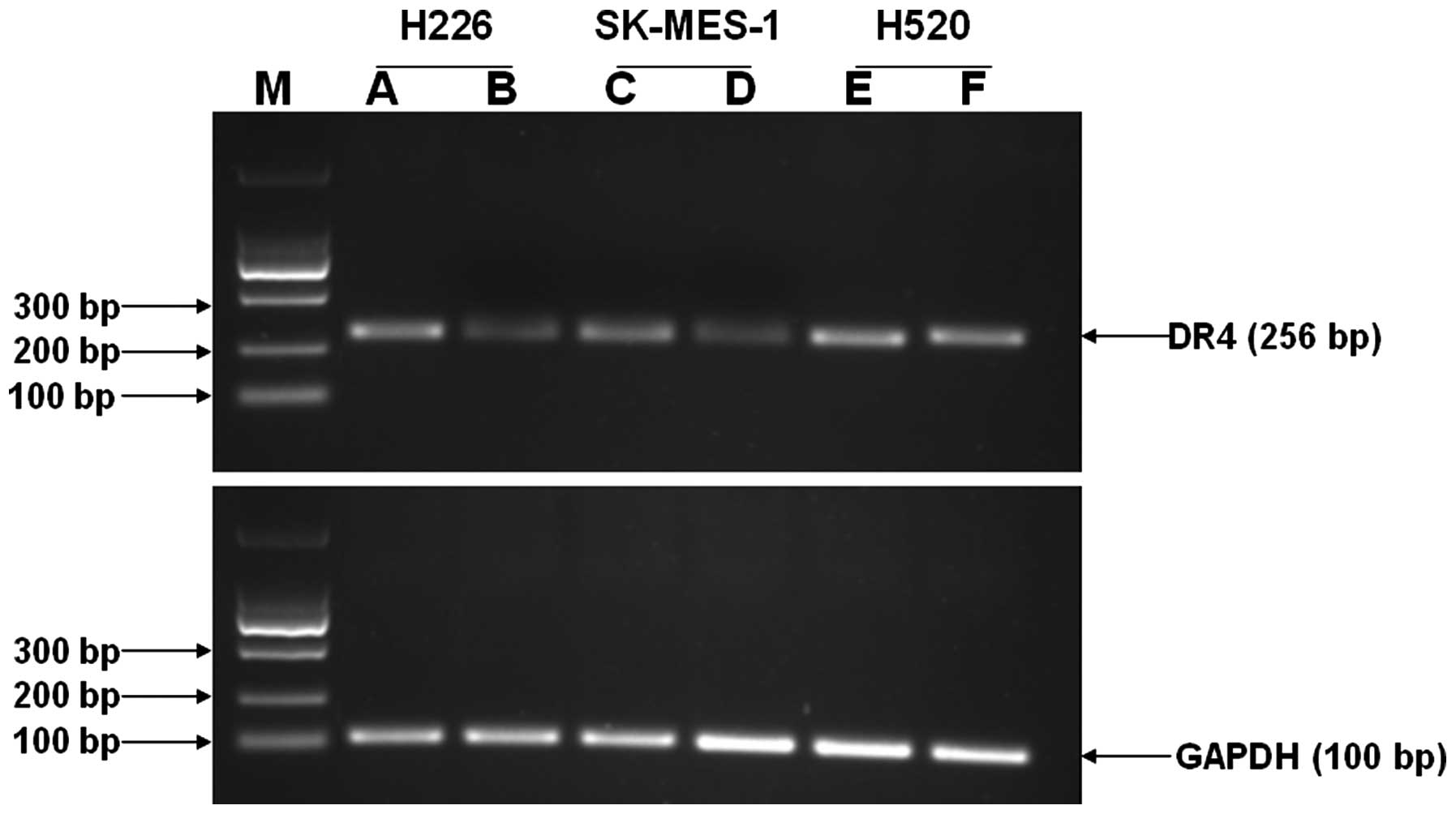
DR4 mRNA expression
The DR4 mRNA relative expression (means ± SD) of
H226, SK-MES-1 and H520 cells prior to and following interference
with 5-azacytidine was identified as 0.245±0.005, 0.899±0.011,
0.139±0.009, 0.528±0.012, 0.789±0.011 and 0.803±0.013,
respectively. The results showed that priro to interference with
5-azacytidine, the DR4 mRNA expression of H226 and SK-MES-1 cells
was low whereas that of H520 cells was high. By contrast, following
interference with 5-azacytidine, the DR4 mRNA expression of H226
and SK-MES-1 cells was significantly enhanced compared with the
previous levels and the differences were statistically significant
(P<0.05). However, the DR4 mRNA expression of H520 cells was
slightly increased compared with the previous levels, and the
differences were not statistically significant (Figs. 5 and 6).
DR4 protein expression
The DR4 protein expression (means ± SD) of H226,
SK-MES-1 and H520 cells prior to and following interference with
5-azacytidine was identified as 0.217±0.019, 0.415±0.031,
0.264±0.021, 0.418±0.036, 0.426±0.028 and 0.419±0.03, respectively.
The results showed that prior to interference with 5-azacytidine,
DR4 protein expression of H226 and SK-MES-1 cells was low whereas
that of H520 cells was high. However, following interference with
5-azacytidine, the DR4 protein expression of H226 and SK-MES-1
cells was significantly enhanced compared with the previous levels
and the differences were statistically significant (P<0.05). By
contrast, the DR4 protein expression of H520 cells did not markedly
change compared with the previous levels and the differences were
not statistically significant (Figs.
7 and 8).
Cell proliferation inhibition rate
Following treatment with TRAIL at different
concentrations for 24 h, H226 and SK-MES-1 cell growth was
inhibited to various degrees, although not significantly. However,
after treatment with 5-azacytidine at the same concentration, the
cell growth was significantly inhibited as compared with the
previous cell growth, and the differences were statistically
significant (P<0.05). The inhibition rates of TRAIL with at the
same concentration against cell proliferation increased with the
extension of the action time (P<0.05) (Tables III and IV, and Figs.
9 and 10).
 | Table IIIProliferation inhibition rate of H226
cells (means ± SD, %). |
Table III
Proliferation inhibition rate of H226
cells (means ± SD, %).
| Time (h) | n | TRAIL concentration
(µg/ml)
|
|---|
| 0.01 | 0.05 | 0.10 | 0.50 | 1.00 | 5.00 |
|---|
| Prior to treatment
with 5-AZ |
| 24 | 6 | 2.23±0.08 | 5.72±0.25 | 7.54±0.25 | 10.69±1.61 | 19.11±1.39 | 31.74±2.88 |
| 48 | 6 | 4.81±0.09 | 9.02±0.95 | 16.57±1.66 | 28.23±2.06 | 37.32±3.61 | 53.87±3.29 |
| Following treatment
with 5-AZ |
| 24 | 6 | 8.15±0.19a | 17.65±1.07a | 25.42±1.46a | 33.04±2.13a | 48.14±3.22a | 69.11±3.67a |
| 48 | 6 | 15.43±1.05b,c | 28.51±1.66b,c | 41.58±2.37b,c | 64.35±3.83b,c | 79.40±3.03b,c | 89.24±4.08b,c |
 | Table IVSK-MES-1 cell proliferation
inhibition rate (means ± SD, %). |
Table IV
SK-MES-1 cell proliferation
inhibition rate (means ± SD, %).
| Time | n | TRAIL concentration
(µg/ml)
|
|---|
| 0.01 | 0.05 | 0.10 | 0.50 | 1.00 | 5.00 |
|---|
| Prior to treatment
with 5-AZ |
| 24 | 6 | 1.51±0.12 | 4.46±0.31 | 6.79±0.41 | 9.89±1.47 | 15.03±2.77 | 27.93±3.35 |
| 48 | 6 | 3.41±0.88 | 8.02±1.03 | 14.67±1.84 | 25.23±2.01 | 32.98±3.11 | 48.69±3.44 |
| Following treatment
with 5-AZ |
| 24 | 6 |
7.78±0.66a |
16.42±1.20a |
22.81±1.94a |
32.54±2.37a |
49.91±3.08a | 65.11±3.798 |
| 48 | 6 | 13.89±1.23b,c | 23.36±1.99b,c | 45.44±2.61b,c | 56.53±3.28b,c | 74.68±3.71b,c | 87.18±4.22b,c |
Cell apoptotic rate
Apoptosis of H226 and SK-MES-1 cells was determined
(Tables V and VI). To determine the effect of each group
on the cell apoptotic rate, a comparison was made of the
5-azacytidine and control groups. The results showed that, the
differences between the groups were not statistically significant,
suggesting that 5 µmol/l 5-AZ has no significant apoptotic
effect on cells. However, the TRAIL and 5-AZ + TRAIL groups induced
apoptotic effects on the cells. This induced apoptotic effect on
the cells in the 5-AZ + TRAIL group was significantly higher than
that of the TRAIL group, and the difference was statistically
significant (P<0.05) (Figs. 11
and 12).
 | Table VEffect of each group on the apoptosis
of H226 cell (means ± SD, %). |
Table V
Effect of each group on the apoptosis
of H226 cell (means ± SD, %).
| Group | n | Early apoptotic
rate | Late apoptotic
rate | Total apoptotic
rate |
|---|
| Negative
control | 3 | 0.41±0.05 | 0.53±0.03 | 0.94±0.10 |
| 5-AZ | 3 | 0.32±0.02a | 0.62±0.06a | 1.02±0.15a |
| TRAIL | 3 | 3.92±0.11b | 3.18±0.19b | 7.10±1.31b |
| 5-AZ+TRAIL | 3 | 3.87±0.23b | 27.33±1.22b,c | 31.20±4.88b,c |
 | Table VIEffect of each group on the apoptosis
of SK-MES-1 cell (means ± SD, %). |
Table VI
Effect of each group on the apoptosis
of SK-MES-1 cell (means ± SD, %).
| Group | n | Early apoptotic
rate | Late apoptotic
rate | Total apoptotic
rate |
|---|
| Negative
control | 3 | 0.29±0.02 | 0.99±0.10 | 1.27±0.15 |
| 5-Aza-CdR | 3 | 0.34±0.03a | 0.94±0.08a | 1.21±0.19a |
| TRAIL | 3 | 4.12±0.15b | 3.78±0.35b | 7.90±1.46b |
| 5-AZ+TRAIL | 3 | 14.86±0.41b,c | 13.37±1.31b,c | 28.23±3.87b,c |
Discussion
For DNA methylation modification, under the
catalysis of methyltransferase, C is integrated with methyl to form
mC, while the integration of methylation mCpG
with DNA methyl-binding domain protein can indirectly block the
binding of transcription initiation elements with the DNA promoter
region, to regulate the gene expression (26–28).
The gene promoter region CpG island is generally found in the
non-methylation status. An abnormally high expression can directly
or indirectly block gene transcription, resulting in silencing or a
function of the cell cycle regulatory gene, DNA repair gene,
apoptosis gene, tumor suppressor gene and other related genes,
which may be one of the main factors inducing tumors (29,30).
In lung carcinoma and other carcinoma cells, the abnormal increase
in the methylation levels of some gene CpG island regions is their
main characteristic (31). For
colorectal cancer, previous findings have shown an abnormal
methylation is a characteristic factor in the regulation of CPT-11
metabolic enzyme change. The presence of methylation in UGT1A1 gene
promoters of colorectal cancer cells is an important mechanism for
silencing of the UGT1A1 gene expression and also a target to
identify the regulatory mechanism for CPT-11 resistance and
reversal resistance (32,33). DNA methylation status is reversible
and the genes with expression silencing or a function due to the
hypermethylation status can restore their expression through the
DNA methyltransferase enzyme inhibitor. Currently, the DNA
methyltransferase inhibitor has made significant progress in the
treatment of hematological malignancies, for example, decitabine,
also known as 5-aza-2′-deoxycytidine, has been approved by the FDA
for the treatment of myelodysplastic syndrome and acute leukemia,
and has achieved satisfactory therapeutic effect. The drug at a low
dose can maximize the inhibitory effect of DNA demethylation, while
the drug at a high dose shows cytotoxicity. Therefore, drug
treatment at s low dose can maximize the therapeutic effect and the
most significant finding was that it can effectively remove DNA
methylation, to re-express many inactivated genes.
TRAIL, a new member of the superfamily of TNF, has
been recently identified, and TRAIL-induced apoptosis is not
dependent on p53 status. Unlike radio- and chemotherapy-induced
apoptosis mechanisms, TRAIL has some value for
chemotherapy-resistant tumors. As a result, TRAIL has been
investigated as a potential antineoplastic agent. However,
tolerance is a major obstacle to TRAIL-induced apoptosis and it has
been identified that this type of apoptosis may be associated with
the hypermethylation of DR4 gene promoters (15–19).
However, whether alteration of DR4 gene promoter methylation status
affects DR4 expression to increase the tumor cell apoptosis induced
by TRAIL remains to be determined. Thus, the focus of this study
was on lung squamous carcinoma and whether the DR4 gene promoter
methylation status affected the TRAIL-induced apoptosis in lung
squamous carcinoma cells.
The results showed that the probability of DR4 gene
methylation was relatively high, at 80.6%, among patients with lung
squamous carcinoma while the methylation status did not correlate
with the clinicopathological characteristics of patients with lung
squamous carcinoma (P>0.05). To confirm the relationship between
methylation status and DR4 expression, immunohistochemistry was
employed to detect DR4 expression for the 36 patients. The result
showed that 58.3% of the patients with lung squamous carcinoma had
a low DR4 expression, which may be associated with their gene
promoter methylation. At the same time, the prognosis of the
patients with lung squamous carcinoma exhibiting a high DR4
expression was lower than that of the patients with a low
expression.
To determine the effect of DR4 gene promoter
methylation status on the TRAIL-induced apoptosis of lung squamous
carcinoma cells, we conducted in vitro experiments. DR4 gene
promoter region CpG islands of H226 and SK-MES-1 cells exhibited a
positive methylation status and the expression of mRNA and proteins
were low, whereas the DR4 gene promoter region CpG islands of H520
cells exhibited a non-methylation status and the expression of
their mRNA and proteins was high. Following treatment with
5-Aza-CdR, DR4 genes of H226 and SK-MES-1 exhibited a
non-methylation status and the expression of their mRNA and
proteins increased significantly (P<0.05), whereas H520 cells in
the non-methylation status did not markedly alter. MTT assay and
flow cytometry confirmed that H226 and SK-MES-1 cells are
TRAIL-insensitive. Following treament with 5-Aza-CdR, the
sensitivity of these two lung squamous carcinoma cells to TRAIL
significantly increased compared with the previous sensitivity
identified (P<0.05). The findings suggest that DR4 gene promoter
methylation may downregulate the expression level of the DR4 gene,
resulting in decreased sensitivity to TRAIL and affecting the
efficacy of TRAIL on lung squamous carcinoma.
In conclusion, a low dose of 5-Aza-CdR is capable of
reversing the methylation status and may improve the efficacy of
TRAIL on lung squamous carcinoma. A low dose of 5-Aza-CdR has no
toxicity, while TRAIL has a high selectivity and high efficiency
induction of apoptosis in a variety of carcinoma cells, which also
has no significant toxicity on normal tissues. Therefore, the
combination of 5-Aza-CdR and TRAIL may be a novel therapeutic
strategy for the treatment of lung squamous carcinoma and is
expected to be useful in the treatment of lung squamous carcinoma.
However, the present study results are limited to the in
vitro experiment and only involve the DR4 gene, while the
reason for TRAIL resistance being affected may be associated with
the change of molecules associated with the tumor necrosis pathway
apoptosis (such as BAX, Bcl-2, Bcl-XL, and caspase-3,8,9).
Therefore, to examine the treatment effect of 5-Aza-CdR combined
with TRAIL in lung squamous carcinoma, animal experiments and stage
I/II clinical tests should be carried out to verify the results and
whether the change of the methylation status of associated
molecules has an impact on the effect of TRAIL-induced lung
squamous carcinoma apoptosis may be detected.
References
|
1
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Azzoli CG, Baker S Jr, Temin S, Pao W,
Aliff T, Brahmer J, Johnson DH, Laskin JL, Masters G, Milton D, et
al American Society of Clinical Oncology: American Society of
Clinical Oncology Clinical Practice Guideline update on
chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol.
27:6251–6266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Zhao J, Zhu W, Gou H, Cao D, Yang
Y, Huang Y and Yi C: Synergistic effect of subtoxic-dose cisplatin
and TRAIL to mediate apoptosis by down-regulating decoy receptor 2
and up-regulating caspase-8, caspase-9 and Bax expression on
NCI-H460 and A549 Cells. Iran J Basic Med Sci. 16:710–718.
2013.PubMed/NCBI
|
|
5
|
Stegehuis JH, de Wilt LH, de Vries EG,
Groen HJ, de Jong S and Kruyt FA: TRAIL receptor targeting
therapies for non-small cell lung cancer: Current status and
perspectives. Drug Resist Updat. 13:2–15. 2010. View Article : Google Scholar
|
|
6
|
Luster TA, Carrell JA, McCormick K, Sun D
and Humphreys R: Mapatumumab and lexatumumab induce apoptosis in
TRAIL-R1 and TRAIL-R2 antibody-resistant NSCLC cell lines when
treated in combination with bortezomib. Mol Cancer Ther. 8:292–302.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jung Y-H, Heo J, Lee YJ, Kwon TK and Kim
Y-H: Quercetin enhances TRAIL-induced apoptosis in prostate cancer
cells via increased protein stability of death receptor 5. Life
Sci. 86:351–357. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schaefer U, Voloshanenko O, Willen D and
Walczak H: TRAIL: A multifunctional cytokine. Front Biosci.
12:3813–3824. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dillon CP, Oberst A, Weinlich R, Janke LJ,
Kang TB, Ben-Moshe T, Mak TW, Wallach D and Green DR: Survival
function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Reports.
1:401–407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maldonado ME, Bousserouel S, Gossé F,
Lobstein A and Raul F: Implication of NF-κB and p53 in the
expression of TRAIL-death receptors and apoptosis by apple
procyanidins in human metastatic SW620 cells. Biomedica.
30:577–586. 2010. View Article : Google Scholar
|
|
11
|
Haag C, Stadel D, Zhou S, Bachem MG,
Möller P, Debatin KM and Fulda S: Identification of c-FLIPL and
c-FLIPS as critical regulators of death receptor-induced apoptosis
in pancreatic cancer cells. Gut. 60:225–237. 2010. View Article : Google Scholar
|
|
12
|
Falschlehner C, Emmerich CH, Gerlach B and
Walczak H: TRAIL signalling: Decisions between life and death. Int
J Biochem Cell Biol. 39:1462–1475. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Earel JK Jr, VanOosten RL and Griffith TS:
Histone deacetylase inhibitors modulate the sensitivity of tumor
necrosis factor-related apoptosis-inducing ligand-resistant bladder
tumor cells. Cancer Res. 66:499–507. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Papageorgiou A, Lashinger L, Millikan R,
Grossman HB, Benedict W, Dinney CP and McConkey DJ: Role of tumor
necrosis factor-related apoptosis-inducing ligand in
interferon-induced apoptosis in human bladder cancer cells. Cancer
Res. 64:8973–8979. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee KH, Lim SW, Kim HG, Kim DY, Ryu SY,
Joo JK, Kim JC and Lee JH: Lack of death receptor 4 (DR4)
expression through gene promoter methylation in gastric carcinoma.
Langenbecks Arch Surg. 394:661–670. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bae SI, Cheriyath V, Jacobs BS, Reu FJ and
Borden EC: Reversal of methylation silencing of Apo2L/TRAIL
receptor 1 (DR4) expression overcomes resistance of SK-MEL-3 and
SK-MEL-28 melanoma cells to interferons (IFNs) or Apo2L/TRAIL.
Oncogene. 27:490–498. 2008. View Article : Google Scholar
|
|
17
|
Suzuki M, Shigematsu H, Shivapurkar N,
Reddy J, Miyajima K, Takahashi T, Gazdar AF and Frenkel EP:
Methylation of apoptosis related genes in the pathogenesis and
prognosis of prostate cancer. Cancer Lett. 242:222–230. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Horak P, Pils D, Haller G, Pribill I,
Roessler M, Tomek S, Horvat R, Zeillinger R, Zielinski C and
Krainer M: Contribution of epigenetic silencing of tumor necrosis
factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL
resistance and ovarian cancer. Mol Cancer Res. 3:335–343. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eramo A, Pallini R, Lotti F, Sette G,
Patti M, Bartucci M, Ricci-Vitiani L, Signore M, Stassi G, Larocca
LM, et al: Inhibition of DNA methylation sensitizes glioblastoma
for tumor necrosis factor-related apoptosis-inducing
ligand-mediated destruction. Cancer Res. 65:11469–11477. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keating GM: Azacitidine: A review of its
use in higher-risk myelodysplastic syndromes/acute myeloid
leukaemia. Drugs. 69:2501–2518. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cataldo VD, Cortes J and Quintás-Cardama
A: Azacitidine for the treatment of myelodysplastic syndrome.
Expert Rev Anticancer Ther. 9:875–884. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kaminskas E, Farrell A, Abraham S, Baird
A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, et
al FDA: Approval summary: Azacitidine for treatment of
myelodysplastic syndrome subtypes. Clin Cancer Res. 11:3604–3608.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vaitkienė P, Skiriutė D, Skauminas K and
Tamašauskas A: Associations between TFPI-2 methylation and poor
prognosis in glioblastomas. Medicina (Kaunas). 48:345–349.
2012.
|
|
24
|
Dong SW, Ma L, Xu N, Yan HQ, Liu HY, Li YW
and Zhang P: Research on the reactivation of Syk expression caused
by the inhibition of DNA promoter methylation in the lung cancer.
Neoplasma. 58:89–95. 2011. View Article : Google Scholar
|
|
25
|
Lin CT, Lai HC, Lee HY, Lin WH, Chang CC,
Chu TY, Lin YW, Lee KD and Yu MH: Valproic acid resensitizes
cisplatin-resistant ovarian cancer cells. Cancer Sci. 99:1218–1226.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Avraham A, Cho SS, Uhlmann R, Polak ML,
Sandbank J, Karni T, Pappo I, Halperin R, Vaknin Z, Sella A, et al:
Tissue specific DNA methylation in normal human breast epithelium
and in breast cancer. PLoS One. 9:e918052014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan H and Sun J: Methylation status of
WWOX gene promoter CpG islands in epithelial ovarian cancer and its
clinical significance. Biomed Rep. 1:375–378. 2013.
|
|
28
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan S, Sun C, Wei X, Li Y and Wu Y, Yan Z,
Feng F, Wang J and Wu Y: Quantitative assessment of lung cancer
associated with genes methylation in the peripheral blood. Exp Lung
Res. 39:182–190. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kupčinskaitė-Noreikienė R, Skiecevičienė
J, Jonaitis L, Ugenskienė R, Kupčinskas J, Markelis R, Baltrėnas V,
Sakavičius L, Semakina I, Grižas S, et al: CpG island methylation
of the MLH1, MGMT, DAPK, and CASP8 genes in cancerous and adjacent
noncancerous stomach tissues. Medicina (Kaunas). 49:361–366.
2013.
|
|
31
|
Balgkouranidou I, Liloglou T and Lianidou
ES: Lung cancer epigenetics: Emerging biomarkers. Biomarkers Med.
7:49–58. 2013. View Article : Google Scholar
|
|
32
|
Xie F-W, Peng Y-H, Wang W-W, Chen X, Chen
X, Li J, Yu ZY and Ouyang XN: Influence of UGT1A1 gene methylation
level in colorectal cancer cells on the sensitivity of the
chemotherapy drug CPT-11. Biomed Pharmacother. 68:825–831. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xie F-W, Peng Y-H, Chen X, Chen X, Li J,
Yu ZY, Wang WW and Ouyang XN: Regulation and expression of aberrant
methylation on irinotecan metabolic genes CES2, UGT1A1 and GUSB in
the in-vitro cultured colorectal cancer cells. Biomed Pharmacother.
68:31–37. 2014. View Article : Google Scholar : PubMed/NCBI
|